Abstract
Plasminogen activator inhibitor-2 (PAI-2) is well documented as an inhibitor of the extracellular serine proteinase urokinase-type plasminogen activator (uPA) and is expressed in activated monocytes and macrophages, differentiating keratinocytes, and many tumors. Here we show that PAI-2 has a novel intracellular function as a retinoblastoma protein (Rb)-binding protein. PAI-2 colocalized with Rb in the nucleus and inhibited the turnover of Rb, which led to increases in Rb protein levels and Rb-mediated activities. Although PAI-2 contains an LXCXE motif, Rb binding was primarily mediated by the C-D interhelical region of PAI-2, which was found to bind to the C pocket of Rb. The C-D interhelical region of PAI-2 contained a novel Rb-binding motif, termed the PENF homology motif, which is shared by many cellular and viral Rb-binding proteins. PAI-2 expression also protected Rb from the accelerated degradation mediated by human papillomavirus (HPV) E7, leading to recovery of Rb and inhibition of E6/E7 mRNA expression. Protection of Rb by PAI-2 begins to explain many of the diverse, uPA-independent phenotypes conferred by PAI-2 expression. These results indicate that PAI-2 may enhance Rb's tumor suppressor activity and suggest a potential therapeutic role for PAI-2 against HPV-transformed lesions.
Plasminogen activator inhibitor-2 (PAI-2) is well documented as an inhibitor of urokinase-type plasminogen activator (uPA) (31), an extracellular serine proteinase that cleaves plasminogen to plasmin. Plasmin facilitates cell migration and invasion by degrading fibrin and activating metalloproteinases. PAI-2 is a major product of activated monocytes and macrophages, differentiating keratinocytes, and placental trophoblasts. PAI-2 is also expressed in many tumors and glial cells and is inducible in endothelial cells and fibroblasts (1, 31). PAI-2 was one of the first identified members of a unique and growing subclass of serine proteinase inhibitors (serpins), now called ovalbumin-like serpins (ov-serpins). Ov-serpins lack a typical amino-terminal secretion sequence, are often found to reside intracellularly, and generally contain a nonconserved loop between their C and D helices, which often contribute to activities distinct from protease inhibition (4, 45). PAI-2 contains a unique C-D interhelical region, which is larger than any identified to date in the ov-serpin family (12, 45). A growing number of studies (2, 6, 11, 12, 26, 39, 40, 44, 45, 49, 54, 55) have suggested that PAI-2, like other ov-serpins (45), may have an intracellular function distinct from its extracellular role as a uPA inhibitor, since the consequences of PAI-2 expression are often difficult to associate with uPA inhibition. PAI-2 expression by cancer cells has been associated repeatedly with a lower incidence of metastases and an improved prognosis (17, 34, 37, 53), whereas expression of PAI-1, also an effective inhibitor of uPA, largely fails to correlate with an improved prognosis. PAI-2 expression during keratinocyte differentiation is also difficult to link with uPA inhibition, since the extracellular uPA is normally restricted to the basal and suprabasal keratinocytes (31), whereas PAI-2 is usually intracellular and largely restricted to the differentiating squamous epithelial cells (6, 31). Although PAI-2 is a major product of inflammatory monocytes and macrophages, the evidence that these cells effectively produce the secreted glycosylated form of PAI-2 has been equivocal (40). PAI-2 expression has also been shown to confer resistance to apoptosis (11, 55) and regulate transcription (2, 44), activities that often could not be mimicked by addition of extracellular PAI-2 (2, 11, 44). Furthermore, both cells stably expressing a PAI-2 mutant protein lacking amino acids 66 to 84 within the C-D interhelical region of PAI-2 (C-D PAI-2) and cells expressing PAI-2 protein containing a mutation (Arg380 to Ala380) in the P1 residue of the reactive site loop (RSL) fail to reproduce many of the phenotypes described above (11, 12).
Here we report that PAI-2 has a novel intracellular function as an inhibitor of retinoblastoma protein (Rb) degradation. Rb is a ubiquitous regulator of transcription involved in many cellular activities including cell cycle control, apoptosis, differentiation, and tumor suppression (21, 33). One of the best-described activities of hypophosphorylated Rb is the binding of the transcription factor E2F-1 and recruitment of histone deacetylases, causing inhibition of E2F-1-mediated transcription and cell cycle arrest in G1 (21, 33). Rb is also targeted by a number of viral oncoproteins, including human papillomavirus (HPV) E7, which accelerate the degradation of Rb (5, 22, 42). The elevation of Rb protein levels and Rb functions by PAI-2 begins to explain some of the diverse and often conflicting data on the biological activity of PAI-2 and illustrates a novel activity for an intracellular serpin.
MATERIALS AND METHODS
Cell culture.
The HeLa, A2/7, S1a, and S1b cell lines were maintained and/or generated as described previously (11). Briefly, HeLa cells were transfected with plasmid DNA and selected with 800 μg of G418 (Life Technologies, Inc.) ml−1. Individual lines derived by limiting dilution were routinely maintained in 300 μg of G418 ml−1. S1a and S1b expressed 132 ± 8 and 28 ± 1.3 ng of PAI-2 per mg−1 of total protein, respectively (11). HeLa cell lines stably expressing the C-D interhelical mutant (with residues 65 to 87 deleted) and RSL mutant (Arg380 to Ala) of PAI-2 were generated as described previously (11, 12), except that cells were transfected using Genejammer (Stratagene, La Jolla, Calif.). Jurkat T-cells (ATCC no. TIB-152) were transfected with pRcCMV-PAI-2, pRcCMV-PAI-2Δ65/87, pRcCMV-PAI-2Ala380, or the control plasmid pRcCMV (11, 12) by electroporation. All transfectants were selected in 800 μg of Geneticin (ICN Biochemicals, Costa Mesa, Calif.) ml−1 and were cloned by limiting dilution. Jurkat PAI-2a and PAI-2b expressed approximately 60 and 100 ng of PAI-2 mg−1, respectively (data not shown). KJD (a kind gift from P.G. Parsons, Queensland Institute of Medical Research [QIMR]) and U937 (ATCC no. CRL-1593.2) were maintained as previously described (44).
Western blot analysis.
Western blot analysis used antibodies against HPV 18 E7 (N-19) (Santa Cruz Biotechnology, Santa Cruz, Calif.), PAI-2 (polyclonal antibodies from American Diagnostica, Greenwich, Conn., and Biotech Australia Pty. Ltd, North Ryde, Australia), Rb (G3-245) (BD PharMingen, Heidelberg, Germany), p53 (D0-1) (Santa Cruz), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Santa Cruz), actin (C-11) (Santa Cruz), horseradish peroxidase-conjugated secondary antibodies (Silenus, Melbourne, Australia), and an ECL chemiluminescent detection system (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). Total-cell, nuclear, and cytoplasmic extracts were generated as described previously (25, 27). Protein concentrations were determined by a BCA-200 protein assay kit (Pierce, Rockford, Ill.), and 15 μg was separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and transferred to Hybond-C nitrocellulose membranes (Amersham). For E7 detection, 50 μg of cell protein was separated on 4 to 20% gradient gels (Gradipore, North Ryde, Australia) run at 150 V for 1 h.
Quantitative real-time reverse transcription-PCR (RT-PCR).
cDNA was prepared from 106 cells as described La Linn et al. (32). PCR analysis used the following nucleotide primers (from Genset, La Jolla, Calif.): for Rb, 5′-GCTAGCCTATCTCCGGCTAAA-3′ and 5′-CTGGAAAAGGGTCCAGATGA-3′; for E7, 5′-GCTGAACCACAACGTCACAC-3′ and 5′-GGTCGTCTGCTGGAGCTTTCT-3′; for cyclin A, 5′-CCCGATGCTTGTCAGATACT-3′ and 5′-ATTTGCAGTTAGCAGCCCTA-3′; for dihydrofolate reductase (DHFR), 5′-AGACCTGGTTCTCCATTCC-3′and 5′-TGTGGAGGTTCCTTGAGTTC-3′; for GAPDH, 5′-GGTCGGTGTGAACGGATTT-3′ and 5′-GTCGTTGATGGCAACAATCT-3′. The amplification reaction mixture of 20 μl contained 0.1 μg of randomly primed cDNA, 0.5 μM each primer pair, 2× Platinum Quantitation PCR Supermix-UDG (Gibco BRL), and 10× SYBR green (Molecular Probes, Eugene, Oreg.). Cycling conditions were as follows: one cycle of 96°C for 2 min, followed by 35 cycles of 96°C for 15 s, 56°C for 15 s, and 72°C for 15 s. Real-time PCR was performed using a Rotogene PCR machine (Corbett Research, Mortlake, Australia). PCR products were visualized with SYBR green dye and analyzed with Rotogene Real-Time Analysis software (Corbett Research). Quantitation was based on a standard curve established using dilutions of parental cell line GAPDH cDNA.
siRNA inhibition of Rb transcription.
S1a cells were grown to 50% confluency, and 200 μM double-stranded small inhibitory RNA (siRNA) (Proligo, La Jolla, Calif.) was transfected by using the Oligofectamine transfection reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, Calif.). The sequences of the Rb siRNA were 5′-ACAGAAGAACCUGAUUUUAUU-3′ (sense) and 5′-UAAAAUCAGGUUCUUCUGUUU-3′ (antisense). The control siRNA sequence was 5′-UAUCGAAUUUCAUUGAAAGCA-3′) (sense) and 5′-UGCUUUCAAUGAAAUUCGAUA-3′ (antisense). Whole-cell lysates were analyzed for Rb, E7, and actin protein levels at 72 h posttransfection by Western blotting as described above.
Confocal immunofluorescence microscopy.
Cells were seeded onto 13-mm coverslips and grown overnight to 80% confluency. U937 cells were treated for 16 h with 25 ng of phorbol myristate acetate (PMA; Sigma) ml−1. All cell lines were washed twice with phosphate-buffered saline (PBS) and fixed in −20°C methanol for 2 min. Antibodies diluted in RPMI 1640-1% fetal bovine serum (FBS) were added in the following sequence and incubated for 30 min at room temperature, followed by five PBS washes and a 30-min block in RPMI 1640-10% FBS: (i) goat anti-PAI-2 (1/500), (ii) rhodamine-conjugated rabbit anti-goat antibody (1/500) (Chemicon, Temecula, Calif.), (iii) mouse anti-Rb (1/500) (G3-245), and (iv) fluorescein isothiocyanate (FITC)-conjugated sheep anti-mouse immunoglobulin G (IgG) absorbed against human and rabbit IgG (1/500) (Silenus). Images were viewed using the Zeiss Axiovert 135 microscope and CAS (confocal assistant imaging software). Dual lasers with excitation at FITC (488 nm) and rhodamine (568 nm) were used, and any cross-detection of the fluorophores was removed by filters and adjustment of gain levels.
Coimmunoprecipitation of Rb with anti-PAI-2 antibody.
Immunoprecipitations were performed using a Seize Primary Immunoprecipitation kit (Pierce). An anti-PAI-2 polyclonal antibody (200 μg) (American Diagnostica) was immobilized on beads and incubated at 4°C overnight with 2 mg of nuclear or cytoplasmic cell fractions generated as described previously (25, 27), except that nuclear pellets were lysed in 500 μl of LICHT buffer (27). Bound proteins were washed three times in LICHT buffer (27), eluted as described in the kit protocol, and detected by Western blotting as described above. U937 cells were treated with lipopolysaccharide (25 ng ml−1) for 48 h.
GST-binding assays.
Glutathione S-transferase (GST) fusion proteins, Rb379-928, Rb379-928-706F, Rb379-928-del (exon 21), p107385-1068, and p130414-1135 were expressed in Escherichia coli XL-10 Blue (Stratagene). C-pocket Rb constructs (50) were kindly supplied by J. Y. Wang, University of California, and the inserts were cloned into the GST fusion vector p9EX-2TK (Amersham). pRcCMV-PAI-2, pRcCMV-CD PAI-2 (11), and pCDNA3-HDAC-3 (52) were in vitro transcribed (IVT) using the TnT Quick coupled transcription-translation system (Promega, Madison, Wis.) in the presence of [35S]methionine (Amersham). Equal amounts of IVT proteins were determined by exposure to film and densitometry of the correct molecular weight bands for PAI-2 and C-D PAI-2. Equal amounts of the following proteins were determined by Coomassie gel staining: GST-Rb, GST-p130, and GST-p107; GST-Rb, GST-(706F), and GST-Rb(del exon 21); GST-Rb, Rb-SE, Rb-SEΔ, and Rb-13S. Binding assays were performed as described previously(27).
Peptide inhibition of HDAC-3-Rb binding.
Each peptide (150 μM) (Mimotopes Pty. Ltd., Clayton, Victoria, Australia) was incubated with GST-Rb379-928 in 200 μl of LICHT buffer (27) for 1 h at 4°C prior to the addition of 35S-labeled IVT histone deacetylase 3 (HDAC-3). After 1 h of incubation at 4°C, the beads were washed and analyzed (27). The control peptide had the sequence NAVTPMTFAAKTSCGFMQQ.
Analysis of Rb turnover.
Exponentially growing cells were pulsed for 1 h at 37°C under 5% CO2 in cysteine- and methionine-free RPMI 1640 (Gibco BRL) containing 10% FBS supplemented with 200 μCi of Promix-35S (Amersham Pharmacia Biotech) ml−1. At the indicated times, whole-cell lysates were extracted and incubated with 250 ng of an anti-Rb antibody (G3-245) for 16 h at 4°C. The anti-Rb antibody was precipitated by adding 10 μl of 50% protein G-Sepharose beads (Sigma) and incubating for 2 h at 4°C. The beads were washed in NP-40 lysis buffer (27), and released proteins were resolved by SDS-PAGE, visualized by autoradiography, and quantified by scanning densitometry.
Cell cycle analysis.
Cells were stained with propidium iodide (Sigma) and analyzed with a fluorescence-activated cell sorter (FACScalibur; Becton Dickinson) and ModFitLT V2 software (Verity Software House Inc.).
RESULTS
PAI-2 expression in HeLa cells results in posttranscriptional recovery of Rb protein levels.
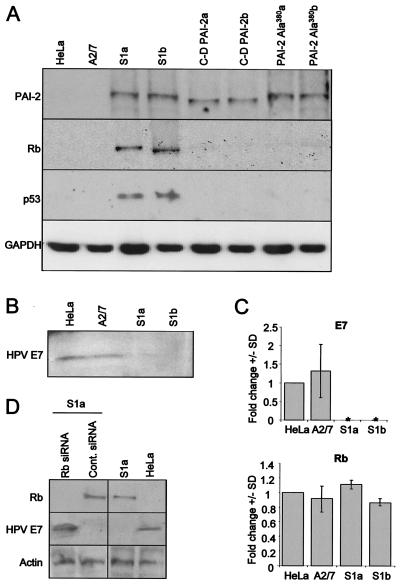
HeLa cells express the HPV-18 oncoproteins E6 and E7, which normally target p53 and Rb, respectively, for accelerated proteolytic degradation, resulting in low to undetectable levels of these proteins (19). Thus, as expected, Western blot analysis of parental HeLa cells and the antisense PAI-2 transfection control (A2/7) showed no expression of Rb and p53 protein (Fig. 1A). In contrast, HeLa cells stably expressing PAI-2 (11) showed substantial levels of both Rb and p53 protein (Fig. 1A, S1a and S1b). HeLa cell lines stably expressing the C-D interhelical mutant of PAI-2 (C-D PAI-2a and C-D PAI-2b) or the RSL mutant of PAI-2 (PAI-2 Ala380a and PAI-2 Ala380b) failed to show significant Rb or p53 protein levels (Fig. 1A), demonstrating that the RSL and C-D interhelical loop of PAI-2 are both required for restoration of Rb and p53 protein levels in HeLa cells.
FIG. 1.
PAI-2 expression in HeLa cells results in posttranscriptional recovery of Rb protein. (A) Western blot analysis of lysates from the parental HeLa line cells (HeLa), HeLa cells transfected with antisense-PAI-2 (A2/7), HeLa cells lines stably expressing PAI-2 (S1a and S1b), HeLa cells stably expressing the PAI-2 C-D interhelical mutant (C-D PAI-2a and C-D PAI-2b), and HeLa cells stably expressing the RSL mutant of PAI-2 (PAI-2 Ala380a and PAI-2 Ala380b). Protein lysates were probed using antibodies specific for PAI-2, Rb, p53, and GAPDH. (B) Western blot analysis of protein lysates from the parental HeLa line cells (HeLa), HeLa cells transfected with antisense-PAI-2 (A2/7), and HeLa cells lines stably expressing PAI-2 (S1a and S1b) by using antibodies specific for the E7 protein from HPV-18. (C) Quantitative real-time RT-PCR expression analysis of E7 (top) and Rb (bottom) mRNA levels. Rb and E7 cDNAs from HeLa cells and A2/7, S1a, and S1b cells were quantified by PCR based on a standard curve established by using dilutions of GAPDH cDNA prepared from the parental HeLa line. Data from three experiments are expressed as the mean fold change ± the standard deviation compared with parental HeLa cells. *, expression levels were undetectable. (D) Treatment of S1a cells with siRNA specific for Rb and a scrambled control (Cont.) siRNA. S1a cells were treated with siRNA, and whole-cell extracts were immunoblotted for Rb, E7, and actin. Control untreated S1a and HeLa cells were run in parallel.
The restored Rb and p53 protein levels in S1a and S1b cells suggested that E6 and E7 were no longer active in these PAI-2-expressing cells. Immunoblotting revealed that E7 protein expression was significantly reduced in S1a and S1b cells (Fig. 1B). Furthermore, real-time RT-PCR showed that levels of mRNA encoding E7 were also substantially reduced in these cells (Fig. 1C, E7). E6 and E7 are cotranscribed (19); thus, these experiments indicate that PAI-2 expression in HeLa cells resulted in loss of mRNAs encoding both E6 and E7. In contrast, quantitative real-time RT-PCR analysis showed that Rb mRNA levels were not significantly affected by PAI-2 expression in HeLa cells (Fig. 1C, Rb), demonstrating that the PAI-2-mediated increase in Rb protein levels was a consequence of posttranscriptional events.
Salcedo et al. (41) reported that Rb expression could inhibit E6 and E7 transcription. To determine whether the PAI-2-mediated suppression of E6 and E7 mRNAs was due to the recovery of Rb expression, S1a cells were treated with siRNA specific for Rb. As expected, this treatment significantly reduced Rb protein expression in these cells (Fig. 1D, first lane, top panel). Importantly, this reduction in Rb protein levels was associated with recovery of E7 protein expression (Fig. 1D, first lane, middle panel) in these PAI-2-expressing HeLa cells. Thus, the PAI-2-mediated recovery of Rb, and not PAI-2 expression, appeared to be responsible for suppression of E6 and E7 transcription.
Rb and PAI-2 colocalize in the nucleus.
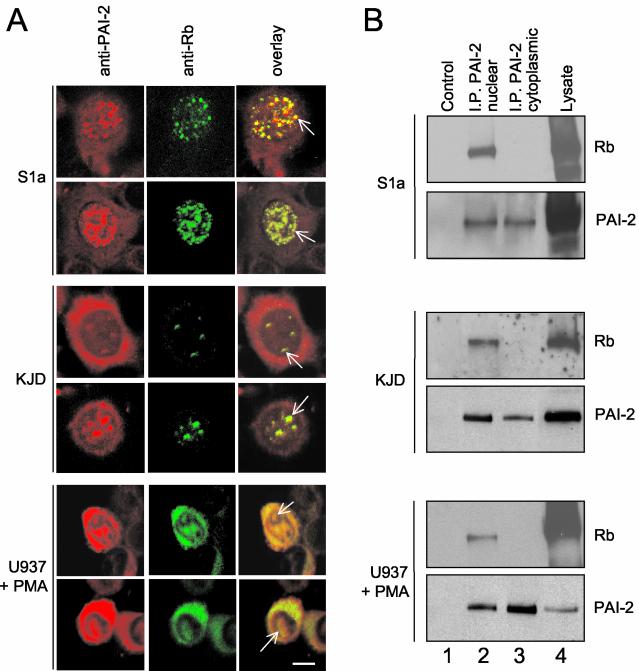
Given that PAI-2 is a serpin and that restoration of Rb was dependent on the RSL of PAI-2 (Fig. 1A), we postulated that PAI-2 might inhibit the proteolytic degradation of Rb. However, for PAI-2 to inhibit Rb degradation, PAI-2 would need to be localized in the nucleus, where Rb generally resides, and PAI-2 has traditionally been reported as a cytoplasmically localized protein (31). By use of confocal immunofluorescence, three cell lines were examined for the subcellular localization of PAI-2 and Rb: (i) S1a cells, which express PAI-2 via a stably integrated plasmid, (ii) KJD keratinocytes (simian virus 40 large T antigen-transformed keratinocytes), which constitutively express endogenous PAI-2, and (iii) phorbol ester-activated U937 cells (a leukemia line from the monocyte lineage), which express PAI-2 after activation. The latter two cell lines were chosen to represent two major physiological producers of PAI-2 in vivo: keratinocytes and monocytes/macrophages. As expected, PAI-2 could be clearly detected in the cytoplasm of all the cells (Fig. 2A). Importantly, PAI-2 was also found in the nuclei of all cells and was found to colocalize with Rb in dappled or punctate staining patterns, suggesting association of PAI-2 with Rb (Fig. 2A, overlay). Isotype-matched control antibodies used with rhodamine- or FITC-conjugated secondary antibodies showed no significant staining (data not shown).
FIG. 2.
PAI-2 colocalizes with, and binds to, Rb in vivo. (A) Colocalization of Rb and PAI-2 shown by indirect immunofluorescent confocal laser microscopy. S1a, KJD, and phorbol ester-activated U937 cells were dually labeled with antibodies against PAI-2 (rhodamine TRITC, red) and Rb (FITC, green). Overlay of the two images shows yellow staining where colocalization has occurred; arrows indicate colocalization in the nucleus Bar, 50 μm. (B) Immunoprecipitation of PAI-2 from nuclear (I.P. PAI-2 nuclear) and cytoplasmic (I.P. PAI-2 cytoplasmic) fractions from S1a, KJD, and activated U937 cells by using an anti-PAI-2 antibody, followed by electrophoresis and Western blotting of the immunoprecipitates with anti-Rb and anti-PAI-2 antibodies (top and bottom panels in each set, respectively). Control (lane 1) represents Western blotting of nuclear immunoprecipitates where the anti-PAI-2 antibody was omitted. Lysate (lanes 4) shows Western blotting of nuclear lysates.
Anti-PAI-2-antibody coimmunoprecipitates Rb.
To ascertain whether PAI-2 associates with Rb in vivo, PAI-2 was immunoprecipitated from nuclear and cytoplasmic lysates of HeLa S1a, KJD, and activated U937 cells. The immunoprecipitates were then analyzed by Western blotting using anti-Rb and anti-PAI-2 antibodies. For all cell lines tested, the anti-PAI-2 antibody coimmunoprecipitated Rb from the nuclear fractions (Fig. 2B, top panel in each set, lane 2) but not from the cytoplasmic fractions (Fig. 2B, top panel in each set, lane 3). As expected, the anti-PAI-2 antibody immunoprecipitated PAI-2 from both nuclear and cytoplasmic fractions of all the cell lines (Fig. 2B, bottom panel in each set, lanes 2 and 3). These data demonstrated that nuclear PAI-2 associates with Rb in cells expressing PAI-2 from a transgene and in cells that are physiological producers of PAI-2.
The cytoplasmic expression of Rb in activated U937 cells (Fig. 2A, U937 + PMA) may reflect the diffusion of Rb into the cytoplasm during metaphase and anaphase (46). The failure of PAI-2 to coimmunoprecipitate Rb from the cytoplasm of these U937 cells (Fig. 2B, U937 + PMA, top panel, lane 3) indicates that PAI-2 does not bind cytoplasmic Rb.
PAI-2 binds Rb.
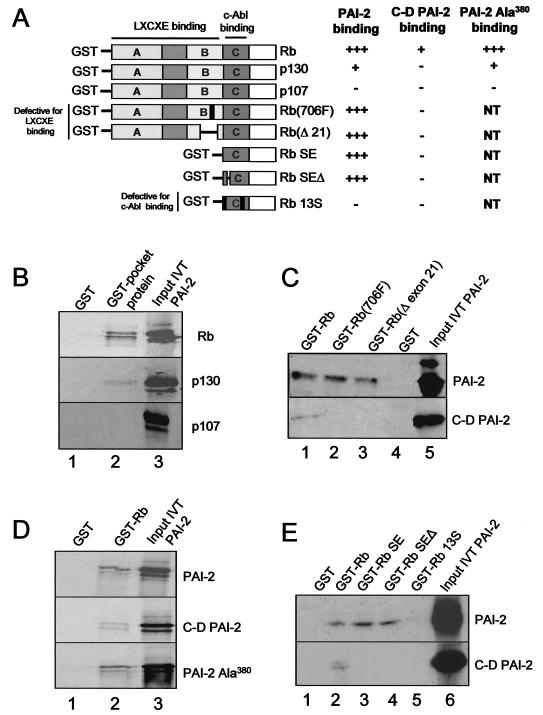
Colocalization and immunoprecipation experiments indicated that PAI-2 and Rb could interact in vivo. To determine if Rb and the related pocket proteins p130 and p107 bind to PAI-2, in vitro GST pulldown experiments were undertaken. p130 and p107 are members of the Rb pocket family of proteins that have distinct but overlapping functions with Rb (21). Equal amounts of the pocket proteins, expressed as GST fusion proteins, were employed in in vitro pulldown binding assays in the presence of equal amounts of IVT PAI-2. IVT PAI-2 was able to bind GST-Rb; however, PAI-2 bound much less efficiently to GST-p130, and binding to GST-p107 was not detected (Fig. 3B, lane 2). No nonspecific binding of IVT PAI-2 to the GST control was observed (Fig. 3B, lane 1). These data demonstrated that PAI-2 binds Rb but failed to bind p107 (summarized in Fig. 3A).
FIG. 3.
Involvement of the PAI-2 C-D interhelical region and the C-pocket of Rb in Rb-PAI-2 binding. (A) Summary of data presented in panels B to E. +++, strong binding; +, weak binding; NT, not tested. (B) PAI-2 binds Rb and p130 but not p107. An in vitro GST binding assay used IVT, 35S-radiolabeled PAI-2 and the GST fusion proteins Rb379-928, p107385-1068, and p130414-1135. Input IVT PAI-2 (lane 3) was incubated with glutathione-agarose beads bound to either GST alone (lane 1), GST-Rb379-928 (Rb), GST-p107385-1068 (p107), or GST-p130414-1135 (p130). After washing, bead-bound, 35S labeled PAI-2 was resolved by SDS-PAGE and exposed to X-ray film (lane 2). (C) Binding of PAI-2 and the C-D interhelical deletion mutant of PAI-2 to Rb and Rb LXCXE mutants. Input IVT PAI-2 (lane 5), in the form of either 35S-labeled IVT PAI-2 (top panel) or a 35S-labeled IVT C-D interhelical deletion mutant of PAI-2 (C-D PAI-2) (bottom panel), was incubated with glutathione-agarose beads bound to either GST-Rb379-928 (lane 1), GST-Rb(706F) (Rb379-928 with a mutation in position 706, which disrupts LXCXE binding) (lane 2), or GST-Rb(del exon 21) (Rb379-928 with a deletion in exon 21, which also disrupts the LXCXE binding site) (lane 3), or GST alone (GST) (lane 4). After washing, bead-bound, 35S-labeled PAI-2 or C-D PAI-2 was resolved by SDS PAGE and exposed to X-ray film. (D) The RSL mutant of PAI-2 retains binding to Rb. Input IVT PAI-2 (lane 3, top panel), a C-D interhelical deletion mutant of PAI-2 (C-D PAI-2) (middle panel), or the RSL mutant of PAI-2 (PAI-2Ala380) was incubated with GST-Rb379-928. After washing, the bead-bound, 35S-labeled PAI-2 proteins were resolved by SDS-PAGE and exposed to X-ray film (lane 2). (E) PAI-2 binds to the C-pocket of Rb. Input 35S-labeled IVT PAI-2 (top panel, lane 6) or 35S-labeled C-D PAI-2 (bottom panel, lane 6) was incubated with glutathione-agarose beads bound to either GST (lane 1), GST-Rb379-928 (lane 2), GST-SE (lane 3), GST-SEΔ (lane 4), or GST-13S (lane 5). SE and GST-SEΔ bind c-Abl, but 13S does not. After washing, bead-bound, 35S-labeled PAI-2 or C-D PAI-2 deletion mutant PAI-2 was resolved by SDS-PAGE and exposed to X-ray film.
Disruption of the LXCXE binding pocket of Rb does not affect PAI-2 binding.
Many cellular and viral Rb-binding proteins contain an LXCXE motif that is used by these proteins to bind to the A/B pocket of Rb (21, 33). PAI-2 also contains a conserved LXCXE motif (159LECAE163) located on a small exposed peptide loop. To analyze the role of this LXCXE motif in the binding of PAI-2 to Rb, two Rb mutants with disrupted LXCXE-binding pockets were used. These mutants, Rb(706F) (3) and Rb(del exon 21), fail to bind proteins that are reliant on their LXCXE motifs for binding to Rb but retain binding to some proteins that bind Rb independently of LXCXE (10).
Equal amounts of GST-Rb, GST-Rb(706F), and GST-Rb(del exon 21) were used in in vitro pulldown binding assays in the presence of equal amounts of IVT PAI-2. PAI-2 bound GST-Rb (Fig. 3C, lane 1, top panel) and also bound with similar efficiency to GST-Rb(706F) and GST-Rb(del exon 21) (Fig. 3C, lanes 2 and 3, top panel). (No binding of PAI-2 to GST was seen [Fig. 3C, lane 4].) This finding (summarized in Fig. 3 A) demonstrated that the LXCXE binding pocket of Rb was not crucial for the binding of PAI-2 to Rb.
The C-D interhelical loop of PAI-2 is important for Rb binding.
It has been reported previously that the C-D interhelical region of PAI-2 is important for the intracellular activity of PAI-2 (12), and it has been shown that HeLa cells stably expressing C-D PAI-2 failed to exhibit recovery of Rb protein levels (Fig. 1). To test the role of the C-D interhelical region in Rb binding, the experiments above were performed in parallel by using IVT C-D PAI-2 instead of IVT PAI-2. Identical amounts of GST-Rb, GST Rb(706F), and Rb(del exon 21), and similar amounts of IVT C-D PAI-2 and IVT PAI-2, were used. C-D PAI-2 bound GST-Rb much less efficiently than wild-type PAI-2 (Fig. 3C, lane 1) and, in contrast to wild-type PAI-2, failed to show any significant binding to GST-Rb(706F) or GST-Rb(del exon 21) (Fig. 3C, bottom panel, lanes 2 and 3). Some binding of C-D PAI-2 to Rb could be detected (Fig. 3C, bottom panel, lane 1), and this binding was lost for the Rb pocket mutants (Fig. 3C, bottom panel, lanes 2 and 3), indicating that LXCXE-dependent binding might make a small contribution to the PAI-2 Rb interaction. C-D PAI-2 showed no detectable binding to p130 and p107 (data not shown).
In summary (Fig. 3A), removal of the C-D interhelical region, rather than disruption of the LXCXE binding pocket on Rb, had the greatest impact on Rb-PAI-2 binding, illustrating that binding of PAI-2 to Rb was primarily dependent on the PAI-2 C-D interhelical region. This largely LXCXE-independent binding of PAI-2 to Rb suggested that the C-D interhelical region is involved in binding Rb outside the A/B pocket LXCXE binding domain.
The RSL mutant of PAI-2 retains binding to Rb.
Figure 1 illustrated that the RSL mutant of PAI-2 (PAI-2 Ala380) failed to elevate the expression level of Rb in HeLa cells. To determine whether binding to Rb was affected by this mutation, equal amounts of GST and GST-Rb were mixed with similar amounts of IVT PAI-2, IVT C-D PAI-2, and IVT PAI-2 Ala380 (as determined by densitometry of the correct molecular weight band [Fig. 3D, lane 3]). IVT C-D PAI-2 again bound less efficiently to GST-Rb than IVT PAI-2, but IVT PAI-2 Ala380 binding to GST-Rb was similar to that seen for IVT PAI-2 (Fig. 3D, lane 2). These data demonstrate that the Ala380 mutation does not affect the binding of PAI-2 to Rb (Fig. 3A) and suggest that PAI-2 binding to Rb is not sufficient for protection of Rb. p130 and p107 binding to PAI-2 Ala380 was also similar to that seen for wild-type PAI-2 (data not shown).
PAI-2 binds the C-pocket of Rb.
Like PAI-2, the nuclear c-Abl tyrosine kinase has been shown to bind Rb, but not p107, through a predominantly LXCXE-independent mechanism (50). c-Abl was reported to bind the C-pocket of Rb (Rb768-928), and the binding site within Rb768-928 was mapped by using a series of mutants. Wild-type Rb768-928 (referred to as Rb-SE) and Rb768-928 with a deletion in region 785-806 (referred to as Rb-SEΔ) retained c-Abl binding, whereas Rb-13S (Rb768-928 incorporating a series of mutations) completely lost the ability to bind c-Abl (50). When PAI-2 was tested for binding to these Rb C-pocket mutants, PAI-2 showed the same pattern of binding to Rb-SE, Rb-SEΔ, and Rb-13S as was reported for c-Abl (Fig. 3E, top panel) (50). PAI-2 was able to bind the C-pocket Rb (GST-Rb SE) as efficiently as Rb (GST-Rb) (Fig. 3E, lanes 2 and 3). PAI-2 was also able to bind the C-pocket mutant Rb-SEΔ, but not Rb-13S (Fig. 3E, lanes 4 and 5). C-D PAI-2 failed to bind the C-pocket of Rb (Fig. 3E, bottom panel, lanes 3, 4, and 5) but retained some LXCXE-dependent binding to Rb (as seen in Fig. 3C) (Fig. 3E, bottom panel, lane 2). These data (summarized in Fig. 3 A) demonstrate that the C-D interhelical region of PAI-2 binds the C-pocket of Rb at the same site as c-Abl, a binding region distinct from the A/B pocket, which contains the LXCXE binding site.
The PAI-2 C-D interhelical region reveals a novel consensus motif shared by many Rb-binding proteins.
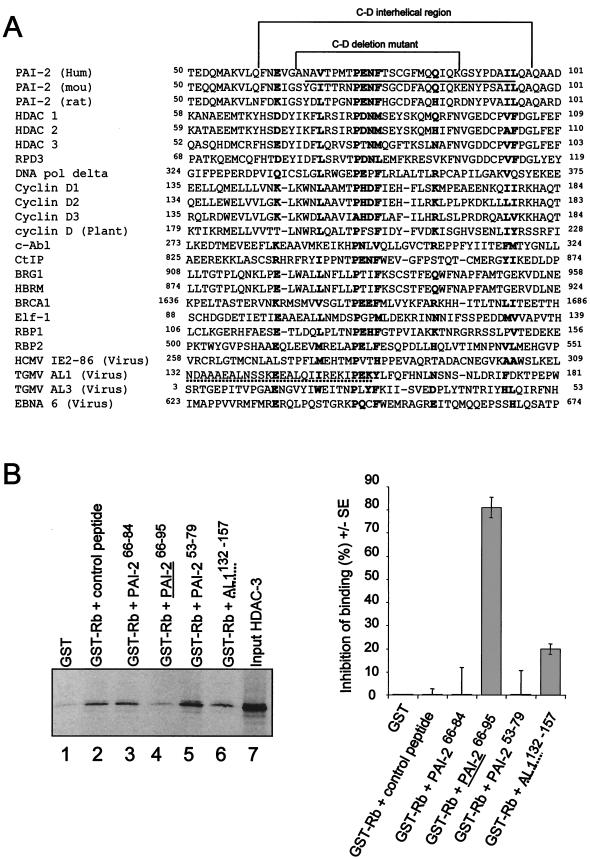
The largely LXCXE-independent Rb binding of both PAI-2 (Fig. 3B and C) and c-Abl (50) is analogous to the situation found for a number of other Rb-binding proteins where Rb binding has also been reported to be LXCXE independent (see the legend to Fig. 4A). When the amino acid sequences of these Rb-binding proteins were searched for regions with homology to the PAI-2 C-D interhelical region, a conserved motif centered around the 73PENF76 sequence of PAI-2 emerged. Additional conserved residues at positions E62, V68, Q84, I94, and L95 within the PAI-2 interhelical region were also seen (Fig. 4A). This alignment demonstrated the existence of a new Rb-binding motif, termed the PENF homology motif, which is shared by many cellular and viral Rb-binding proteins. A group of Rb-binding proteins for which LXCXE-independent binding has not been reported was also found to share the new motif; these were HBRM, cyclins D2 and D3, HDAC-2 (10), Elf-1 (48), and RBP1 and -2 (15).
FIG.4.
A new Rb-binding motif, the PENF homology region. (A) Alignment of Rb-binding proteins with the C-D interhelical region of PAI-2 reveals a conserved motif (boldfaced). Rb-binding proteins for which Rb binding has been reported to be LXCXE independent include HDAC-1 (16) and -3 (15), yeast RPD3 (28), DNA polymerase delta (DNA pol delta) (30), cyclin D1 (35), c-Abl (50), Ctip, BRG1 (10), BRCA-1 (14), TGMV AL3 (43) and AL1 (29), Epstein-Barr virus nuclear protein 6 (EBNA6) (36), and human cytomegalovirus (HCMV) IE2 86 (18). LXCXE-independent binding has not been reported for the Rb-binding proteins HBRM, cyclin D2 and -3, HDAC-2 (10), Elf-1 (48), and RBP1 and -2 (15). The sequence representing the C-D interhelical region and the sequence deleted in the PAI-2 C-D interhelical mutant are indicated. The peptide sequence that is able to inhibit HDAC-3 binding to Rb379-928 is underlined (PAI-266-95; see panel B). The proposed geminivirus TGMV AL1 Rb-binding site (29) is shown with a dashed underline. The alignments were facilitated by using MENE software, available at the BioNavigator bioinformatics website (www.bionavigator.com.au). (B) Inhibition of HDAC-3 binding to Rb379-928 by using synthetic peptides based on the PAI-2 C-D interhelical region. 35S-labeled IVT HDAC-3 was incubated with the indicated peptides, and the mixture was added to glutathione-agarose beads bound to GST or GST-Rb379-928. After washing, bead-bound, 35S-labeled HDAC-3 was resolved by SDS-PAGE and exposed to X-ray film. This experiment was repeated three times, and scanning densitometry was used to quantify resolved HDAC-3 bands. The mean percent inhibition from the three separate experiments is shown on the right; percent inhibition of binding refers to percent reduction in band intensity compared with that of GST-Rb plus control peptide. The inhibitory PAI-266-95 peptide is underlined, and the TGMV AL1 peptide sequence, AL1132-157, is shown with a dashed underline, in both the sequence and the gel.
To further investigate the role of the PENF homology motif in Rb binding, a series of overlapping peptides spanning the PAI-2 C-D interhelical region were tested for their ability to inhibit the binding of HDAC-3 to GST-Rb in pulldown experiments. HDAC-3 does not contain a LXCXE motif and binds Rb through an LXCXE-independent mechanism (9). Based on the alignment above (Fig. 4A), we postulated that HDAC-3 utilizes its PENF homology motif for Rb binding. Of the peptides tested, the PAI-266-95 peptide (Fig. 4B, lane 4) substantially inhibited HDAC-3 binding to Rb, showing an ≈80% reduction in the amount of HDAC-3 pulled down by GST-Rb relative to the control peptide (Fig. 4B, lane 2). Neither of the shorter PAI-2 C-D interhelical peptides (PAI-266-84 and PAI-253-79) was able to inhibit HDAC-3 binding (Fig. 4B, lanes 3 and 5). A peptide representing the proposed Rb-binding site of the AL1 protein of tomato golden mosaic virus (TGMV) (29) (AL1132-157), which contained part of the PENF homology motif, partially inhibited the binding of HDAC-3 to Rb (Fig. 4B, lane 6). These experiments demonstrated that the binding of HDAC-3 to Rb could be inhibited by a PAI-2-derived peptide spanning the PENF homology region, further supporting the role of the PENF homology region in Rb binding by viral and mammalian proteins.
PAI-2-expressing Jurkat cells also show posttranscriptional elevation of Rb protein levels.
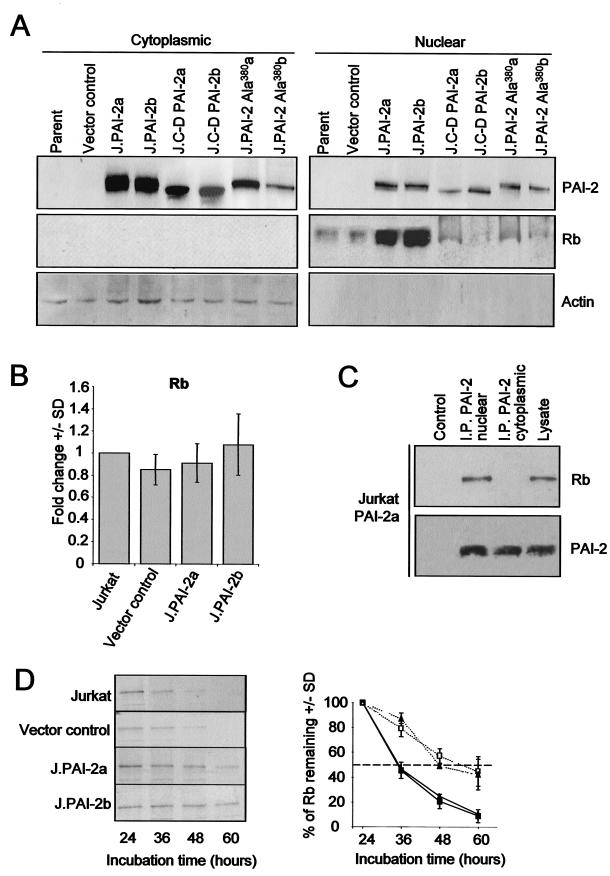
The striking PAI-2-mediated recovery of Rb may be exaggerated in HeLa cells by the presence of E7, which normally targets Rb for accelerated proteolytic degradation (19). To investigate PAI-2-mediated effects on Rb expression in cells devoid of confounding viral Rb-binding oncoproteins, Jurkat T cells stably expressing wild-type PAI-2 (J.PAI-2a and J.PAI-2b), the C-D interhelical mutant of PAI-2 (J.C-D PAI-2a/b), and the RSL mutant of PAI-2 (J.PAI-2Ala380a/b) were generated. Western blot analysis of nuclear and cytoplasmic extracts from the Jurkat lines again showed (i) that PAI-2 was present in the nucleus, (ii) that PAI-2 expression resulted in substantial elevation of nuclear Rb protein levels compared with the control lines (the parental Jurkat and a vector control line), and (iii) that transfection with C-D PAI-2 or PAI-2 Ala380 did not result in the elevation of Rb levels (Fig. 5A). Furthermore, as was observed for PAI-2-expressing S1a and S1b cell lines, quantitative real-time RT-PCR of the Jurkat lines again showed that Rb mRNA levels were not significantly affected by wild-type PAI-2 expression (Fig. 5B). In addition, anti-PAI-2 antibody was again able to immunoprecipate Rb from nuclear, but not cytoplasmic, fractions of cells expressing wild-type PAI-2 (Fig. 5C). These data demonstrated that in the absence of HPV oncoproteins, PAI-2 again associated with nuclear Rb, and PAI-2 expression again led to substantial posttranscriptional increases in Rb protein levels. Furthermore, this increase in Rb levels required that PAI-2 have an intact C-D interhelical loop and RSL.
FIG.5.
Increases in Rb protein levels in PAI-2-expressing Jurkat cells. (A) Western blot analysis of nuclear and cytoplasmic proteins extracted from the parental Jurkat cell line (Parent), a vector control Jurkat line (Vector control), Jurkat lines stably expressing wild-type PAI-2 (J.PAI-2a and J.PAI-2b), the C-D interhelical mutant of PAI-2 (J.C-D PAI-2a/b), or the RSL mutant of PAI-2 (J.PAI-2 Ala380a/b). Nuclear and cytoplasmic cell lysates were probed using antibodies specific for PAI-2, Rb, and actin. The low level of the cytoplasmic protein actin in the nuclear extract confirmed the low level of cytoplasmic proteins contaminating these extracts. (B) Quantitative real-time RT-PCR expression analysis of Rb mRNA levels in Jurkat control and PAI-2-expressing lines. Rb cDNA prepared from parental Jurkat cells, Jurkat vector control cells, and PAI-2-expressing Jurkat lines (J.PAI-2a and -b) was performed using GAPDH as a standard. Data from three experiments are expressed as the mean fold change (± standard deviation) compared with parental Jurkat cells. (C) Immunoprecipitation of PAI-2 from nuclear (I.P. PAI-2 nuclear) and cytoplasmic (I.P. PAI-2 cytoplasmic) fractions from PAI-2-expressing Jurkat lines by using an anti-PAI-2 antibody, followed by electrophoresis and Western blotting of the immunoprecipitates with anti-Rb (top panel) and anti-PAI-2 (bottom panel) antibodies. Control and lysate lanes are as for Fig. 2B. (D) Increased Rb protein stability in PAI-2-expressing Jurkat cells. Rb turnover was analyzed by a pulse-chase procedure using the control parental Jurkat cell line (solid squares), the vector control Jurkat cell line (solid triangles), and the PAI-2-expressing Jurkat lines J.PAI-2a (open squares) and J.PAI-2b (open triangles). Cells were metabolically labeled with [35S]methionine-cysteine and washed, and at the indicated times, cell lysates were prepared and immunoprecipitated with an antibody specific for Rb. Immunoprecipitated Rb was resolved by SDS-PAGE and detected by autoradiography. Scanning densitometry was used to quantify the percentage of Rb remaining. (Left) Autoradiogram from one representative experiment. (Right) Graph showing the mean percentage of Rb remaining ± standard deviation as determined from three separate experiments.
PAI-2 inhibits Rb degradation.
The PAI-2-mediated posttranscriptional increase in Rb protein levels in HeLa and Jurkat cells, and the requirement for an intact PAI-2 RSL for elevation of Rb levels (see Fig. 1A and 5A), suggested that PAI-2 might protect Rb from proteolytic degradation. To investigate whether PAI-2 expression inhibits Rb degradation, pulse-chase experiments were performed in control and PAI-2-expressing Jurkat cells. (HeLa cell lines could not be analyzed, because Rb was undetectable in HeLa and A2/7 cells.) The half-life of Rb in control Jurkat cell lines was found to be approximately 12 h, whereas the half-life of Rb in PAI-2-expressing Jurkat lines was increased to >24 h (Fig. 5D). This finding suggested that PAI-2 expression inhibits Rb protein degradation or turnover, with the degradation presumably mediated by cellular proteases.
PAI-2 expression elevates Rb-mediated activities.
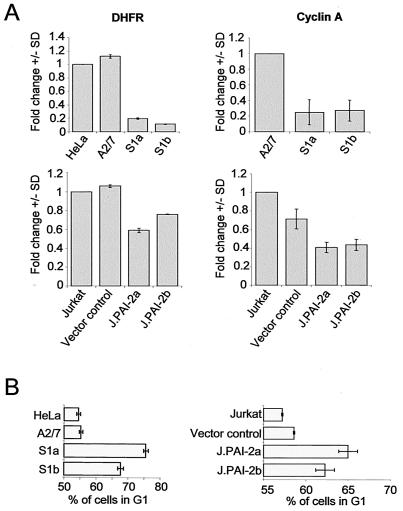
One of the best-described activities of hypophosphorylated Rb is inhibition of E2F-1-mediated transcription and cell cycle arrest in G1 (21, 33). To determine whether the PAI-2-mediated increase in Rb protein levels in Jurkat and HeLa cells influenced E2F-1-mediated transcription, mRNA levels for E2F-1-dependent genes and cell cycle were analyzed. Quantitative real-time RT-PCR analysis showed reduced levels of mRNAs encoding the E2F-1-dependent genes, DHFR, and cyclin A in both HeLa and Jurkat cell lines expressing PAI-2 compared with control cell lines (Fig. 6A). The percentage of cells in the G1 phase of the cell cycle was also higher in PAI-2-expressing cells than in control cell lines (Fig. 6B). These experiments showed that PAI-2-mediated increases in Rb protein levels resulted in repression of E2F-1 transcription, as evidenced by the reduction in mRNA levels for E2F-1-dependent genes and the elongated G1 phase of the cell cycle.
FIG. 6.
Changes in E2F-1-dependent transcription and G1 arrest in PAI-2-expressing cells. (A) Quantitative real-time RT-PCR analysis of E2F-1-dependent gene expression. DHFR and cyclin A cDNAs from parental HeLa cells, antisense PAI-2 (A2/7), PAI-2-expressing HeLa lines (S1a and S1b), parental Jurkat cells, vector control Jurkat cells, and PAI-2-expressing Jurkat lines (J.PAI-2a and -b) were quantified by PCR using GAPDH as a standard. Data from three experiments are expressed as the mean fold change (± standard deviation) compared with parental HeLa and Jurkat cells. P < 0.0001 by the Student's t test comparing DHFR levels in HeLa and A2/7 cells with those in S1a and S1b cells; for cyclin A in the HeLa lines, P < 0.001; for DHFR levels in Jurkat cells and the vector control versus PAI-2a and -b lines, P < 0.008; and for cyclin A in the same Jurkat lines, P < 0.003. (B) Cell cycle analysis of PAI-2-expressing and control cell lines. Cells were analyzed using propidium iodide staining and fluorescence-activated cell sorter analysis to determine the percentage of cells in the G1 phase of the cell cycle. The mean of three experiments ± standard deviation is shown. By Student's t test comparing control and PAI-2-expressing cells, P < 0.0002 for PAI-2-expressing HeLa cells and P < 0.05 for PAI-2-expressing Jurkat lines.
It should be noted that the latter activities (and those shown in Fig. 1 and 5) were not a consequence of PAI-2 overexpression. The levels of PAI-2 expressed in the transfected HeLa and Jurkat cell lines ranged from 28 to 132 ng mg−1 of protein (12) (data not shown). These levels are at the lower end of the range of PAI-2 protein levels found in primary PAI-2-expressing cells; for instance, synovial fibroblasts express 23 ± 2 ng mg−1 (20), keratinocytes express 4,424 ± 1,592 ng mg−1 (6), and monocytes express up to 810 ± 75 ng mg−1 (40).
DISCUSSION
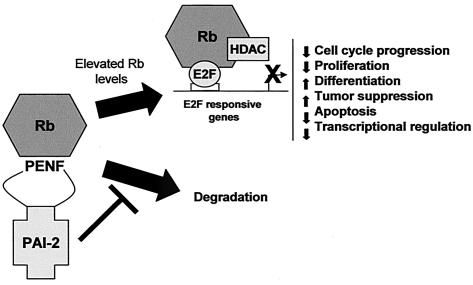
This paper describes a novel function for PAI-2 as an intracellular Rb-binding protein, which inhibits Rb protein turnover, leading to increases in Rb-mediated activities (Fig. 7). PAI-2 was found to colocalize with Rb in the cell nucleus, and an anti-PAI-2 antibody was shown to coimmunoprecipitate Rb from nuclear fractions of cells that physiologically produce PAI-2. Furthermore, PAI-2 bound to the C-pocket of Rb via a novel Rb-binding motif shared by the C-D interhelical region of PAI-2 and many mammalian and viral Rb-binding proteins. PAI-2 expression in HeLa and Jurkat cell reduced Rb degradation, resulting in increased Rb protein levels and Rb-mediated activities, such as promotion of G1 cell cycle arrest and inhibition of E2F-1-mediated transcription. Finally, the ability of PAI-2 to inhibit Rb degradation was shown to be dependent on both the C-D interhelical region and the RSL of PAI-2. This new activity for PAI-2 supports the view expressed in a large body of literature that PAI-2 likely has an additional intracellular function distinct from uPA inhibition (2, 4, 6, 11, 12, 40, 44, 45, 54, 55).
FIG. 7.
Proposed intracellular Rb protection activity of PAI-2. PAI-2 binds Rb via the PENF homology motif and inhibits the activity of unknown proteases that degrade Rb. Rb turnover is reduced and Rb protein levels rise, resulting in enhancement of Rb-mediated activities. Rb can associate with many transcription factors, although the best characterized is association of Rb with E2F-1, which results in inhibition of E2F-1-dependent gene transcription. Inhibition of E2F-1-dependent transcription is associated with decreased cell cycle progression, reduced proliferation, an increase in tumor suppression activity, promotion of differentiation, inhibition of apoptosis, and transcriptional regulation. PAI-2 expression has been shown to promote similar phenotypes in several systems.
PAI-2 binds to the C-pocket of Rb via the PENF homology motif.
Ov-serpins show a high degree of structural homology; however, the nonconserved C-D interhelical regions of several ov-serpins have been shown to confer unique activities individual to particular ov-serpins (45). We propose that the C-D interhelical region of PAI-2 contains a previously undescribed Rb-binding motif, the PENF homology motif, which is shared by many cellular and viral Rb-binding proteins but not by other ov-serpins. This PENF homology motif likely represents the LXCXE-independent Rb-binding site within the large pocket region of Rb (comprising A, B, and C pockets) proposed by several groups (9, 10, 14, 16, 18, 28-30, 35, 36, 43). The full motif conforms to the consensus Z(X4-5)J(X4)PZZJ(X6-7)Z(X8-9)JJ (where Z represents intermediate or large polar residues, J represents large hydrophobic residues, and X represents any amino acid) with up to 4 mismatches allowed (Fig. 4A). A motif with a similar format, described as Y(X7)E(X3)DLF, has been proposed to be involved in the binding of E2F-1 and NF-IL-6 (C/EBPβ) to Rb (8). The PENF homology region appears to be involved in binding Rb in the C-pocket region, in a region distinct from the E2F and LXCXE binding domains (Fig. 3A and D). This observation is consistent with the literature describing the importance of the Rb C-pocket region in growth suppression and differentiation (35, 50).
Inhibition of Rb degradation by PAI-2.
The ability of PAI-2 expression to elevate Rb protein levels in HeLa cells, the failure of the RSL mutant of PAI-2 to achieve this elevation of Rb levels, and the increased half-life of Rb in PAI-2-expressing Jurkat cells all suggest that PAI-2 might be inhibiting a proteinase that targets Rb for degradation. Although Rb is thought to be degraded by the proteasomal pathway (47), it is unlikely that PAI-2 directly inhibits the proteinase activity of the proteasome. The ability of PAI-2 Ala380 to bind Rb, but not to protect Rb from degradation, also suggests that protection of Rb by PAI-2 is not simply due to PAI-2 blocking ubiquitinylation or E7 binding. A number of proteinases are known to be active in the nucleus and are known to target Rb; these include calpain-1 (24), caspases (7), and Spase (23). We have preliminary in vitro evidence (not shown) that PAI-2 is able to inhibit the cleavage of a small fragment from Rb by the cysteine proteinase calpain-1; an observation consistent with the location of a predicted PEST sequence with a high score (+14.93) in the N terminus of Rb (38). We are currently exploring whether calpains and/or other proteases are the targets of PAI-2 inhibition in vivo, and we are investigating the possibility that proteasomal degradation of Rb may be preceded by a separate cleavage event (22) that is inhibited by nuclear PAI-2.
PAI-2-mediated recovery of Rb and transcriptional repression of HPV oncogenes E6 and E7.
The ability of PAI-2 expression to repress transcription of the HPV E6 and E7 oncogenes in HeLa cells (Fig. 1) appeared to be dependent on the PAI-2-mediated recovery of Rb (Fig. 1D), an observation consistent with the report of Salcedo et al. (41), who also showed that Rb expression could repress E6 and E7 transcription. Thus, PAI-2 protects Rb from the accelerated degradation mediated by E7 (5, 22, 42), and the increase in Rb protein levels leads to inhibition of E6 and E7 transcription and a reduction in E6 and E7 protein expression. This would then result in recovery of the other E6- and E7-targeted proteins, such as p53 (Fig. 1) and ISGF3 (2). We are currently characterizing the Rb-dependent process responsible for repression of E6 and E7 transcription and are investigating how PAI-2 might block E7 activity. The ability of PAI-2 to reverse the activities of the HPV oncogene products suggests a potential therapeutic role for PAI-2 or PAI-2 derivatives in the treatment of papillomavirus-transformed lesions.
There is no direct evidence to show that PAI-2 inhibits uPA in vivo, and several studies on the physiological role of PAI-2 have inferred a uPA-independent intracellular function for PAI-2 (13, 26, 39, 49, 51, 55). The proposed new Rb-associated activity for PAI-2 (Fig. 7) may explain many of the diverse phenotypes described for cells and tissues expressing PAI-2 (11, 12, 17, 31, 34, 37, 53). Both the Rb family of proteins and PAI-2 are associated with tumor suppression (17, 21, 31, 33, 34, 37, 53), promotion of differentiation (21, 31, 33, 49, 54), inhibition of apoptosis (11, 21, 33, 44), and transcriptional regulation (12, 21, 33) (Fig. 1 and 6). The data also suggest the potential for PAI-2-based therapeutics for modulation of Rb activities or inhibition of viral Rb-binding proteins.
Acknowledgments
We thank Biotech Australia for the gift of recombinant PAI-2 and the anti-PAI-2 antibody. Thanks also go to Joy Gardner, Stephanie Kosmala, Kathy Buttigieg, Julie Muddiman, and Victoria Mann for assistance during the project.
This project was funded by the Queensland Institute of Medical Research Trust, the Australian Centre for International and Tropical Health and Nutrition, and the Queensland Cancer Fund. G.A.D. is supported by a University of Queensland Graduate School award. R.W.J. is a Wellcome Trust Senior Research Fellow.
G.A.D. and T.M.A. contributed equally to this work.
REFERENCES
- 1.Akiyama, H., K. Ikeda, H. Kondo, M. Kato, and P. L. McGeer. 1993. Microglia express the type 2 plasminogen activator inhibitor in the brain of control subjects and patients with Alzheimer's disease. Neurosci. Lett. 164:233-235. [DOI] [PubMed] [Google Scholar]
- 2.Antalis, T. M., M. La Linn, K. Donnan, L. Mateo, J. Gardner, J. L. Dickinson, K. Buttigieg, and A. Suhrbier. 1998. The serine proteinase inhibitor (serpin) plasminogen activation inhibitor type 2 protects against viral cytopathic effects by constitutive interferon alpha/beta priming. J. Exp. Med. 187:1799-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bignon, Y. J., J. Y. Shew, D. Rappolee, S. L. Naylor, E. Y. Lee, J. Schnier, and W. H. Lee. 1990. A single Cys706 to Phe substitution in the retinoblastoma protein causes the loss of binding to SV40 T antigen. Cell Growth Differ. 1:647-651. [PubMed] [Google Scholar]
- 4.Bird, C. H., E. J. Blink, C. E. Hirst, M. S. Buzza, P. M. Steele, J. Sun, D. A. Jans, and P. I. Bird. 2001. Nucleocytoplasmic distribution of the ovalbumin serpin PI-9 requires a nonconventional nuclear import pathway and the export factor Crm1. Mol. Cell. Biol. 21:5396-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, S. N., D. E., Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 6.Braungart, E., V. Magdolen, and K. Degitz. 2001. Retinoic acid upregulates the plasminogen activator system in human epidermal keratinocytes. J. Investig. Dermatol. 116:778-784. [DOI] [PubMed] [Google Scholar]
- 7.Chau, B. N., H. L. Borges, T. T. Chen, A. Masselli, I. C. Hunton, and J. Y. Wang. 2002. Signal-dependent protection from apoptosis in mice expressing caspase-resistant Rb. Nat. Cell Biol. 4:757-765. [DOI] [PubMed] [Google Scholar]
- 8.Chen, P. L., D. J. Riley, S. Chen-Kiang, and W. H. Lee. 1996. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc. Natl. Acad. Sci. USA 93:465-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahiya, A., M. R. Gavin, R. X. Luo, and D. C. Dean. 2000. Role of the LXCXE binding site in Rb function. Mol. Cell. Biol. 20:6799-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dick, F. A., E. Sailhamer, and N. J. Dyson. 2000. Mutagenesis of the pRB pocket reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol. Cell. Biol. 20:3715-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickinson, J. L., E. J. Bates, A. Ferrante, and T. M. Antalis. 1995. Plasminogen activator inhibitor type 2 inhibits tumor necrosis factor alpha-induced apoptosis. Evidence for an alternate biological function. J. Biol. Chem. 270:27894-27904. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson, J. L., B. J. Norris, P. H. Jensen, and T. M. Antalis. 1998. The C-D interhelical domain of the serpin plasminogen activator inhibitor-type 2 is required for protection from TNF-α induced apoptosis. Cell Death Differ. 5:163-171. [DOI] [PubMed] [Google Scholar]
- 13.Dougherty, K. M., J. M. Pearson, A. Y. Yang, R. J. Westrick, M. S. Baker, and D. Ginsburg. 1999. The plasminogen activator inhibitor-2 gene is not required for normal murine development or survival. Proc. Natl. Acad. Sci. USA 96:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan, S., R. Yuan, Y. X. Ma, J. Xiong, Q. Meng, M. Erdos, J. N. Zhao, I. D. Goldberg, R. G. Pestell, and E. M. Rosen. 2001. Disruption of BRCA1 LXCXE motif alters BRCA1 functional activity and regulation of RB family but not RB protein binding. Oncogene 20:4827-4841. [DOI] [PubMed] [Google Scholar]
- 15.Fattaey, A. R., K. Helin, M. S. Dembski, N. Dyson, E. Harlow, G. A. Vuocolo, M. G. Hanobik, K. M. Haskell, A. Oliff, and D. Defeo-Jones. 1993. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene 8:3149-3156. [PubMed] [Google Scholar]
- 16.Ferreira, R., L. Magnaghi-Jaulin, P. Robin, A. Harel-Bellan, and D. Trouche. 1998. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc. Natl. Acad. Sci. USA 95:10493-10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foekens, J. A., F. Buessecker, H. A. Peters, U. Krainick, W. L. van Putten, M. P. Look, J. G. Klijn, and M. D. Kramer. 1995. Plasminogen activator inhibitor-2: prognostic relevance in 1012 patients with primary breast cancer. Cancer Res. 55:1423-1427. [PubMed] [Google Scholar]
- 18.Fortunato, E. A., M. H. Sommer, K. Yoder, and D. H. Spector. 1997. Identification of domains within the human cytomegalovirus major immediate-early 86-kilodalton protein and the retinoblastoma protein required for physical and functional interaction with each other. J. Virol. 71:8176-8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodwin, E. C., and D. DiMaio. 2000. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc. Natl. Acad. Sci. USA 97:12513-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton, J. A., J. Wojta, M. Gallichio, K. McGrath, and E. L. Filonzi. 1993. Contrasting effects of transforming growth factor-β and IL-1 on the regulation of plasminogen activator inhibitors in human synovial fibroblasts. J. Immunol. 151:5154-5161. [PubMed] [Google Scholar]
- 21.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed] [Google Scholar]
- 22.Helt, A. M., and D. A. Galloway. 2003. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis 24:159-169. [DOI] [PubMed] [Google Scholar]
- 23.Irving, J. A., S. S. Shushanov, R. N. Pike, E. Y. Popova, D. Bromme, T. H. Coetzer, S. P. Bottomley, I. A. Boulynko, S. A. Grigoryev, and J. C. Whisstock. 2002. Inhibitory activity of a heterochromatin-associated serpin (MENT) against papain-like cysteine proteinases affects chromatin structure and blocks cell proliferation. J. Biol. Chem. 277:13192-13201. [DOI] [PubMed] [Google Scholar]
- 24.Jang, J. S., S. J. Lee, Y. H. Choi, P. M. Nguyen, J. Lee, S. G. Hwang, M. L. Wu, E. Takano, M. Maki, P. A. Henkart, and J. B. Trepel. 1999. Posttranslational regulation of the retinoblastoma gene family member p107 by calpain protease. Oncogene 18:1789-1796. [DOI] [PubMed] [Google Scholar]
- 25.Janicke, R. U., F. H. Lee, and A. G. Porter. 1994. Nuclear c-Myc plays an important role in the cytotoxicity of tumor necrosis factor alpha in tumor cells. Mol. Cell. Biol. 14:5661-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen, P. J., T. Yang, D. W. Yu, M. S. Baker, B. Risse, T. T. Sun, and R. M. Lavker. 2000. Serpins in the human hair follicle. J. Investig. Dermatol. 114:917-922. [DOI] [PubMed] [Google Scholar]
- 27.Johnstone, R. W., W. Wei, A. Greenway, and J. A. Trapani. 2000. Functional interaction between p53 and the interferon-inducible nucleoprotein IFI 16. Oncogene 19:6033-6042. [DOI] [PubMed] [Google Scholar]
- 28.Kennedy, B. K., O. W. Liu, F. A. Dick, N. Dyson, E. Harlow, and M. Vidal. 2001. Histone deacetylase-dependent transcriptional repression by pRB in yeast occurs independently of interaction through the LXCXE binding cleft. Proc. Natl. Acad. Sci. USA 98:8720-8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong, L. J., B. M. Orozco, J. L. Roe, S. Nagar, S. Ou, H. S. Feiler, T. Durfee, A. B. Miller, W. Gruissem, D. Robertson, and L. Hanley-Bowdoin. 2000. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krucher, N. A., A. Zygmunt, N. Mazloum, S. Tamrakar, J. W. Ludlow, and M. Y. Lee. 2000. Interaction of the retinoblastoma protein (pRb) with the catalytic subunit of DNA polymerase delta (p125). Oncogene 19:5464-5470. [DOI] [PubMed] [Google Scholar]
- 31.Kruithof, E. K., M. S. Baker, and C. L. Bunn. 1995. Biological and clinical aspects of plasminogen activator inhibitor type 2. Blood 86:4007-4024. [PubMed] [Google Scholar]
- 32.La Linn, M., J. Gardner, D. Warrilow, G. A. Darnell, C. R. McMahon, I. Field, A. D. Hyatt, R. W. Slade, and A. Suhrbier. 2001. Arbovirus of marine mammals: a new alphavirus isolated from the elephant seal louse, Lepidophthirus macrorhini. J. Virol. 75:4103-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura, M., H. Konno, T. Tanaka, Y. Maruo, N. Nishino, K. Aoki, S. Baba, S. Sakaguchi, Y. Takada, and A. Takada. 1992. Possible role of plasminogen activator inhibitor 2 in the prevention of the metastasis of gastric cancer tissues. Thromb. Res. 65:709-719. [DOI] [PubMed] [Google Scholar]
- 35.Pan, W., S. Cox, R. H. Hoess, and R. H. Grafstrom. 2001. A cyclin D1/cyclin-dependent kinase 4 binding site within the C domain of the retinoblastoma protein. Cancer Res. 61:2885-2891. [PubMed] [Google Scholar]
- 36.Parker, G. A., T. Crook, M. Bain, E. A. Sara, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen (EBNA) 3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene 13:2541-2549. [PubMed] [Google Scholar]
- 37.Praus, M., K. Wauterickx, D. Collen, and R. D. Gerard. 1999. Reduction of tumor cell migration and metastasis by adenoviral gene transfer of plasminogen activator inhibitors. Gene Ther. 6:227-236. [DOI] [PubMed] [Google Scholar]
- 38.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267-271. [PubMed] [Google Scholar]
- 39.Risse, B. C., N. M. Chung, M. S. Baker, and P. J. Jensen. 2000. Evidence for intracellular cleavage of plasminogen activator inhibitor type 2 (PAI-2) in normal epidermal keratinocytes. J. Cell. Physiol. 182:281-289. [DOI] [PubMed] [Google Scholar]
- 40.Ritchie, H., and N. A. Booth. 1998. Secretion of plasminogen activator inhibitor 2 by human peripheral blood monocytes occurs via an endoplasmic reticulum-Golgi-independent pathway. Exp. Cell Res. 242:439-450. [DOI] [PubMed] [Google Scholar]
- 41.Salcedo, M., E. Garrido, L. Taja, and P. Gariglio. 1995. The retinoblastoma gene product negatively regulates cellular or viral oncogene promoters in vivo. Arch. Med. Res. 26:157-162. [PubMed] [Google Scholar]
- 42.Scheffner, M., K. Munger, J. M. Huibregtse, and P. M. Howley. 1992. Targeted degradation of the retinoblastoma protein by human papillomavirus E7-E6 fusion proteins. EMBO J. 11:2425-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Settlage, S. B., A. B. Miller, W. Gruissem, and L. Hanley-Bowdoin. 2001. Dual interaction of a geminivirus replication accessory factor with a viral replication protein and a plant cell cycle regulator. Virology 279:570-576. [DOI] [PubMed] [Google Scholar]
- 44.Shafren, D. R., J. Gardner, V. H. Mann, T. M. Antalis, and A. Suhrbier. 1999. Picornavirus receptor down-regulation by plasminogen activator inhibitor type 2. J. Virol. 73:7193-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silverman, G. A., P. I. Bird, R. W. Carrell, F. C. Church, P. B. Coughlin, P. G. Gettins, J. A. Irving, D. A. Lomas, C. J. Luke, R. W. Moyer, P. A. Pemberton, E. Remold-O'Donnell, G. S. Salvesen, J. Travis, and J. C. Whisstock. 2001. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 276:33293-33296. [DOI] [PubMed] [Google Scholar]
- 46.Szekely, L., E. Uzvolgyi, W. Q. Jiang, M. Durko, K. G. Wiman, G. Klein, and J. Sumegi. 1991. Subcellular localization of the retinoblastoma protein. Cell Growth Differ. 2:287-295. [PubMed] [Google Scholar]
- 47.von Willebrand, M., E. Zacksenhaus, E. Cheng, P. Glazer, and R. Halaban. 2003. The tyrphostin AG1024 accelerates the degradation of phosphorylated forms of retinoblastoma protein (pRb) and restores pRb tumor suppressive function in melanoma cells. Cancer Res. 63:1420-1429. [PubMed] [Google Scholar]
- 48.Wang, C. Y., B. Petryniak, C. B. Thompson, W. G. Kaelin, and J. M. Leiden. 1993. Regulation of the Ets-related transcription factor Elf-1 by binding to the retinoblastoma protein. Science 260:1330-1335. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Y., and P. J. Jensen. 1998. Regulation of the level and glycosylation state of plasminogen activator inhibitor type 2 during human keratinocyte differentiation. Differentiation 63:93-99. [DOI] [PubMed] [Google Scholar]
- 50.Whitaker, L. L., H. Su, R. Baskaran, E. S. Knudsen, and J. Y. Wang. 1998. Growth suppression by an E2F-binding-defective retinoblastoma protein (RB): contribution from the RB C pocket. Mol. Cell. Biol. 18:4032-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams, D. L., B. Risse, S. Kim, D. Saunders, S. Orlin, M. S. Baker, P. J. Jensen, and R. M. Lavker. 1999. Plasminogen activator inhibitor type 2 in human corneal epithelium. Investig. Ophthalmol. Vis. Sci. 40:1669-1675. [PubMed] [Google Scholar]
- 52.Yang, W. M., Y. L. Yao, J. M. Sun, J. R. Davie, and E. Seto. 1997. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 272:28001-28007. [DOI] [PubMed] [Google Scholar]
- 53.Yoshino, H., Y. Endo, Y. Watanabe, and T. Sasaki. 1998. Significance of plasminogen activator inhibitor 2 as a prognostic marker in primary lung cancer: association of decreased plasminogen activator inhibitor 2 with lymph node metastasis. Br. J. Cancer 78:833-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, H., F. Maurer, and R. L. Medcalf. 2002. Plasminogen activator inhibitor type 2: a regulator of monocyte proliferation and differentiation. Blood 99:2810-2818. [DOI] [PubMed] [Google Scholar]
- 55.Zhou, H. M., I. Bolon, A. Nichols, A. Wohlwend, and J. D. Vassalli. 2001. Overexpression of plasminogen activator inhibitor type 2 in basal keratinocytes enhances papilloma formation in transgenic mice. Cancer Res. 61:970-976. [PubMed] [Google Scholar]