Abstract
Machado-Joseph disease is caused by an expansion of a trinucleotide CAG repeat in the gene encoding the protein ataxin-3. We investigated if ataxin-3 was a proteasome-associated factor that recognized ubiquitinated substrates based on the rationale that (i) it is present with proteasome subunits and ubiquitin in cellular inclusions, (ii) it interacts with human Rad23, a protein that may translocate proteolytic substrates to the proteasome, and (iii) it shares regions of sequence similarity with the proteasome subunit S5a, which can recognize multiubiquitinated proteins. We report that ataxin-3 interacts with ubiquitinated proteins, can bind the proteasome, and, when the gene harbors an expanded repeat length, can interfere with the degradation of a well-characterized test substrate. Additionally, ataxin-3 associates with the ubiquitin- and proteasome-binding factors Rad23 and valosin-containing protein (VCP/p97), findings that support the hypothesis that ataxin-3 is a proteasome-associated factor that mediates the degradation of ubiquitinated proteins.
The genetic basis for Machado-Joseph disease (MJD) is an expansion of a trinucleotide CAG repeat near the C terminus of the gene encoding ataxin-3, a cytoplasmic protein whose normal function is unknown (24, 25). This autosomal dominant disorder, also known as spinocerebellar ataxia type 3 (SCA3), is a common inherited ataxia and is characterized by the expansion of a polyglutamine tract (40). Unaffected individuals have 10 to 40 glutamine repeat lengths, whereas MJD patients exhibit 55 to 84 expanded repeat lengths (36), with a significant correlation between the number of repeats and disease severity (13, 24). Polyglutamine expansion presumably leads to an altered, misfolded domain within the protein. Despite the fact that ataxin-3 is ubiquitously expressed throughout the body, pathology occurs only in the brain, where ataxin-3 accumulates in inclusions, along with other proteins including molecular chaperones and components of the ubiquitin-proteasome degradation pathway (37, 42). The presence of ubiquitin or ubiquitinated species in inclusions suggests that alterations in the ubiquitin-proteasome degradation system may contribute to the etiology of this disease, whose clinical presentation includes impaired walking, coordination, and speech.
The ubiquitin-proteasome pathway is the principal mechanism for the turnover of short-lived and damaged proteins in eukaryotic cells (16). The 26S proteasome consists of a 20S proteolytic core that is capped at one or both ends by the 19S regulatory particle. Studies of the polyglutamine disease spinocerebellar ataxia type 1 (SCA1) revealed a redistribution of the proteasome into intranuclear aggregates formed by the disease protein, ataxin-1 (8). More recently, it was determined by immunohistochemistry that only a fraction of examined intranuclear inclusions from MJD patients were immunopositive for antibodies directed against subunits of the 20S catalytic core, whereas subunits of 19S regulatory particles were found in the majority of inclusions (42). The apparent dissociation of the primary subcomplexes of the proteasome suggests that a perturbation in the proteasomal machinery that degrades misfolded and damaged proteins, in addition to important regulatory molecules, could contribute to the pathology in MJD patients.
The 20S proteolytic core can degrade unfolded peptides in the absence of ATP and ubiquitin (11). Degradation of properly folded, misfolded, or damaged proteins by the 26S proteasome, on the other hand, requires the presence of a multiubiquitin chain that is conjugated to the substrate for recognition by the proteasome (5, 43, 47) and is ATP dependent in the unfolding and translocation of substrates by the six “AAA” ATPases present in the 19S particle (30). Subunit S5a/Rpn10, one of 18 “core” subunits of the 19S regulatory cap, has been implicated in the recognition of multiubiquitin chains (12, 52). In yeast, this subunit is dispensable for viability (45), consistent with the presence of other multiubiquitin chain recognition factors (28). An investigation of the composition and regulation of the 26S proteasome in budding yeast revealed approximately 24 proteasome-interacting proteins that had not been previously detected in association with the “core” set of proteasome subunits (46, 48). The proteins identified, which may be conserved in humans, could perform a regulatory role or could exist only in a certain subset of cells.
One such protein that may play a role as a “shuttle factor” for translocating proteins to the proteasome for degradation is Rad23. Rad23 can bind ubiquitin (4, 7), multiubiquitin chains (39, 50), and multiubiquitinated proteins (34) through its ubiquitin-associated (UBA) domains (4, 6, 31, 38, 39). Human Rad23 has been shown to interact with both the proteasome subunit S5a (Rpn10) (19) and ataxin-3 (49) through its N-terminal ubiquitin-like (Ubl) domain. Another such protein that may perform a role as a multiubiquitin chain-targeting factor, required for the degradation of certain ubiquitin-proteasome substrates, is valosin-containing protein (VCP) (9, 10). VCP is a mammalian homolog of the yeast cell cycle division protein Cdc48p and of p97 in Xenopus laevis (14, 27). These proteins are members of a highly conserved AAA family of ATPases associated with a variety of cellular activities, including the control of cell cycle division, membrane fusion, vesicle-mediated transport, and the ubiquitin-proteasome degradation pathway (35). Cdc48p contributes to ubiquitin-mediated proteolysis by the ubiquitin-fusion degradation (UFD) pathway in yeast (14) and has been identified as a proteasome-interacting protein (46) that is necessary for the dislocation of endoplasmic reticulum degradation substrates (21). Similarly, VCP has been shown to copurify and coimmunoprecipitate with the 26S proteasome, to have ATPase activity, and to be a multiubiquitin chain-targeting factor required for degradation by the ubiquitin-proteasome pathway (10). VCP/p97 has been found in abnormal protein aggregates (18), is shown here and elsewhere to bind ataxin-3 (26), and has been identified as a modulator of polyglutamine-induced neurodegeneration (17).
The physical interactions between ataxin-3 and Rad23, as well as those between ataxin-3 and VCP, provide compelling links for a role of ataxin-3 in the proteolytic system. We demonstrate here that ataxin-3 interacts with ubiquitinated proteins, can bind the proteasome, and when the gene harbors an expanded repeat length, can interfere with the degradation of a well-characterized test substrate. Building on earlier studies, we suggest a model in which the delivery of ubiquitinated substrates by Rad23 to ataxin-3 requires VCP. The activity of VCP is consistent with that of an “uncoupling factor” that transfers ubiquitinated substrates from Rad23 to ataxin-3. An important implication of these studies is that the delivery of ubiquitinated substrates to the proteasome might not occur through a passive interaction with substrate-linked multiubiquitin chains. We propose that substrate targeting may involve a system of regulated trafficking that requires the ATP-dependent uncoupling activity of regulatory molecules to transfer substrates from the shuttle-factors to specific proteasome subunits.
MATERIALS AND METHODS
Strains and plasmids.
Blood was extracted from MJD patients who gave informed consent. Total RNA was extracted, and cDNA was prepared by using Superscript II (Invitrogen, Carlsbad, Calif.). The SCA3 gene encoding ataxin-3 was amplified from cDNA with gene-specific primers and cloned into pCR2.1-TOPO (Invitrogen). The product was verified by DNA sequencing. Two clones (Q20 and Q79 repeat lengths) were chosen. Wild-type and expanded genes were amplified by PCR and cloned into pCBGST1 (2) to generate pCBGST1-ataxin-3Q20 and pCBGST1-ataxin-3Q79. This construct makes use of the metallothionein CUP1 promoter, which is inducible by the presence of copper ions. Under copper-limiting conditions the basal level of expression from the CUP1 promoter is low. Thioredoxin-ataxin-3Q20 was generated in Escherichia coli by using the pBAD/TOPO ThioFusion expression system (Invitrogen). A plasmid expressing Pre1-FLAG was provided by J. Dohmen. A plasmid for expression of glutathione S-transferase (GST)-VCP in E. coli was provided by L. Samelson. This construct has a Factor Xa (Pharmacia) site for the cleavage and preparation of VCP. The gene for VCP was amplified from this plasmid by PCR and cloned into pCBGST1 to generate pCBGST1-VCP for expression in yeast. A plasmid for the expression of GST-UBA1 (of yeast Rad23) in E. coli was provided by L. Chen. GST-hHR23B was purified from E. coli. A plasmid expressing PGAL1::Rpn8-V5 was purchased from Invitrogen. The expression and purification of Ubc2 have been described previously (34). E1 (yeast), histone H2B, and ubiquitin were purchased from Boston Biochem (Cambridge, Mass.), Roche (Indianapolis, Ind.), and Sigma (St. Louis, Mo.), respectively.
Antibodies.
Monoclonal antibodies to Rpt1, S5a, ubiquitin (FK1), and the 20S particle (subunits α1, -2, -3, -5, -6, and -7) were purchased from Affiniti (Exeter, United Kingdom). FLAG- and GST-specific monoclonal antibodies and a ubiquitin-specific polyclonal antibody were purchased from Sigma. Monoclonal anti-thioredoxin (anti-Thio) and anti-V5 antibodies were purchased from Invitrogen. An anti-VCP/p97 monoclonal antibody was purchased from Research Diagnostics (Flanders, N.J.) and showed no cross-reactivity with Cdc48. A monoclonal antibody against c-Myc was purchased from Clontech (Palo Alto, Calif.). Monoclonal anti-β-galactosidase (anti-β-Gal) antibodies were purchased from Promega (Madison, Wis.) and Sigma (2.5 μg/ml for immunoprecipitations). Antibodies were used at the dilutions recommended by the manufacturers: 1/5,000 for anti-Rpt1; 1/1,000 for the monoclonal antibodies against S5a, ubiquitin (FK1), the 20S particle, FLAG, and GST; 1/500 for the polyclonal antibody against ubiquitin; 1/5,000 for anti-Thio and anti-V5; 1/1,000 for anti-VCP/p97 and anti-c-Myc; and 1/5,000 and 2.5 μg/ml (for immunoprecipitations) for anti-β-Gal. Anti-ataxin-3 polyclonal antibodies were generated against Thio-ataxin-3Q20 (Pocono Rabbit Farm and Laboratory, Canadensis, Pa.), affinity purified, and used at a dilution of 1/1,000 for Western blotting. Anti-mouse and anti-rabbit secondary antibodies were obtained from Chemicon (Temecula, Calif.). Recombinant protein A (rPA)-Sepharose beads were obtained from Repligen (Cambridge, Mass.).
Cell culture.
HEK293T and NT2 (American Type Culture Collection, Manassas, Va.) cells were cultured at 37°C in minimal essential medium (MEM) with 10% heat-inactivated horse serum supplemented with 1.0 mM sodium pyruvate, 0.1 mM MEM nonessential amino acids, and 1.5 g of sodium bicarbonate (Gibco BRL, Rockville, Md.)/liter in a humidified atmosphere with 5% CO2. All pellets were harvested by trypsin dissociation, washed twice in phosphate-buffered saline, and frozen in liquid nitrogen.
Preparation of protein extracts and immunological methods.
To demonstrate the in vivo interaction of ataxin-3 with ubiquitinated species, yeast strains expressing GST, GST-ataxin-3Q20, or GST-ataxin-3Q79 were grown to late-logarithmic phase in synthetic medium, pelleted, and frozen at −70°C. As positive controls, FLAG-Rpn10 (29), FLAG-Rad23 (41), and GST-Rad23 (41) were similarly grown and harvested. Yeast cells were suspended in buffer A (50 mM HEPES [pH 7.5], 150 mM NaCl, 5 mM EDTA, and 1% Triton X-100 with the addition of protease inhibitors [including Pefabloc SC, leupeptin, aprotinin, antipain, pepstatin, and chymostatin]) and lysed by glass bead disruption. For gel filtration, protease assays, and isolation of the intact 26S proteasome, 4 mM ATP was included in buffer A. Extracts were normalized to equal protein concentrations and volumes and were applied directly to either glutathione Sepharose 4B (Pharmacia, Piscataway, N.J.) or anti-FLAG-M2 affinity agarose (Sigma), depending on the protein to be isolated. Beads were washed three times with buffer A, and the bound proteins were released by boiling and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, and subjected to immunoblotting using appropriate antibodies.
NT2 and 293T cells were lysed in buffer M (50 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 4 mM ATP, 0.5% NP-40, 10 μg [each] of leupeptin, aprotinin, and pepstatin/ml, 1 mM phenylmethylsulfonyl fluoride) with sonication followed by centrifugation at 14,000 × g. Immunoprecipitations from mammalian cell lysates were performed using antibodies coupled to rPA-Sepharose (Repligen). Equal protein concentrations were used for immunoprecipitation and pulldown experiments.
An equimolar mixture of di-, tri-, and tetraubiquitin [(Ub)2-4], a mix of (Ub)2-7, or tetraubiquitin [(Ub)4] alone (all from Affiniti) was used in experiments to examine the interaction between ataxin-3 and multiubiquitin chains. Following pulldowns, beads were washed extensively with buffer A. Bound proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, and subjected to immunoblotting with antibodies against ubiquitin (Sigma or Affiniti).
Protease assays.
Total-cell lysates from both yeast and mammalian cells were prepared as described above with the addition of 4 mM ATP, normalized to equal protein contents, and assayed for chymotrypsin-like protease activity by using the fluorogenic substrate Suc-LLVY-AMC (Boston Biochem) (32) with a TD-700 fluorometer (Turner Designs, Inc., Sunnyvale, Calif.). Lysates (containing 5 mg of protein) were also separated by gel filtration chromatography using a Superose 6 10/30 HR column (Pharmacia) in a buffer containing 25 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM ATP, and 10% glycerol. One-milliliter fractions were collected, and equal volumes were tested for protease activity and examined by SDS-PAGE.
Protein stability.
Pulse-chase measurements were performed as described previously (34). Briefly, yeast cells expressing GST-ataxin-3Q20 or GST-ataxin-3Q79 were grown at 30°C to mid-log phase and labeled with [35S]methionine and [35S]cysteine (Perkin-Elmer, Boston, Mass.) for 5 min. The cells were washed in a chase medium containing excess unlabeled methionine and cysteine and 0.5 mg of the translation inhibitor cycloheximide/ml, samples were withdrawn at intervals, and proteins were prepared for pulldowns with glutathione Sepharose 4B (Pharmacia). To examine the effects of wild-type and mutant ataxin-3 on the degradation of a well-characterized test substrate, Ub-Pro-β-Gal was expressed in yeast cells containing either GST, GST-ataxin-3Q20, or GST-ataxin-3Q79, followed by the pulse-chase procedures described above. The stability of a control substrate (Met-β-Gal) was also measured. Protein concentrations were normalized to 10% trichloroacetic acid-insoluble 35S, and proteins were prepared for immunoprecipitation with ∼2.5 μg of a monoclonal antibody against β-Gal (Sigma)/ml. The immunoprecipitates were washed three times in buffer A plus 0.1% SDS and then subjected to SDS-8% PAGE. Because these test substrates display biphasic degradation, half-life measurements were performed during the initial phase (0 to 10 min) of the assay by a conventional method as described by Bachmair et al. (1).
RESULTS
Ataxin-3 interacts with multiubiquitin chains and ubiquitinated proteins in vitro and in vivo.
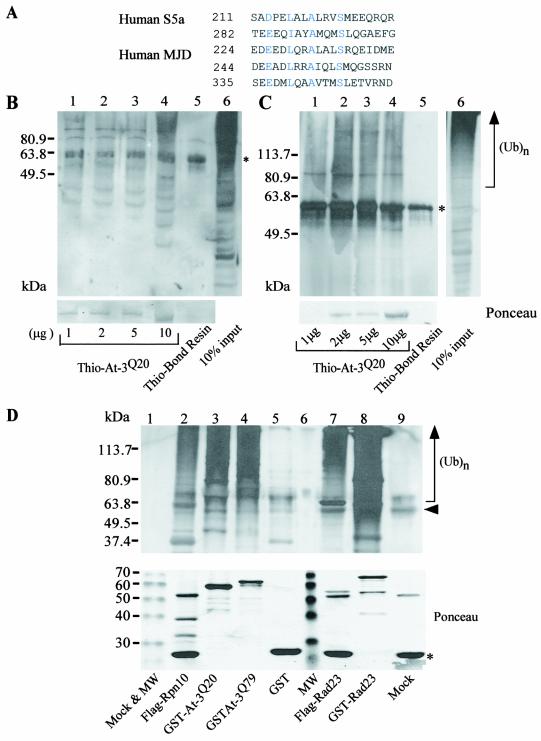
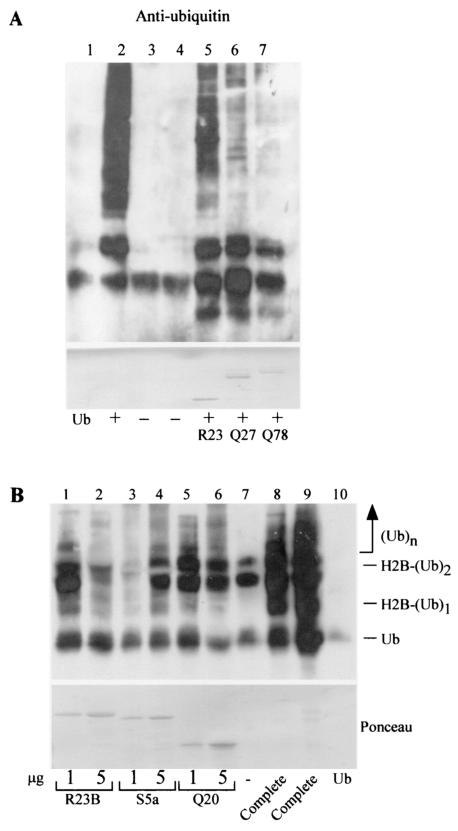
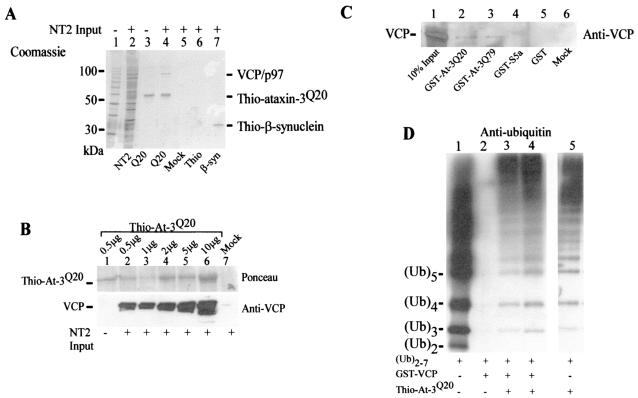
Sequence alignment revealed significant similarity between ataxin-3 and S5a in a conserved ubiquitin-interacting motif (UIM) which is known to recognize multiubiquitin chains (hydrophobic amino acid-X-X-Ala-X-X-X-Ser-X-X-acidic amino acid) (20) (Fig. 1A). Therefore, we examined whether ataxin-3 could bind ubiquitinated proteins. cDNA was prepared from lymphocytes derived from patients with normal and expanded alleles of ataxin-3, and DNA was amplified by PCR. We anticipated that studies with yeast and human cells would yield similar results, because the ubiquitin-proteasome system is one of the most conserved pathways in eukaryotic evolution, and the sequence of ubiquitin is ∼90% identical across species. Therefore, we expressed and purified the fusion protein Thio-ataxin-3Q20 from E. coli on Thio-Bond resin (Invitrogen), applied total yeast and NT2 cell extracts to the Thio-ataxin-3Q20 matrix, and detected an interaction with high-molecular-weight ubiquitin- cross-reacting material (Fig. 1B and C, respectively). We also constructed GST fusion proteins of ataxin-3Q20 and the expanded form, ataxin-3Q79, for expression in yeast cells. To examine the interaction with ubiquitinated proteins in vivo, we purified GST (Fig. 1D, lane 5), GST-ataxin-3Q20 (lane 3), and GST-ataxin-3Q79 (lane 4) from yeast cells and separated the precipitated proteins by SDS-PAGE. As positive controls, FLAG-Rpn10 (Fig. 1D, lane 2), FLAG-Rad23 (lane 7), and GST-Rad23 (lane 8), which have been shown to interact with multiubiquitinated proteins, were also expressed and purified accordingly. The resolved proteins were transferred to nitrocellulose membranes and then incubated with antibodies against ubiquitin (Fig. 1D). We determined that both ataxin-3Q20 and ataxin-3Q79 are associated with ubiquitinated proteins, as has been observed for other multiubiquitin chain-binding proteins, such as Rad23 and Rpn10. A small fraction of the ataxin-3 protein was conjugated to one or two ubiquitin moieties, as evidenced by a slight shift in the electrophoretic mobilities of Ub-GST-ataxin-3Q20 and Ub-GST-ataxin-3Q79 (Fig. 2A, lanes 2 and 3), which cross-reacted weakly with anti-GST antibodies. In addition, anti-ataxin-3 antibodies showed a pattern very similar to that of anti-GST antibodies (data not shown). However, the bulk of high-molecular-weight ubiquitin-cross-reacting material that was associated with GST-ataxin-3Q20 and GST-ataxin-3Q79 (Fig. 2A, lanes 5 and 6) represented other ubiquitinated cellular proteins, as indicated by a lack of material reactive against anti-GST antibodies in the high-molecular-weight regions of the gel which cross-reacted with ubiquitin-specific antibodies (Fig. 2A).
FIG. 1.
A sequence motif (UIM) in S5a that can bind ubiquitinated proteins is present in ataxin-3. (A) Alignment of conserved regions of S5a and ataxin-3. The position of the amino acid residue at the N terminus of the conserved UIM sequence is shown on the left. Amino acid residues in blue are identical or highly conserved. (B) Ataxin-3 can bind ubiquitinated proteins in yeast cell extracts. Protein extracts were prepared from a wild-type yeast strain, and 1 mg was incubated with various amounts of Thio-ataxin-3Q20 that was purified from E. coli. After extensive washing, the proteins that were bound to Thio-ataxin-3 were separated in an SDS-12% polyacrylamide gel, transferred to nitrocellulose filters (Bio-Rad), and incubated with ubiquitin-specific antibodies (Sigma). The immunoblot was developed with enhanced chemiluminescence (Dupont, NEN). Lanes 1 to 4 contain, respectively, 1, 2, 5, and 10 μg of Thio-ataxin-3Q20 on affinity beads. Lane 5, control in which Thio-Bond resin was incubated with a yeast cell extract. A sample of the yeast cell extract (10% input) is shown in lane 6. The positions of molecular size standards are shown in the left margin. Asterisks in panels B and C indicate a nonspecific band (from Thio-Bond resin) that cross-reacted with anti-ubiquitin antibodies. Ponceau staining shows the amount of ataxin-3 protein used in the pulldown assay. At-3, ataxin-3. (C) Ataxin-3 can bind ubiquitinated proteins from NT2 human cell extracts. Protein extracts were prepared from NT2 cells, applied to various amounts of Thio-ataxin-3Q20 immobilized onbeads, and examined as described for panel B, but in an SDS-10% polyacrylamide gel. Lanes are as in panel B, except that lane 6 corresponds to 10% input of NT2 cell extract. Ubiquitinated proteins are indicated by the designation (Ub)n in the right margin. Ponceau staining shows the amount of ataxin-3 protein used in the pulldown assay. (D) Ataxin-3 interacts with ubiquitinated proteins in vivo in yeast cells. Protein extracts were prepared from strains expressing FLAG-Rpn10, GST-ataxin-3Q20, GST-ataxin-3Q79, GST, FLAG-Rad23, or GST-Rad23 from the copper-inducible CUP1 promoter. Fusion proteins were isolated either by pulldown with glutathione Sepharose (Amersham Pharmacia Biotech) or by immunoprecipitation with FLAG-agarose (Sigma). The bound proteins were separated in an SDS-10% polyacrylamide gel and transferred to nitrocellulose membranes, and ubiquitinated proteins were detected. Lanes 1 and 9, mock reactions in which an extract derived from wild-type yeast that did not overexpress any of the fusion proteins was applied to either glutathione Sepharose or FLAG-agarose, respectively; lane 2, FLAG-Rpn10; lane 3, GST-ataxin-3Q20; lane 4, GST-ataxin-3Q79; lane 5, GST; lane 6, molecular weight (MW) markers; lane 7, FLAG-Rad23; lane 8, GST-Rad23. For lanes 2, 7, and 9, an arrowhead indicates the immunoglobulin heavy chain. Asterisk, immunoglobulin light chain detected as an ∼23-kDa band in lanes 2, 7, and 9 in the lower panel. Ponceau staining shows the purified proteins prior to incubation with anti-ubiquitin antibodies, with theoretical molecular sizes as follows: for Flag-Rpn10, ∼30 kDa; for GST-ataxin-3Q20, ∼69 kDa; for GST-ataxin-3Q79, ∼78 kDa; for GST, ∼28 kDa; for FLAG-Rad23, ∼43 kDa; and for GST-Rad23, ∼70 kDa. Note that apparent molecular sizes as determined from SDS-PAGE are slightly different from those determined theoretically.
FIG. 2.
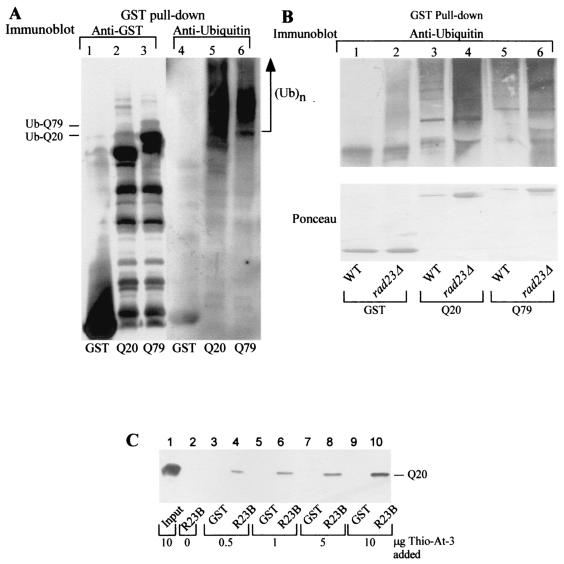
Ataxin-3 interacts with ubiquitinated proteins in wild-type and rad23Δ yeast strains. (A) The high-molecular-weight ubiquitin-cross-reacting material that is purified with GST-ataxin-3Q20 and GST-ataxin-3Q79 does not represent ubiquitinated ataxin-3. Protein extracts were prepared from yeast strains expressing either GST (lanes 1 and 4), GST-ataxin-3Q20 (Q20) (lanes 2 and 5), or GST-ataxin-3Q79 (Q79) (lanes 3 and 6) and were purified on glutathione Sepharose. The precipitated proteins were washed with buffer A, separated by SDS-10% PAGE, transferred to nitrocellulose membranes, and exposed to either GST-specific antibodies (lanes 1 to 3) or ubiquitin-specific antibodies (lanes 4 to 6). The anti-GST immunoblot demonstrates that the majority of high-molecular-weight ubiquitin-cross-reacting material is not a ubiquitinated GST-ataxin-3 species. The GST-ataxin-3 constructs appear to be conjugated to one or two ubiquitins (Ub-Q79 and Ub-Q20 in the left margin) based on the reduced mobility of GST-ataxin-3Q20 and GST-ataxin-3Q79. The bands are consistent with the addition of one ubiquitin moiety. In contrast, incubation of the same blot with anti-ubiquitin antibodies demonstrates that a large quantity of ubiquitinated proteins migrating at a high molecular weight is associated with the ataxin-3 constructs. The reaction against anti-GST and anti-ataxin-3 (data not shown) antibodies demonstrated that the high-molecular-weight ubiquitin-cross-reacting material is not ubiquitinated GST-ataxin-3. The designations Q20 and Q79 refer to full-length ataxin-3 proteins. (B) Protein extracts were prepared from wild-type (WT) or rad23Δ yeast strains expressing either GST (lanes 1 and 2), GST-ataxin-3Q20 (Q20) (lanes 3 and 4), or GST-ataxin-3Q79 (Q79) (lanes 5 and 6). Equal amounts of extract were applied to glutathione Sepharose to purify GST or the GST-ataxin-3 fusion proteins. Interacting ubiquitinated proteins were detected with ubiquitin-specific antibodies. Ubiquitinated proteins were bound to both GST-ataxin-3 constructs purified from wild-type and rad23Δ strains. Ponceau staining shows the amounts of proteins purified. (C) Ataxin-3 interacts with hHR23B. GST-hHR23B and Thio-ataxin-3Q20 were purified from E. coli. Varying amounts of Thio-ataxin-3Q20 (indicated below the gel) were incubated with either 1 μg of GST (lanes 3, 5, 7, and 9) or 2 μg of GST-hHR23B (R23B) (lanes 4, 6, 8, and 10). Interaction between GST-hHR23B and Thio-ataxin-3Q20 was detected by immunoblotting using anti-Thio antibodies (Invitrogen). Lane 1, 10 μg of Thio-ataxin-3Q20 alone; lane 2, GST-hHR23B without incubation with Thio-ataxin-3Q20. The position of Thio-ataxin-3Q20 (Q20) is indicated in the right margin.
We wanted to confirm that the interactions of the ataxin-3 proteins with ubiquitinated proteins were not dependent on the presence of Rad23, since the human homologs of the yeast DNA repair protein Rad23 (hHR23A and hHR23B) have been shown to bind ataxin-3 (49). Both GST-ataxin-3Q20 and GST-ataxin-3Q79 showed strong interactions with ubiquitinated proteins in rad23Δ cells, comparable to those observed in wild-type cells based on the amount of ataxin-3 that was recovered (see the Ponceau stain of the immunoblot) from each strain (Fig. 2B, lanes 3 and 4 for GST-ataxin-3Q20 and lanes 5 and 6 for GST-ataxin-3Q79). We confirmed the results reported by other investigators, that hHR23B interacts with ataxin-3 (Fig. 2C), by pulldown experiments using the purified proteins GST-hHR23B and Thio-ataxin-3Q20. Briefly, GST-hHR23B was purified from E. coli by using glutathione Sepharose 4B. A 20-fold concentration range (∼0.5 to 10 μg) of purified Thio-ataxin-3Q20 was incubated with either 1 μg of GST (molar ratio of GST to Thio-ataxin-3Q20, ∼4:1 to 1:6) or 2 μg of GST-hHR23B (molar ratio of GST-hHR23B to Thio-ataxin-3Q20, ∼3:1 to 1:7) purified on beads. No interaction of Thio-ataxin-3Q20 with GST (Fig. 2C, lanes 3, 5, 7, and 9) was found, whereas an interaction was observed between GST-hHR23B and Thio-ataxin-3Q20 over the entire concentration range of Thio-ataxin-3Q20 examined (lanes 4, 6, 8, and 10).
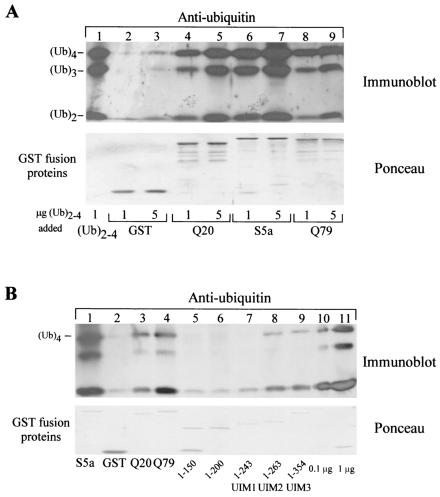
We purified GST-ataxin-3Q20 and GST-ataxin-3Q79 from yeast cells with glutathione Sepharose, washed the matrices with 1 M NaCl and 0.1% SDS to remove nonspecifically bound proteins, equilibrated them in buffer A, added a mixture of (Ub)2, (Ub)3, and (Ub)4 to the matrix, and determined that both GST-ataxin-3Q20 and GST-ataxinQ79could bind ubiquitin chains, like GST-S5a (Fig. 3A). In order to determine which domain(s) of ataxin-3 is responsible for the binding to ubiquitin chains, truncated derivatives of ataxin-3Q20 were generated by PCR. As above, these constructs were expressed and purified from yeast cells with glutathione Sepharose, washed with 1 M NaCl and 0.1% SDS to remove nonspecifically bound proteins, and equilibrated in buffer A. (Ub)4 (1 μg) was added to the purified proteins, and we determined, as expected, that the interaction with (Ub)4 is dependent on the presence of the UIM domains. We consistently observed a very weak interaction for the construct harboring only the first UIM motif (Fig. 3B, lane 7; band not readily visible). Addition of UIM2 (Fig. 3B, lane 8) and UIM3 (lane 9) greatly enhanced the affinity of ataxin-3 for (Ub)4, although none of the truncated constructs exhibited an affinity comparable to that observed for the full-length ataxin-3 protein (lanes 3 and 4), suggesting that the affinity for (Ub)4 chains may require multiple UIM domains or may be influenced by the proper folding of the full-length protein.
FIG. 3.
Ataxin-3 can interact with multiubiquitin chains. (A). GST, GST-ataxin-3Q20, and GST-ataxin-3Q79 were bound to glutathione Sepharose 4B. GST-S5a was purified on glutathione agarose (Affiniti). One and five micrograms of an equimolar mixture of (Ub)2-4 (Affiniti) were incubated with the beads, and bound proteins were resolved on an SDS-10% polyacrylamide gel, transferred to nitrocellulose membranes, and detected with ubiquitin-specific antibodies. Lanes: 2 and 3, GST; 4 and 5, GST-ataxin-3Q20; 6 and 7, GST-S5a; 8 and 9, GST-ataxin-3Q79. One microgram of (Ub)2-4 was added to lanes 1, 2, 4, 6, and 8, while 5 μg was added to lanes 3, 5, 7, and 9. Ponceau staining shows that the protein levels were similar. (B) UIMs of ataxin-3 are necessary for binding to ubiquitin chains. GST-S5a was purified on glutathione agarose (Affiniti). GST, full-length GST-ataxin-3Q20 (Q20), full-length GST-ataxin-3Q79 (Q79), and GST-ataxin-3 truncation proteins were purified on glutathione Sepharose, and equal amounts were incubated with 1 μg of (Ub)4 (Affiniti). The interactions were determined by immunoblotting with ubiquitin-specific antibodies (Sigma). Lane 1, GST-S5a; lane 2, GST; lane 3, GST-ataxin-3Q20; lane 4, GST-ataxin-3Q79; lane 5, GST-ataxin-31-150; lane 6, GST-ataxin-31-200; lane 7, GST-ataxin-31-243 (UIM1); lane 8, GST-ataxin-31-263 (UIM2); lane 9, GST-ataxin-31-354 (UIM3); lane 10, 100 ng of (Ub)4; lane 11, 1 μg of (Ub)4. Ponceau staining shows that equal amounts of GST proteins were present.
Ataxin-3 associates with the proteasome.
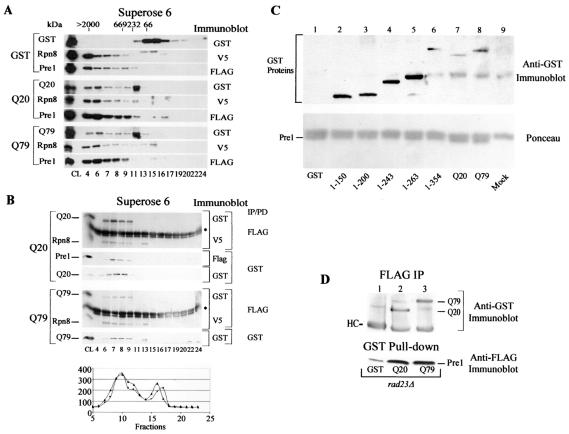
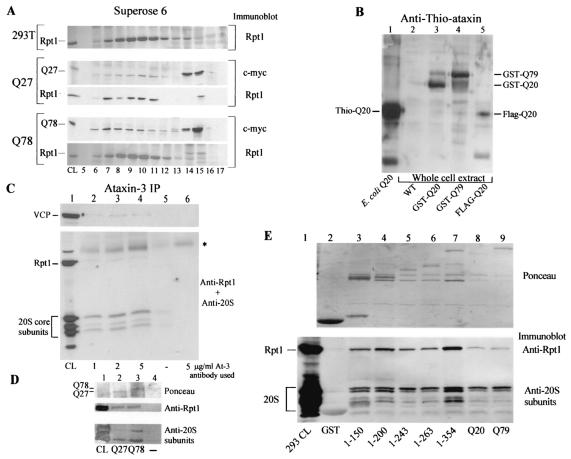
To examine the interaction of wild-type and mutant ataxin with the proteasome, GST, GST-ataxin-3Q20, and GST-ataxin-3Q79 were expressed in yeast cells that harbored Rpn8-V5 and Pre1-FLAG (which are epitope-tagged subunits of the 19S and 20S particles, respectively). Total protein extracts were prepared and resolved by gel filtration chromatography using a Superose 6 HR 10/30 column. Fractions were analyzed by immunoblotting to identify the tagged proteasome subunits and to examine proteasome-specific activity (Fig. 4). We detected GST in fractions corresponding to low-molecular-weight proteins (∼30 to 60 kDa; fractions 15 to 19), in contrast to Pre1-FLAG and Rpn8-V5, which were detected almost exclusively in fractions corresponding to the void volume and high-molecular-weight regions of the chromatogram (Fig. 4A, top set of panels, fractions 6 to 8). The occurrence of Pre1-FLAG and Rpn8-V5 in the same fractions suggested the presence of the intact 26S proteasome. In striking contrast to GST, both GST-ataxin-3Q20 and GST-ataxin-3Q79 were detected in the fractions that contained Pre1-FLAG and Rpn8-V5, suggesting an association with the proteasome (Fig. 4A, middle and bottom sets of panels, respectively). We also detected the 19S subunit, Rpt1/Cim5, in these fractions (data not shown).
FIG. 4.
Ataxin-3 interacts with the 26S proteasome. (A) Ataxin-3 cofractionates with yeast proteasome subunits Pre1 and Rpn8 in a Superose 6 10/30 HR column. (Top set of panels) Fractions of protein extracts that contain GST, Pre1-FLAG, and Rpn8-V5. (Center set of panels) Fractions of protein extracts that contain GST-ataxin-3Q20 (Q20), Pre1-FLAG, and Rpn8-V5. (Bottom set of panels) Fractions of protein extracts that contain GST-ataxin-3Q79 (Q79), Pre1-FLAG, and Rpn8-V5. Proteins from fractions were precipitated with 10% trichloroacetic acid, washed with 100% acetone, separated by SDS-10% PAGE, transferred to nitrocellulose membranes, and incubated with appropriate antibodies. Specific proteins that were detected are indicated in the left margin. Antibodies that were used for detection in immunoblots are given in the right margin. Fraction numbers are given below the bottom gel. (B) Ataxin-3 can be immunoprecipitated with the proteasome. The upper set of panels represents the fractions of separated extracts containing ataxin-3Q20, and the lower set represents ataxin-3Q79. Fractions were incubated with FLAG-agarose or glutathione Sepharose to purify Pre1-FLAG or GST-ataxin-3, respectively. Proteins that were coprecipitated were then separated in an SDS-10% polyacrylamide gel. The immunoprecipitations (IP) or pulldowns (PD) and the subsequent immunoblots that were performed are indicated in the right margin. Fraction numbers are given below the bottom gel. GST-ataxin-3Q20 and Rpn8-V5 were both copurified with Pre1-FLAG (top set, first panel). Similarly, Pre1-FLAG was detected in association with GST-ataxin-3Q20 (top set, second panel). Immunoblotting with anti-GST antibodies confirmed that similar amounts of GST-ataxin-3Q20 and GST-ataxin-3Q79 were recovered (top set, third panel, and bottom set, second panel). Pre1-FLAG was immunoprecipitated from fractions that contained GST-ataxin-3Q79, and both ataxinQ79 and Rpn8-V5 could be detected. Asterisks indicate a cross-reaction against the immunoglobulin heavy chain. The graph shows chymotrypsin-like protease activities determined in the gel filtration fractions (▪, Q20; ▴, Q79) by using the fluorogenic substrate Suc-Leu-Leu-Val-Tyr-AMC (10 μM). The y axis represents arbitrary fluorescence signal units for raw fluorescence readings (with the value for the blank subtracted). The x axis represents the fractions assigned. Maximal activity was detected in fractions 6 to 13. The activity detected in fractions 15 to 17 may represent an undetermined protein that has chymotryptic activity. (C) Residues 1 to 150 of ataxin-3 are sufficient for the interaction with the proteasome. Protein extracts containing GST, GST-ataxin-3, or truncated constructs were prepared from yeast cells that also expressed Pre1-FLAG. Pre1-FLAG was immunoprecipitated with FLAG-agarose (Sigma), and Ponceau staining showed that equivalent amounts were purified from each extract (bottom panel). Lane 1, GST; lane 2, GST-ataxin-31-150; lane 3, GST-ataxin-31-200; lane 4, GST-ataxin-31-243 (UIM1); lane 5, GST-ataxin-31-263; lane 6, GST-ataxin-31-354; lane 7, full-length GST-ataxin-3Q20; lane 8, full-length GST-ataxin-3Q79; lane 9, mock reaction in which a wild-type yeast cell extract was applied to FLAG-agarose. The immunoblot filter was incubated with anti-GST antibodies, and all the ataxin-3 proteins were detected (top panel). (D) Rad23 is not necessary for the interaction of ataxin-3 with the proteasome. GST (lane 1), GST-ataxin-3Q20 (lane 2), or GST-ataxin-3Q79 (lane 3) was expressed in wild-type and rad23Δ yeast strains that expressed Pre1-FLAG. Pre1-FLAG or GST-ataxin-3 fusion proteins were purified with FLAG-agarose or glutathione Sepharose, respectively. Pre1-FLAG was immunoprecipitated, and the interaction between GST-ataxin-3 and the proteasome was determined by immunoblotting with anti-GST antibodies (top panel). The immunoglobulin heavy chain is indicated in the left margin (HC). The bottom panel shows that in a reciprocal experiment, extracts were incubated with glutathione Sepharose, and GST or GST-ataxin-3 was purified. Immunoblotting with anti-FLAG antibodies confirmed the interaction between ataxin-3 and the proteasome. A small amount of Pre1-FLAG interacted nonspecifically with glutathione Sepharose (lane 1).
To further explore the idea that ataxin-3 may associate with the proteasome, we incubated Superose 6 gel filtration fractions with either FLAG-agarose (to precipitate Pre1-FLAG) or glutathione Sepharose (to isolate GST-ataxin-3Q20 and GST-ataxin-3Q79) in order to determine if the presence of ataxin-3 in the fractions that contained proteasome subunits reflected a genuine interaction. For instance, it was conceivable that GST-ataxin-3Q20 or GST-ataxin-3Q79 had aggregated or was associated with another large complex. The precipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, and incubated with specific antibodies. We found that GST-ataxin-3Q20 and GST-ataxin-3Q79 coimmunoprecipitated with Pre1-FLAG (Fig. 4B, top panels, each set), although the amount of ataxin-3Q79 associated with Pre1 was less than that observed for ataxin-3Q20. In the reciprocal experiment, Pre1-FLAG was recovered in association with GST-ataxin-3Q20, demonstrating that the proteins were in the same complex (Fig. 4B, middle panel, top set). We consistently copurified Pre1-FLAG in association with GST-ataxin-3Q79, although the signal was always weak and not readily reproducible on X-ray film (data not shown).
We investigated if the amount of GST-ataxin-3Q79 that was purified on glutathione Sepharose was less than the amount of GST-ataxin-3Q20 that was recovered. A Western blot of the corresponding GST pulldowns for GST-ataxin-3Q20 and GST-ataxin-3Q79 performed with anti-GST antibodies (Fig. 4B, bottom panels for each set) demonstrated that similar amounts of these two ataxin-3 proteins were purified. As described above, the amount of GST-ataxin-3Q79 that was immunoprecipitated with Pre1-FLAG was less than that observed for GST-ataxin-3Q20 (Fig. 4B, top panels for each set). Equivalent levels of Rpn8-V5 were recovered in association with Pre1-FLAG. This finding indicates that comparable levels of the intact 26S proteasome were present in strains that expressed either GST-ataxin-3Q20 or GST-ataxin-3Q79 (top panel, each set). Since similar levels of GST-ataxin-3 proteins were recovered in GST pulldowns, we conclude that ataxin-3Q79 interacts more weakly with the proteasome. GST alone could not coprecipitate Pre1-FLAG or Rpn8-V5 (data not shown). As expected, protease activity was detected in the fractions that contained Pre1-FLAG (Fig. 4B). The protease activity observed in fractions 15 to 17 probably represents a protein in yeast cell lysates that demonstrates a chymotrypsin-like activity.
GST-ataxin-3 truncation constructs were expressed in yeast cells that also harbored plasmids for the expression of Pre1-FLAG and Rpn8-V5 in order to identify the domain that is necessary for the interaction of ataxin-3 with the proteasome. We demonstrated that the N-terminal residues 1 to 150 are sufficient for interaction with the proteasome (Fig. 4C, lane 2). Immunoprecipitations of Pre1-FLAG with FLAG-agarose showed that GST-ataxin-3 constructs could be coprecipitated (Fig. 4C, top panel). The amounts of Pre1 that were purified with FLAG-agarose were equivalent in all the immunoprecipitation reactions (Fig. 4C, bottom panel). The relative amounts of full-length ataxin-3Q20 and ataxin-3Q79 that were immunoprecipitated with Pre1 were less than those observed for the truncated ataxin-3 constructs. This result suggests that the C-terminal residues may affect ataxin-3-proteasome interaction. Experiments performed with rad23Δ cells demonstrated that the association of ataxin-3 with the proteasome is independent of Rad23 (Fig. 4D).
GST, GST-ataxin-3Q20, and GST-ataxin-3Q79 were also expressed in yeast cells that harbored Pup1-HA, a 20S catalytic β-subunit. Pup1-HA could be coimmunoprecipitated with the ataxin-3 proteins but not with GST, and in the reciprocal experiment, only the ataxin-3 proteins were recovered with Pup1-HA (data not shown).
We confirmed these results in 293T cells with Myc-tagged ataxin-3Q27 and ataxin-3Q78 (cell pellets kindly provided by R. Pittman). Protein lysates were separated by gel filtration chromatography, and the fractions were analyzed for the presence of ataxin-3 and Rpt1 (a component of the 19S particle). As was observed in yeast cells, both ataxin-3 constructs cofractionated with the proteasome (Fig. 5A, fractions 6 to 11). As anticipated, protease activity was detected in the fractions in which ataxin-3 and Rpt1 comigrated, indicating the presence of the 20S particle (data not shown).
FIG. 5.
(A) Myc-tagged ataxin-3 cofractionates with the proteasome in 293T cell extracts. The association of ataxin-3Q27 and ataxin-3Q78 with the proteasome was examined. The top panel shows the fractionation of Rpt1 (an ATPase subunit in the 19S regulatory particle) in untransfected 293T cells. A pair of panels shows the cofractionation of ataxin-3Q27 with Rpt1 (Q27) (middle panels). Similarly, ataxin-3Q78 is present in fractions that contain Rpt1 (Q78) (lower panels). Fraction numbers are given at the bottom, and the antibodies used are given in the right margin. Specific proteins detected are shown in the left margin. (B) Validation of polyclonal ataxin-3 antibodies in an SDS-12% polyacrylamide gel. Polyclonal antibodies were generated against Thio-ataxin-3Q20 that was purified from E. coli and affinity purified before use. Lane 1 contains purified Thio-ataxin-3Q20; lanes 2 to 5 contain control yeast extract, GST-ataxin-3Q20, GST-ataxin-3Q79, and FLAG-ataxin-3Q20, respectively. The immunoblot was incubated with affinity-purified anti-ataxin-3 antibodies, and proteins of the expected sizes were detected. (C) VCP, Rpt1, and 20S core subunits (α1, -2, -3, -5, -6, and -7) can be coimmunoprecipitated with ataxin-3 from 293T cell extracts. Two milligrams of cell lysate was applied to rPA-Sepharose beads (Repligen) that were coupled with 1, 2, and 5 μg of an affinity purified anti-Thio-ataxin-3Q20 antibody/ml (lanes 2 to 4, respectively). Lane 1, 10% input, total 293T cell extract; lane 5, 293T cell extract applied to rPA-Sepharose; lane 6, anti-Thio-ataxin-3Q20 antibody coupled to rPA-Sepharose beads with no extract applied. The 20S core showed a very weak nonspecific binding to the rPA-Sepharose beads (lane 5). The asterisk in the right margin indicates a reaction against the immunoglobulin heavy chain. (D) Both 19S (Rpt1) and 20S proteasome (α1, -2, -3, -5, -6, and -7) subunits can be purified with MBP-ataxin-3Q27 (lane 2) and MBP-ataxin-3Q79 (lane 3). Two milligrams of cell lysate was applied to beads that contained the MBP-ataxin-3 proteins. Immunoblotting showed that two bands, representing the 20S subunits at approximately 29 and 32 kDa, were detected. Similarly, Rpt1 was detected in lanes that contained MBP-ataxin-3 (lanes 2 and 3). Lane 1 contains 10% of the input extract, while lane 4 represents a reaction in which 2 mg of the extract was applied to amylose beads (lacking MBP-ataxin-3 proteins). (E) GST, GST-ataxin-3 truncation constructs, GST-ataxin-3Q20, and GST-ataxin-3Q79 were purified from yeast cells and incubated with 1 mg of 293T cell extract. Lane 1, 293T whole-cell extract (10% input); lane 2, GST; lane 3, GST-ataxin-31-150; lane 4, GST-ataxin-31-200; lane 5, GST-ataxin-31-243 (UIM1); lane 6, GST-ataxin-31-263; lane 7, GST-ataxin-31-354; lane 8, full-length GST-ataxin-3Q20 (Q20); lane 9, full-length GST-ataxin-3Q79 (Q79). The top panel shows Ponceau staining of the amounts of GST or GST-fusion proteins used in the reactions. The bottom panel shows that reactions with antibodies against Rpt1 and 20S core subunits were consistent with the in vivo experiments that were performed in yeast cells. Specifically, the GST-ataxin-3 construct harboring only residues 1 to 150 was sufficient for interaction with both 19S (Rpt1) and 20S (α1, -2, -3, -5, -6, and -7) proteasome subunits.
Antibodies generated against Thio-ataxin-3Q20 expressed in E. coli were used in various immunoprecipitation experiments in mammalian cells. Validation of these antibodies is demonstrated in Fig. 5B. As can be seen, anti-ataxin-3 antibodies do not cross-react with proteins in yeast whole-cell extracts but could detect GST-ataxin-3Q20, GST-ataxin-3Q79, and FLAG-ataxin-3Q20 in wild-type yeast cells that expressed these proteins. Human Rpt1 and the 20S core subunits (α1, -2, -3, -5, -6, and -7) could be immunoprecipitated with endogenous ataxin-3 (Fig. 5C) from 293T cells by using these antibodies. (In addition, as discussed below, VCP can be immunoprecipitated with endogenous ataxin-3). Furthermore, fusions of ataxin-3 to maltose-binding protein (MBP-ataxin-3Q27 and MBP-ataxin-3Q78, provided by R. Pittman) could interact with endogenous 19S (Rpt1) and 20S (α1, -2, -3, -5, -6, and -7) subunits in NT2 human protein extracts, providing further evidence that ataxin-3 interacts with the intact 26S proteasome in human cells (Fig. 5D). Thio-ataxin-3Q20 could also be used to pull down proteasome subunits from human cell extracts (data not shown). In addition, we purified GST-ataxin-3 truncation constructs, GST-ataxin-3Q20, and GST-ataxin-3Q79 (Fig. 5E) from yeast cells with glutathione Sepharose and incubated them with protein extracts from 293T cells. As observed in in vivo experiments in yeast cells, the GST-ataxin-3 truncation construct harboring residues 1 to 150 was sufficient for interaction with the human 19S (Rpt1) and 20S (α1, -2, -3, -5, -6, and -7) proteasome subunits.
Ataxin-3, harboring an expanded glutamine repeat length, interferes with protein degradation in yeast cells.
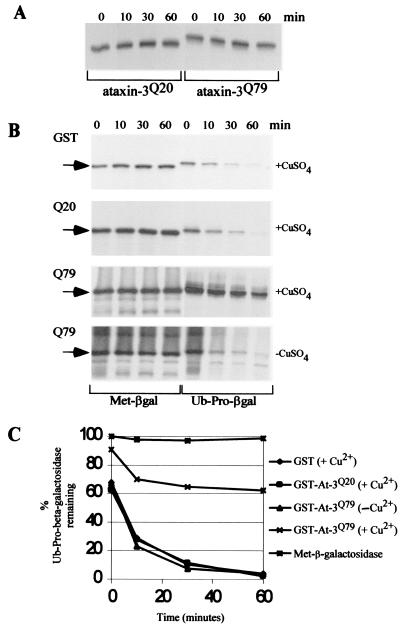
We considered the possibility that ataxin-3 might be targeted for degradation by the ubiquitin-proteasome system, since a failure to effectively regulate its levels could contribute to MJD. Alternatively, the association of ataxin-3 with the 26S proteasome could influence the stability of other cellular proteins, and the potential for mutant alleles to cause deleterious effects is evident. We examined the stability of both GST-ataxin-3Q20 and GST-ataxin-3Q79 by pulse-chase methods in yeast cells and determined that they were stable proteins (Fig. 6A). However, when we examined the stability of Ub-Pro-β-Gal, a well-characterized proteolytic test substrate (23), we found that it was stabilized by GST-ataxin-3Q79 (Fig. 6B), with an estimated half-life of >40 min, compared to half-lives of 12 and 11 min in the presence of GST and GST-ataxin-3Q20, respectively. The half-life for this substrate was derived over the initial 10 min of the assay, since the kinetics of degradation are biphasic, with an early phase that occurs quite rapidly (Fig. 6C). In addition, we observed a significant elevation of the initial amount of the substrate, Ub-Pro-β-Gal, which remained in cells that overexpressed GST-ataxin-3Q79. This phenomenon, termed the “zero point” effect, reflects a very rapid rate of decay in this initial phase of the chase (22, 44). Therefore, the increased level of Ub-Pro-β-Gal at time zero in cells that expressed ataxin-3Q79 is significant (Fig. 6C). In contrast, the amount of Ub-Pro-β-Gal at time zero in cells expressing ataxin-3Q20 or GST is much lower than the amount of Met-β-Gal detected at this time point. With prolonged incubation, degradation of Ub-Pro-β-Gal is observed in the presence of GST-ataxin-3Q79, although it is significantly lower than that observed in the presence of GST or GST-ataxin-3Q20. The half-life of Ub-Pro-β-Gal in a strain that harbored GST-ataxin-3Q79 but was grown in the absence of copper sulfate (and therefore expressed much lower levels of GST-ataxin-3Q79) was approximately 8 min. This result indicates that high-level expression of ataxin-3Q79 can interfere with the degradation of proteolytic substrates. It should be noted that ataxin-3 is highly abundant in neuronal cells. In addition, a determination of the β-Gal activity demonstrated that coexpression of GST-ataxin-3Q79 resulted in a 5- to 6-fold increase in activity, whereas coexpression of GST-ataxin-3Q20 resulted in only a 1.5-fold increase in activity (data not shown). These results provide the first direct evidence that ataxin-3 can affect proteolysis by the ubiquitin-proteasome system, and they are likely to be relevant to human cells, because Ub-Pro-β-Gal is efficiently degraded by the ubiquitin-proteasome system in mammalian cells (15).
FIG. 6.
Ataxin-3 affects proteasome function. (A) Pulse-chase experiments were performed to measure the stability of GST-ataxin-3Q20 and GST-ataxin-3Q79 in yeast cells. Both proteins were found to be stable during a 60-min chase. (B) Similarly, the stabilities of test substrates (Met-β-Gal and Ub-Pro-β-Gal) were determined in yeast cells that also expressed GST, GST-ataxin-3Q20 (Q20), or GST-ataxin-3Q79 (Q79). Met-β-Gal was stable in all three strains. In contrast, Ub-Pro-β-Gal was stabilized in the presence of high levels of GST-ataxin-3Q79 (+CuSO4). GST, GST-ataxin-3Q20, and low-level expression of GST-ataxin-3Q79 (−CuSO4) did not result in stabilization of Ub-Pro-β-Gal. (C) The percentage of Ub-Pro-β-Gal that remained at each time point (relative to the level of Met-β-Gal) was determined. We used the Kodak-one-dimensional imaging system to quantitate levels of Met-β-Gal and Ub-Pro-β-Gal at each time point in cells that expressed high levels of GST, GST-ataxin-3Q20, or GST-ataxin-3Q79. The degradation of Ub-Pro-β-Gal was also examined in a yeast strain that expressed lower levels of GST-ataxin-3Q79 (−Cu2+). The relative abundance of Met-β-Gal is also presented. Half-lives were determined over the first 10 min for the biphasic degradation. A zero point “stabilizing” effect is observed for Ub-Pro-β-Gal in yeast cells overexpressing GST-ataxin-3Q79.
Ataxin-3 inhibits the conjugation of multiubiquitin chains on H2B.
Rad23 can bind multiubiquitinated proteins and has been demonstrated to prevent multiubiquitin chain formation on H2B in an in vitro reaction (34). Since we have demonstrated that ataxin-3 can bind ubiquitinated proteins, we considered the possibility that it could also inhibit multiubiquitin chain formation on H2B in a manner similar to that of Rad23. We examined the effect of ataxin-3 in an in vitro ubiquitination reaction that contained purified Ubc2, E1, ubiquitin, and histone H2B (34). The addition of either MBP-ataxin-3Q27 or MBP-ataxin-3Q78 (kindly provided by R. Pittman) to this reaction (Fig. 7A, lanes 6 and 7) consistently inhibited the formation of high-molecular-mass ubiquitin conjugates on H2B. Thio-ataxin-3Q20 had effects similar to those observed for MBP-ataxin-3 (data not shown). In contrast to Rad23 (Fig. 7A, lane 5) or ataxin-3, thioredoxin did not affect multiubiquitin chain assembly (data not shown). To determine if the ability to inhibit chain formation was a property common to proteins that are able to bind ubiquitinated proteins, we performed an in vitro reaction in the presence of GST-hHR23B (Fig. 7B, lanes 1 and 2), GST-S5a (lanes 3 and 4), or Thio-ataxin-3Q20 (lanes 5 and 6). We found that these proteins inhibited the formation of multiubiquitin chains on H2B compared to a reaction in the absence of any additional proteins. While these results are qualitatively reproducible, quantitation of the data is difficult, due to the inherent problem of unequal mixing of matrix-bound proteins. The presence of glutathione Sepharose or Thio-Bond resin does not affect the conjugation reaction (Fig. 7B, lanes 8 and 9, respectively). These findings are consistent with those of previous studies, which suggested that the interaction between ubiquitinated proteins and factors such as Rad23 could interfere with the expansion of multiubiquitin chains (34).
FIG. 7.
Ataxin-3 can inhibit multiubiquitin chain assembly on histone H2B. (A) Addition of recombinant hHR23B (R23) (lane 5), MBP-ataxin-3Q27 (Q27) (lane 6), or MBP-ataxin-3Q78 (Q78) (lane 7) caused a marked reduction in levels of high-molecular-mass ubiquitinated derivatives of H2B. Lane 1, free ubiquitin (Ub); lane 2, complete reaction for in vitro ubiquitination of H2B; lanes 3 and 4, reactions performed in the absence of ATP or the enzymes E1 and E2, respectively. A complete ubiquitination reaction mixture contained 25 μg of ubiquitin, 0.3 μg of E1, 0.3 μg of Ubc2, and 0.5 μg of H2B in reaction buffer (4 mM ATP, 4 mM MgCl2, 40 mM KCl, 36 mM Tris-HCl [pH 8.0]). (B) Inhibition of multiubiquitin chain assembly may be a property common to proteins that bind ubiquitinated proteins. An in vitro ubiquitination reaction was performed as for panel A except that two different amounts of Rad23, S5a, and ataxin-3 were added to the reaction mixture (1 and 5 μg). Lanes 1 and 2, GST-hHR23B (R23B); lanes 3 and 4, GST-S5a (S5a); lanes 5 and 6, Thio-ataxin-3Q20 (Q20); lane 7, reaction performed in the absence of ATP; lane 8, complete reaction performed in the presence of glutathione Sepharose beads; lane 9, complete reaction performed in the presence of Thio-Bond resin; lane 10, free ubiquitin.
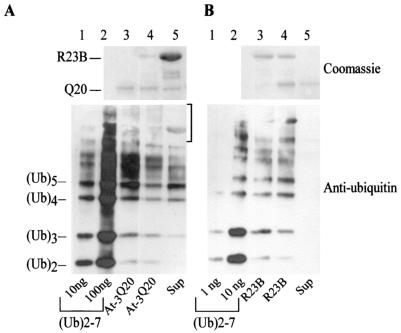
We also examined the abilities of GST-hHR23B and Thio-ataxin-3Q20 to compete for binding to purified multiubiquitin chains (Ub)2-7. We observed that an excess of GST-hHR23B could successfully displace the lower-molecular-weight ubiquitin chains (Fig. 8A, lane 5) that were bound to Thio-ataxin-3Q20 (lane 3). In contrast, a significant amount of higher-molecular-weight ubiquitin chains remained bound to Thio-ataxin-3Q20 (Fig. 8A, lane 4). Ubiquitin chains were not displaced from Thio-ataxin-3Q20 in the absence of any additional protein (data not shown). In the reciprocal experiment, we observed that an excess of Thio-ataxin-3Q20 (Fig. 8B, lane 5) or hHR23B (data not shown) did not displace multiubiquitin chains that had been prebound to GST-hHR23B purified on beads. As expected, some ataxin-3 interacted with GST-hHR23B (Fig. 8B, lane 4, top panel), or with the multiubiquitin chains that were bound to GST-hHR23B, which could explain its inability to displace multiubiquitin chains from Rad23. We determined that in the absence of added ataxin-3, ubiquitinated proteins were not released from hHR23B, even after prolonged incubation.
FIG. 8.
GST-hHR23B can displace short multiubiquitin chains from Thio-ataxin-3Q20. (A) Lanes 1 and 2, 10 and 100 ng of (Ub)2-7, respectively; lane 3, 50% of bound material after purification of Thio-ataxin-3Q20 and incubation with (Ub)2-7 chains; lane 4, as in lane 3, but following incubation with GST-hHR23B; lane 5, supernatant from the reactions described for lane 4, incubated with glutathione Sepharose 4B beads to purify GST-hHR23B. Ubiquitinated material that was associated with each step was detected by immunoblotting. Bracket on the right indicates high-molecular-weight ubiquitin-cross-reacting material in lanes 3 and 4 that was not released into the supernatant by Rad23 (lane 5). (B) Thio-ataxin-3Q20 (lane 5) or GST-hHR23B (data not shown) could not displace multiubiquitin chains from GST-hHR23B (compare lanes 3 and 4, before and after competition) under these experimental conditions. Lanes 1 and 2, 1 and 10 ng of (Ub)2-7, respectively; lane 3, 50% of material associated with GST-hHR23B; lane 4, remaining 50% as in lane 3, but following incubation with Thio-ataxin-3Q20; lane 5, supernatant from the reaction described for lane 4, incubated with Thio-Bond resin to purify Thio-ataxin-3Q20 and associated material. Because Thio-ataxin-3Q20 can bind GST-hHR23B (lane 4, top panel, Coomassie stain) and multiubiquitin chains, in this scenario, it is conceivable that multiubiquitin chains that are displaced from Rad23 bind ataxin-3, which remains associated with Rad23.
Ataxin-3 interacts with VCP/p97.
We consistently detected a Coomassie-stainable band of ∼95 to 100 kDa in association with Thio-ataxin-3Q20 following incubation with extracts prepared from mammalian NT2 and 293T HEK cells (Fig. 9A, lane 4). However, this protein was not observed in pulldowns performed with control beads (Fig. 9A, lane 5), or with Thio or an unrelated protein, Thio-β-synuclein (lanes 6 and 7). This ∼97-kDa band was excised and subjected to tryptic digestion followed by mass spectrometry and was determined to be VCP/p97. VCP has been found in abnormal protein aggregates (18) and can bind ataxin-3 (26). We observed that an increased amount of endogenous VCP could be recovered with larger amounts of Thio-ataxin-3Q20 (Fig. 9B). VCP can also be immunoprecipitated with ataxin-3 from 293T cells (Fig. 5C). We investigated whether there was a difference in binding of VCP by either ataxin-3Q20 or ataxin-3Q79 (Fig. 9C). Both GST-ataxin-3Q20 (Fig. 9C, lane 2) and GST-ataxin-3Q79 (lane 3), but not GST (lane 5) or GST-S5a (lane 4), bound VCP in NT2 cell extracts. No apparent difference was observed in the amount of VCP pulled down with equal amounts of ataxin-3 proteins. The ability of GST-VCP to bind (Ub)2-7 was observed to be weaker (Fig. 9D, lane 2) than the interaction between (Ub)2-7 and ataxin-3 (lane 5). However, preincubation of immobilized GST-VCP with Thio-ataxin-3Q20 resulted in a marked increase in the interaction with (Ub)2-7 (Fig. 9D, lanes 3 and 4), which could reflect the interaction between Thio-ataxin-3Q20 and GST-VCP.
FIG. 9.
Ataxin-3 binds VCP/p97. (A) Proteins bound to ataxin-3 are identified by Coomassie staining following separation by SDS-10% PAGE. Lane 1, molecular size standards (10 kDa); lane 2, NT2 cell extract (10% of input); lane 3, purified Thio-ataxin-3Q20; lane 4, 2 mg of the NT2 cell extract applied to Thio-ataxin-3Q20; lane 5, 2 mg of the NT2 cell extract applied to Thio-Bond resin; lane 6, 2 mg of the NT2 cell extract applied to affinity beads containing Thio; lane 7, 2 mg of the NT2 cell extract applied to affinity beads containing Thio-β-synuclein. (B) Binding of VCP by Thio-ataxin-3Q20. Lane 1, purified Thio-ataxin-3Q20; lanes 2 to 6, 0.5, 1, 2, 5, and 10 μg, respectively, of Thio-ataxin-3Q20 to which 1 mg of the NT2 cell extract was applied; lane 7, mock pulldown with Thio-Bond resin. (Top panel) Thio-ataxin-3Q20 purified on beads and visualized with Ponceau stain. (Bottom panel) VCP interacting with Thio-ataxin-3Q20, detected with antibodies to VCP (Research Diagnostics). (C) GST-ataxin-3Q20 and GST-ataxin-3Q79, but not GST-S5a, bind VCP. Two milligrams of the NT2 cell extract was applied to equal amounts of immobilized GST-ataxinQ20 (lane 2), GST-ataxin-3Q79 (lane 3), or GST-S5a (lane 4). Lane 1, NT2 cell lysate (10% of input); lane 5, GST purified on beads; lane 6, mock pulldown with glutathione Sepharose. VCP/p97 was detected by immunoblotting with anti-VCP antibodies. (See also Fig. 5C for immunoprecipitation of VCP with ataxin-3 from 293T cells by using a polyclonal ataxin-3 antibody.) (D) VCP binds to (Ub)2-7 chains more weakly than ataxin-3. Lane 1, 10 ng of (Ub)2-7; lane 2, GST-VCP (1 μg); lanes 3 and 4, 1 and 2 μg, respectively, of GST-VCP plus Thio-ataxin-3Q20; lane 5, Thio-ataxin-3Q20 (1 μg). All lanes are from the same gel, and the immunoblot was incubated with ubiquitin-specific antibodies. Lane 5 represents a separate pulldown.
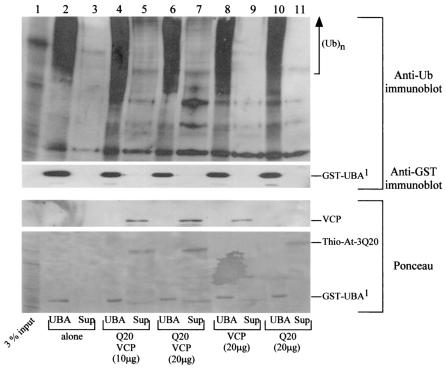
VCP has been reported to be involved in a number of cellular functions. We therefore considered how VCP and ataxin-3 might contribute to the ubiquitin-proteasome degradation pathway. We expressed UBA1 derived from yeast Rad23 as a GST fusion protein in yeast cells. We determined that the presence of both VCP and ataxin-3 was required for displacement of ubiquitinated proteins that were copurified with GST-UBA1 (Fig. 10, lanes 2, 5, and 7), whereas no significant displacement of ubiquitinated proteins was observed in the absence of ataxin-3 and VCP (lane 3) or in the presence of only VCP (lane 9) or ataxin-3 (lane 11). Similarly, in an additional experiment, we isolated FLAG-Rad23, in association with multiubiquitinated proteins from yeast cells (data not shown). In the presence of both VCP and ataxin-3, ubiquitinated proteins that were bound to FLAG-Rad23 were displaced. In contrast, consistent with results described for GST-UBA1, ubiquitinated proteins bound to Rad23 were not displaced in the absence of ataxin-3 and VCP or in the presence of VCP or ataxin-3 alone, suggesting that VCP may promote the transfer of multiubiquitinated proteins from Rad23 to a ubiquitin chain-binding proteasomal protein, such as ataxin-3 (Fig. 11). Ubiquitinated proteins that were bound to either FLAG-Rpn10, GST-ataxin-3Q20, or GST-ataxin-3Q79 were not displaced in the presence of VCP (data not shown), suggesting that there may be some specificity involving Rad23.
FIG. 10.
Multiubiquitinated proteins can be displaced from GST-UBA1 (derived from yeast Rad23) in the presence of ataxin-3 and VCP. A protein extract was prepared from yeast cells that expressed GST-UBA1 of Rad23, and the bound ubiquitinated proteins were purified on glutathione Sepharose. Equal amounts of the GST-UBA1 beads were combined with varying amounts of VCP and ataxin-3. The proteins were incubated for several hours at 4°C, and the supernatant was subjected to 10% trichloroacetic acid precipitation, followed by an acetone wash and examination by SDS-PAGE. (Top panel) The proteins remaining on beads or displaced in the supernatant were resolved by SDS-10% PAGE, transferred to nitrocellulose membranes, and incubated with ubiquitin-specific antibodies. Lane 1, 100 μg of the yeast cell extract (3% of input); lane 2, multiubiquitinated proteins that were copurified with GST-UBA1; lane 3, supernatant (Sup) after incubation of GST-UBA1 without VCP or ataxin-3; lane 4, GST-UBA1 beads after incubation with 10 μg of VCP and 10 μg of ataxin-3; lane 5, supernatant (Sup) from lane 4 after incubation, showing partial displacement of ubiquitinated proteins; lane 6, GST-UBA1 beads after incubation with 20 μg of VCP and 20 μg of ataxin-3; lane 7, supernatant (Sup) from lane 6 after incubation; lane 8, GST-UBA1 beads after incubation with 20 μg of VCP; lane 9, supernatant from reaction in lane 8 after incubation; lane 10, GST-UBA1 beads after incubation with 20 μg of ataxin-3; lane 11, supernatant from reactions in lane 10 after incubation. Note that ubiquitinated proteins were not displaced from GST-UBA1 in the presence of VCP or ataxin-3 alone (lanes 9 and 11). (Second panel) Incubations with anti-GST antibodies showed that GST-UBA1 was not displaced from the beads during the incubation period. (Bottom panels) Ponceau-stained nitrocellulose filters show VCP, Thio-ataxin-3Q20, and GST-UBA1 used in the experiment, and their positions are indicated on the right.
FIG. 11.
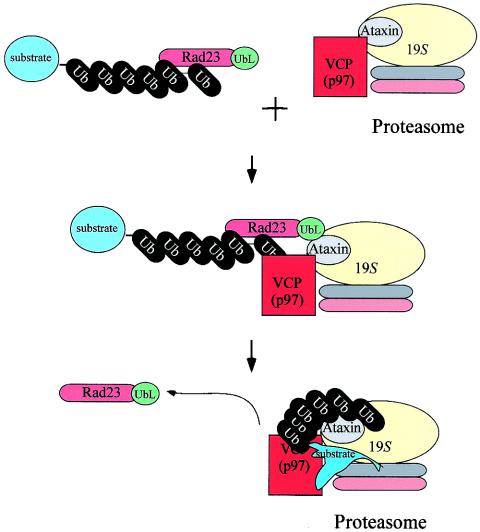
VCP may promote the transfer of multiubiquitinated proteins from Rad23 to ataxin-3. In this model, ataxin-3 is a transiently associated multiubiquitin chain recognition subunit in the proteasome that receives ubiquitinated substrates through the concerted action of VCP and shuttle-factors, such as Rad23. Since Rad23 can bind ataxin-3 and other multiubiquitin chain-interacting proteins, such as S5a, it is likely that substrates can be delivered to different proteasome subunits. Alternatively, completely distinct classes of proteasomes, which contain ataxin-3 or S5a, could be present in different types of cells or exist under various cellular conditions.
DISCUSSION
We propose that ataxin-3 is a transiently associated multiubiquitin chain recognition subunit in the proteasome that receives ubiquitinated substrates through the concerted action of VCP and shuttle factors, such as Rad23. There are several scenarios for this regulated mechanism, and one is depicted in Fig. 11. In this model, step 1 involves the binding of multiubiquitinated proteolytic substrates to Rad23 through its UBA domains, while VCP may form an association with ataxin-3 at the proteasome. Step 2 involves the binding of Rad23 to the proteasome (conceivably an ataxin-3-containing proteasome) through its UbL domain. Step 3 entails the presumed action of VCP in the transfer of multiubiquitinated substrates from Rad23 to ataxin-3. It is conceivable that if ataxin-3 represents a proteasome component that binds ubiquitinated proteins, it would be expected to preferentially bind high-molecular-weight chains rather than chains containing fewer than four ubiquitin moieties. In agreement with this model, we found that low-molecular-weight ubiquitin chains were displaced from ataxin-3 by Rad23, while higher-molecular-weight ubiquitin chains remained preferentially bound to ataxin-3 (Fig. 8).
Genetic and biochemical studies have shown that Rad23 (6, 29, 34) and S5a are required for efficient proteolysis (12, 45, 52). Rad23 can transiently stabilize substrates when it interacts with them. The major fractions (∼90%) of Rad23 and S5a are not associated with the proteasome, suggesting that they play a dynamic role in binding ubiquitinated substrates and translocating them to the proteasome to promote degradation. Similarly, we believe that ataxin-3 plays a positive role in proteolysis. We showed previously that when Rad23 binds a ubiquitinated substrate, it blocks multiubiquitin chain expansion (which results in stabilization) (34). However, subsequent delivery to the proteasome could initiate rapid degradation.
Other proteins, including VCP/p97/Cdc48, have been shown to bind ubiquitin and the proteasome. These proteins, however, contain neither UBA or UIM motifs for binding ubiquitinated proteins nor UbL domains for binding the proteasome. Therefore, based on both functional and sequence similarities, we propose that ataxin-3 is a biochemical counterpart of S5a and not of Rad23.
The AAA family of ATPases, which contain a highly conserved ∼300-amino-acid motif, has been linked to diverse cellular proteolytic functions (35). Members of this group include the ATPases in the proteasome and specific mitochondrial ATP-dependent proteases. The VCP/p97/Cdc48 AAA ATPases play a role in the ubiquitin-proteasome pathway and have been shown to bind the proteasome, interact with ubiquitinated substrates, and affect the degradation of endoplasmic reticulum-specific substrates (9, 10, 14, 21). However, a clear understanding of their biochemical function is lacking. We found that VCP formed a weak interaction with ubiquitin and ubiquitinated substrates, in contrast to the findings of previous studies (10). Other laboratories have also demonstrated little to no interaction of VCP with multiubiquitinated proteins. However, it is likely that the binding partners of VCP/p97 (such as Ufd1/Npl4) form the interaction with ubiquitin chains (33), in a manner analogous to the interaction described here between ataxin-3 and VCP. We propose that the interaction between VCP and ataxin-3 is important for the recognition of ubiquitinated substrates by the proteasome. It is unclear if ataxin-3 and VCP have substrate specificity, as it is possible that they mediate the turnover of most cellular ubiquitinated proteins. We had suspected that an active mechanism might be required for transferring ubiquitinated substrates from Rad23 to the proteasome in vivo, because its interaction with multiubiquitin chains resisted treatment with 1 M NaCl and 2 M urea (data not shown). Should VCP, and Cdc48 in yeast, satisfy this requirement, we could propose a model that raises an important prediction. We anticipate that ataxin-3, lacking one or more UIM domains, will be able to bind VCP, Rad23, and the proteasome but should be unable to receive or stably bind ubiquitinated substrates from Rad23. Studies to test this hypothesis are under way. These findings make a significant contribution toward understanding the function of VCP in the context of ataxin-3 and Rad23.
The ubiquitin-proteasome pathway plays an essential role in controlling the degradation of important regulators of the stress response, cell growth, and differentiation. The presence of cellular deposits that contain ubiquitin and proteasome subunits suggests that a failure in this proteolytic system could, in some instances, underlie the defects in human neurodegeneration (3). However, it has been unclear if protein aggregation is the cause, or a consequence, of the disease state. Specifically, the ubiquitin-proteasome pathway could play an indirect role in neurodegeneration, and the accumulation of aggregated proteins might only reflect a by-product of the disease state. In this regard, the interaction between ataxin-3 and ubiquitinated proteins, as well as between ataxin-3 and the proteasome, reveals a much more direct link between this proteolytic system and MJD. We have found that both normal and expanded forms of ataxin-3 bind ubiquitinated proteins and the proteasome. There is considerable evidence that substrate-proteasome interaction and substrate degradation are temporally distinct steps. Therefore, the stabilization of Ub-Pro-β-Gal by ataxin-3Q79 may be the result of a specific posttargeting defect. For instance, substrate unfolding or channeling into the proteasome may be affected by ataxin-3Q79. The ataxin-3 protein is a ubiquitously expressed cytoplasmic protein whose normal function has been unknown. Based on our studies, we speculate that ataxin-3 plays a role in the recognition of proteolytic substrates by the proteasome.
The pathology associated with expanded alleles of ataxin-3 occurs only in the brain, specifically in neurons, as ubiquitinated intranuclear inclusions. Because the expanded form of ataxin-3 is more toxic for nondividing or postmitotic cells, it may be significant that neurons are postmitotic (51). It is possible that nonneuronal cells have developed mechanisms to cope with the failure of mutant ataxin-3 to maintain efficient degradation of proteolytic substrates. Experiments to examine the wild-type and expanded proteins in NT2 cells that have been differentiated are in progress.
Acknowledgments
These studies were supported in part by grants from the National Institutes of Health (CA83875 to K.M.; AG01047 to E.W.D.-P.), the National Ataxia Foundation (to W.G.J. and K.M.), and the Atran Foundation (to W.G.J.).
We thank R. Pittman and members of his laboratory (University of Pennsylvania) for providing purified ataxin-3 proteins, cell pellets, and thoughtful discussions.
REFERENCES
- 1.Bachmair, A., D. Finley, and A. Varshavsky. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234:179-186. [DOI] [PubMed] [Google Scholar]
- 2.Baniahmad, C., A. Baniahmad, and B. W. O'Malley. 1994. A rapid method combining a functional test of fusion proteins in vivo and their purification. BioTechniques 16:194-196. [PubMed] [Google Scholar]
- 3.Bence, N. F., R. M. Sampat, and R. R. Kopito. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292:1552-1555. [DOI] [PubMed] [Google Scholar]
- 4.Bertolaet, B. L., D. J. Clarke, M. Wolff, M. H. Watson, M. Henze, G. Divita, and S. I. Reed. 2001. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 8:417-422. [DOI] [PubMed] [Google Scholar]
- 5.Chau, V., J. W. Tobias, A. Bachmair, D. Marriott, D. J. Ecker, D. K. Gonda, and A. Varshavsky. 1989. A multi-ubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576-1583. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L., and K. Madura. 2002. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 22:4902-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L., U. Shinde, T. G. Ortolan, and K. Madura. 2001. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings, C. J., M. A. Mancini, B. Antalffy, D. B. DeFranco, H. T. Orr, and H. Y. Zoghbi. 1998. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 19:148-154. [DOI] [PubMed] [Google Scholar]
- 9.Dai, R.-M., E. Chen, D. Longo, C. Gorbea, and C-C Li. 1998. Involvement of valosin-containing protein, an ATPase co-purified with IκBα and 26S proteasome, in ubiquitin-proteasome-mediated degradation of IκBα. J. Biol. Chem. 273:3562-3573. [DOI] [PubMed] [Google Scholar]
- 10.Dai, R.-M., and C-C Li. 2001. Valosin-containing protein is a multi-ubiquitin chain targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 3:740-744. [DOI] [PubMed] [Google Scholar]
- 11.De Mot, R., I. Nagy, J. Walz, and W. Baumeister. 1999. Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol. 7:88-92. [DOI] [PubMed] [Google Scholar]
- 12.Deveraux, Q., V. Ustrell, C. Pickart, and M. Rechsteiner. 1994. A 26S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 269:7059-7061. [PubMed] [Google Scholar]
- 13.Evidente, V. G., K. A. Gwinn-Hardy, J. N. Caviness, and S. Gilman. 2000. Hereditary ataxias. Mayo Clin. Proc. 75:475-490. [DOI] [PubMed] [Google Scholar]
- 14.Ghislain, M., J. Dohmen, F. Levy, and A. Varshavsky. 1996. CDC48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15:4884-4889. [PMC free article] [PubMed] [Google Scholar]
- 15.Gonda, D. K., A. Bachmair, I. Wunning, J. W. Tobias, W. S. Lane, and A. Varshavsky. 1989. Universality and structure of the N-end rule. J. Biol. Chem. 264:16700-16712. [PubMed] [Google Scholar]
- 16.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 17.Higashiyama, H., F. Hirose, M. Yamaguchi, Y. H. Inoue, N. Fujikake, A. Matsukage, and A. Kakizuka. 2002. Identification of ter94, Drosophila VCP, as a modulator of polyglutamine-induced neurodegeneration. Cell Death Differ. 9:264-273. [DOI] [PubMed] [Google Scholar]
- 18.Hirabayashi, M., K. Inoue, K. Tanaka, K. Nakadate, Y. Ohsawa, Y. Kamei, A. H. Popiel, A. Sinohara, A. Iwamatsu, Y. Kimura, Y. Uchiyama, S. Hori, and A. Kakizuka. 2001. VCP/p97 in abnormal protein aggregates, cytoplasmic vacuoles, and cell death, phenotypes relevant to neurodegeneration. Cell Death Differ. 8:977-984. [DOI] [PubMed] [Google Scholar]
- 19.Hiyama, H., M. Yokoi, C. Masutani, K. Sugasawa, T. Maekawa, K. Tanaka, J. H. Hoeijmakers, and F. Hanaoka. 1999. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26S proteasome. J. Biol. Chem. 274:28019-28025. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann, K., and L. Falquet. 2001. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci. 26:347-350. [DOI] [PubMed] [Google Scholar]
- 21.Jarosch, E., C. Taxis, C. Volkwein, J. Bordallo, D. Finley, D. H. Wolf, and T. Sommer. 2002. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4:134-139. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, E. S., B. Bartel, W. Seufert, and A. Varshavsky. 1992. Ubiquitin as a degradation signal. EMBO J. 11:497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, E. S., P. C. M. Ma, I. M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442-17456. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi, Y., T. Okamoto, M. Taniwaki, M. Aizawa, M. Inoue, S. Katayama, H. Kawakami, S. Nakamura, M. Nishimura, I. Akiguchi, et al. 1994. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 8:221-228. [DOI] [PubMed] [Google Scholar]
- 25.Klockgether, T., U. Wullner, A. Spauschus, and B. Evert. 2000. The molecular biology of the autosomal-dominant cerebellar ataxias. Mov. Disord. 15:604-612. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi, T., K. Tanaka, K. Inoue, and A. Kakizuka. 2002. Functional ATPase activity of p97/VCP is required for the quality control of endoplasmic reticulum in neuronally differentiated mammalian PC12 cells. J. Biol. Chem. 277:47358-47365. [DOI] [PubMed] [Google Scholar]
- 27.Koller, K. J., and M. J. Brownstein. 1987. Use of a cDNA clone to identify a supposed precursor protein containing valosin. Nature 325:542-545. [DOI] [PubMed] [Google Scholar]
- 28.Lam, Y. A., T. G. Lawson, M. Velayutham, J. L. Zweier, and C. M. Pickart. 2002. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416:763-767. [DOI] [PubMed] [Google Scholar]
- 29.Lambertson, D., L. Chen, and K. Madura. 1999. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics 153:69-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen, C. N., and D. Finley. 1997. Protein translocation channels in the proteasome and other proteases. Cell 91:431-434. [DOI] [PubMed] [Google Scholar]
- 31.Madura, K. 2002. The ubiquitin-associated (UBA) domain: on the path from prudence to prurience. Cell Cycle 1:235-244. [PubMed] [Google Scholar]
- 32.Merrick, W. C. 1979. Assays for eukaryotic protein synthesis. Methods Enzymol. 60:108-123. [DOI] [PubMed] [Google Scholar]
- 33.Meyer, H. H., Y. Wang, and G. Warren. 2002. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 21:5645-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortolan, T. G., P. Tongaonkar, D. Lambertson, L. Chen, C. Schauber, and K. Madura. 2000. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat. Cell Biol. 2:601-608. [DOI] [PubMed] [Google Scholar]
- 35.Patel, S., and M. Latterich. 1998. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 8:65-71. [PubMed] [Google Scholar]
- 36.Paulson, H. L. 1998. Analysis of triplet repeat disorders: spinocerebellar ataxia type 3/Machado Joseph disease. BIOS Scientific Publishers, Oxford, United Kingdom.
- 37.Paulson, H. L., M. K. Perez, Y. Trottier, J. Q. Trojanowski, S. H. Subramony, S. S. Das, P. Vig, J. L. Mandel, K. H. Fischbeck, and R. N. Pittman. 1997. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 19:333-344. [DOI] [PubMed] [Google Scholar]
- 38.Raasi, S., and C. M. Pickart. 2003. Rad23 ubiquitin-associated domains (UBA) inhibit 26S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem. 278:8951-8959. [DOI] [PubMed] [Google Scholar]
- 39.Rao, H., and A. Sastry. 2002. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 277:11691-11695. [DOI] [PubMed] [Google Scholar]
- 40.Rosenberg, R. 1992. Machado-Joseph disease: an autosomal dominant system degeneration. Mov. Disord. 3:193-203. [DOI] [PubMed] [Google Scholar]
- 41.Schauber, C., L. Chen, P. Tongaonkar, I. Vega, D. Lambertson, W. Potts, and K. Madura. 1998. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391:715-718. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, T., K. S. Lindenberg, A. Krebs, L. Schols, F. Laccone, J. Herms, M. Rechsteiner, O. Riess, and G. B. Landwehrmeyer. 2002. Protein surveillance machinery in brains with spinocerebellar ataxia type 3: redistribution and differential recruitment of 26S proteasome subunits and chaperones to neuronal intranuclear inclusions. Ann. Neurol. 51:302-310. [DOI] [PubMed] [Google Scholar]
- 43.Thrower, J. S., L. Hoffman, M. Rechsteiner, and C. M. Pickart. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19:94-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner, G. C., and A. Varshavsky. 2000. Detecting and measuring cotranslational protein degradation in vivo. Science 289:2117-2120. [DOI] [PubMed] [Google Scholar]
- 45.van Nocker, S., S. Sadis, D. M. Rubin, M. Glickman, H. Fu, O. Coux, I. Wefes, D. Finley, and R. D. Vierstra. 1996. The multi-ubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 16:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verma, R., S. Chen, R. Feldman, D. Schieltz, J. Yates, J. Dohmen, and R. J. Deshaies. 2000. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11:3425-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verma, R., and R. J. DeShaies. 2000. A proteasome howdunit: the case of the missing signal. Cell 101:341-344. [DOI] [PubMed] [Google Scholar]
- 48.Voges, D., P. Zwicki, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015-1068. [DOI] [PubMed] [Google Scholar]
- 49.Wang, G.-H., N. Sawai, S. Kotliarova, I. Kanazawa, and N. Nukina. 2000. Ataxin-3, the MJD1 gene product, interacts with the two human homologs of yeast DNA repair protein RAD23, HHR23A and HHR23B. Hum. Mol. Genet. 9:1795-1803. [DOI] [PubMed] [Google Scholar]
- 50.Wilkinson, C. R., M. Seeger, R. Hartmann-Petersen, M. Stone, M. Wallace, C. Semple, and C. Gordon. 2001. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 3:939-943. [DOI] [PubMed] [Google Scholar]
- 51.Yoshizawa, T., Y. Yamagishi, N. Koseki, J. Goto, H. Yoshida, F. Shibasaki, S. Shoji, and I. Kanazawa. 2000. Cell cycle arrest enhances the in vitro cellular toxicity of the truncated Machado-Joseph disease gene product with an expanded polyglutamine stretch. Hum. Mol. Genet. 9:69-78. [DOI] [PubMed] [Google Scholar]
- 52.Young, P., Q. Deveraux, R. Beal, C. Pickart, and M. Rechsteiner. 1998. Characterization of the two polyubiquitin binding sites in the 26S protease subunit 5a. J. Biol. Chem. 273:5461-5467. [DOI] [PubMed] [Google Scholar]