Abstract
Smad7, an inhibitor of transforming growth factor beta superfamily signaling, is induced by bone morphogenetic protein (BMP) in an inhibitory feedback loop. Here, we identify multiple BMP response elements (BREs) in the Smad7 gene and demonstrate that they function differentially to interpret BMP signals in a cell type-specific manner. Two BREs (BRE-1 and -2) reside in the promoter region. One of these contains several conserved Smad1 and Smad4 binding sites that cooperate to mediate BMP-dependent induction, most likely in the absence of DNA binding partners. The third BRE (I-BRE) resides in the first intron and contains GATA factor binding sites. GATA-1, -5, or -6 is required for strong activation of I-BRE, and we show that they assemble with Smad1 on the I-BRE in living cells. Activation of the I-BRE is mediated by a specific region in GATA-5 and -6 but does not require direct physical interaction with Smad1. Comparison of I-BRE to BRE-1 showed that I-BRE is more responsive to low BMP concentrations. Moreover, analysis by chromatin immunoprecipitation experiments demonstrates that the endogenous I-BRE is occupied more robustly by endogenous Smad1 than is BRE-1. This correlates with regulation of the Smad7 gene, which is induced at lower BMP concentrations in GATA-expressing cell lines compared to non-GATA-expressing lines. These data thus define how cooperative and noncooperative Smad-dependent transcriptional regulation can function to interpret different BMP concentrations.
Members of the transforming growth factor beta (TGF-β) superfamily regulate various cellular processes, including proliferation, differentiation, cell fate determination, apoptosis, adhesion, morphogenesis, and development (reviewed in references 28, 29, 66, and 81). Many of these ligands have been shown to act as morphogens, i.e., they emanate from a localized source, diffuse across tissues to produce a concentration gradient, and induce different transcriptional responses depending on their concentrations (2, 22, 73, 76).
TGF-β family members bind to heteromeric complexes of transmembrane Ser/Thr kinases known as type I and type II receptors (reviewed in references 53 and 83). The type II receptor, a constitutively active kinase, transphosphorylates and activates the type I receptor, which, in turn, phosphorylates Smad proteins. Receptor-regulated Smads (R-Smads) include Smad2 and Smad3, which function in a TGF-β/activin pathway, and Smad1, Smad5, and Smad8, which participate in bone morphogenetic protein (BMP) signaling. The R-Smads are phosphorylated on the last two serines within an SSXS carboxy-terminal motif, which induces association with Smad4, a protein common to the TGF-β/activin and BMP pathways. The resulting R-Smad/Smad4 complex then translocates to the nucleus, where it can directly regulate transcriptional responses to TGF-β family members (reviewed in references 4, 17, and 52).
Smad MH2 domains have transcriptional activity when fused to a heterologous Gal4 DNA binding domain (49) while the MH1 domain can bind DNA directly (16, 41). Smad3 and Smad4 MH1 domains bind to 5′-GnCT, 5′-CAGA, and other GC-rich sequences (16, 45, 68, 85) while the Smad1 MH1 binds a 5′-GCCGNCGC motif (41, 44). Association of Smads with other transcription factors enhances the DNA binding affinity of the MH1 domains and is thought to be key for recruitment of Smads to specific promoter elements, whereas interactions with the coactivator CBP/p300 increase the transcriptional activity of the R-Smad/Smad4 complex (reviewed in reference 17). In addition, Smads can bind corepressors to negatively regulate target gene expression (reviewed in reference 82).
Smad6 and Smad7 (I-Smads) are inhibitors of the TGF-β/activin and BMP pathways (26, 32, 58). Smad6 and Smad7 lack the SSXS phosphorylation site and stably associate with the activated type I receptors, preventing phosphorylation of the R-Smads and mediating interaction between receptors and E3-ubiquitin ligases, Smurf1 and Smurf2, leading to the degradation of receptors (19, 23, 38). Smad6 may also inhibit signaling by competing with R-Smads for Smad4 binding (24) and by acting as a transcriptional corepressor (5). Smad6 and Smad7 are immediate early target genes of TGF-β, activin, and BMP (3, 34, 58, 74). A TGF-β and activin responsive element of the Smad7 promoter has been characterized, and Smad3/4, AP-1, SP1, and TFE3 sites are implicated in TGF-β-dependent regulation of this element (10, 15, 30, 57, 71, 80). Thus, I-Smads can function in a negative feedback loop to shut off TGF-β and BMP signaling.
How BMP regulates Smad7 is unknown. Therefore, we undertook the identification of cis and trans elements that regulate Smad7 expression in response to BMP-7. In this study, we isolated and identified two BMP-7 responsive elements (BREs) in the mouse Smad7 promoter (BRE-1 and -2) and one BRE in the first intron of the Smad7 gene (I-BRE). BMP induction of BRE-1 requires three independent regions of the BRE, which contain Smad1 and Smad4 binding sites. I-BRE, on the other hand, contains conserved binding sites for GATA proteins and is activated by GATA-1, -5, and -6. Directly comparing the activity of BRE-1 to I-BRE revealed that I-BRE is more responsive to low BMP concentrations than is BRE-1. Furthermore, chromatin immunoprecipitation (ChIP) assays of the endogenous elements also showed strong binding of endogenous Smad1 to I-BRE at low BMP concentrations, whereas occupancy of BRE-1 was weaker and required much higher levels of BMP stimulation. This correlates with Smad7 expression, which was induced by lower BMP concentrations in cell lines expressing GATA-5 and -6 than in a cell line lacking these factors. These results suggest that Smad7 expression is induced through multiple BREs that can function in a cell type-specific manner to interpret BMP signals.
MATERIALS AND METHODS
Cloning of the mouse Smad7 promoter.
A 296-bp DNA fragment corresponding to the +1204 to +1498 region of the 5′ untranslated region of the Smad7 cDNA, obtained from an expressed sequence tag database, was used to screen a mouse 129Sv female kidney DNA library, constructed by partial Sau3A digests cloned into the BamH1 site of Lambda Dash 129 (Stratagene). Positive clones containing a 5.4-kbp BamH1 fragment were isolated, and their inserts were subcloned into pBluescript KS (Stratagene).
Cell lines and transfections.
HepG2 cells were cultured in minimal essential medium supplemented with 1% nonessential amino acids, Neuro-2A cells were cultured in F-15 minimal essential medium, and 293T cells and mouse embryonic fibroblasts (MEF) were cultured in Dulbecco's modified essential medium. All media were supplemented with 10% fetal bovine serum. HepG2, Neuro-2A, and 293T cells were transiently transfected by using the calcium phosphate-DNA precipitation method described previously (26), and MEF were transfected by using the FuGENE 6 transfection reagent (Roche).
Luciferase assay.
Fragments of the Smad7 gene were subcloned into the pGL3 promoter luciferase plasmid (Promega), in which the simian virus 40 promoter was replaced with an E1b promoter (1). For BMP-induced luciferase assays, HepG2 and Neuro-2A cells were transiently transfected with the lux reporter plasmids and pCMV-Gal. Cells were seeded at 20% confluency in 24-well plates and transfected overnight with 0.5 μg of DNA per well. To induce the luciferase reporter, cells were treated overnight with the appropriate ligands and lysed, and luciferase activity was measured with the luciferase assay system (Promega) in a Berthold microplate luminometer LB 96V. Galactosidase activity was measured as described previously (1), and data were used to determine the transfection efficiency.
Electrophoretic mobility shift assays.
293T cells transfected with cDNA encoding GATA-5 or -6 were homogenized in 50 mM Tris-HCl (pH 8.0), 25% glycerol, 100 mM KCl, 4 mM dithiothreitol, 2 mM sodium orthovanadate, 10 mM sodium pyrophosphate, and protease inhibitors. Extracts were preincubated on ice for 10 min [20 mM HEPES-KOH (pH 7.9), 20% glycerol, 2 mM EDTA, 2 mM dithiothreitol, 7.2 mM MgCl2, 110-μg/ml poly(dI-dC)] and then incubated for 30 min in the presence of 30,000 cpm of 32P-labeled DNA prior to nondenaturing electrophoresis on a 5% polyacrylamide gel. Probes were prepared by digestion of constructs containing wild-type or mutated I-BRE, and digested fragments were labeled by Klenow fill-in reactions.
DNA and chromatin immunoprecipitation.
DNA and chromatin immunoprecipitations were performed following a modification of the procedure described by Boyd et al. (9). Briefly, protein and DNA were cross-linked by treating HepG2 or 293T cells with 1% formaldehyde for 10 min at room temperature. Glycine was added to a final concentration of 0.125 M, and the cells were incubated for 5 min. Cells were washed twice with ice-cold phosphate-buffered saline containing 1 mM phenylmethylsulfonyl fluoride. Cells were scraped, collected by centrifugation, and resuspended in 1% sodium dodecyl sulfate (SDS), 10 mM EDTA, 50 mM Tris-HCl (pH 8.1), and protease inhibitors. Following a 30-min incubation at 4°C, cells were sonicated five times for 10 s and centrifuged for 10 min at 4°C and 13,000 rpm. The supernatant was diluted in 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl (pH 8.1), and protease inhibitors. Immunoprecipitation was carried out overnight with 2 μg of antibody, and protein A Sepharose was added for 1 h. The beads were washed for 5 min in 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, and 150 mM NaCl (pH 8.1); for 5 min in 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, and 500 mM NaCl (pH 8.1); for 5 min in 0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl (pH 8.1); and twice for 5 min in 10 mM Tris-HCl and 1 mM EDTA (pH 8.1). Protein-DNA complexes were eluted from the beads in 1% SDS and 0.1 M NaHCO3 by incubating twice for 15 min at room temperature. De-cross-linking was carried out by adding NaCl to a final concentration of 200 mM and incubating the eluates at 65°C for 4 h. Proteins were digested for 1 h at 45°C by adding EDTA to a final concentration of 10 mM, Tris-HCl (pH 6.5) to a concentration of 40 mM, and proteinase K to a concentration of 0.04 mg/ml. The DNA was purified by phenol-chloroform treatment and ethanol precipitation. The DNA was analyzed by PCR or real-time PCR by using the ABI Prism 7900HT sequence detection system. Primers used for the analysis are as follows: for BRE-1, CGGGCACACTCGCTTGCTG and CGACGGACTGGCTCTGTCTCG; for I-BRE, TTGGAGGCAGGGCCGCGAGG and CAGACGTGGAGGAGCGGAGC; for the control, ATGGTGACGGGCACCTGTAG and TCCTGGGTTCACACCATTGTC.
For double DNA immunoprecipitation, 293T cells were transfected with cDNA encoding Flag-GATA-5 and Myc-Smad1. Lysates were subjected to immunoprecipitation with 2 μg of Flag antibody, and protein G Sepharose was added for 1 h. Protein-DNA complexes were eluted off the beads by incubation with 0.5-mg/ml Flag peptide (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA [pH 7.4]) and subjected to immunoprecipitation with 2 μg of Smad1 antibody.
Reverse transcription (RT).
Total RNA from HepG2 and Neuro-2A cells was extracted by using Trizol reagent (Gibco BRL). The mRNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase, and cDNA levels were analyzed with the ABI Prism 7900HT sequence detection system. Primers used to detect Smad7 cDNA are GGCTGTGTTGCTGTGAATCTTACG and TTGAGAAAATCCATCGGGTATCTG, and those used to detect hypoxanthine phosphoribosyltransferase cDNA are AAACAATGCAAACTTTGCTTTCC and GGTCCTTTTCACCAGCAAGCT.
Oligonucleotides.
Oligonucleotides used to subclone, by PCR, fragments of the Smad7 gene into the pGL3 vector and regions of GATA-5 in pCDNA3 are described in Table 1.
TABLE 1.
Oligonucleotides used for PCR in this study
| Subclone | Forward oligonucleotide(s) (source), sequence(s) | Reverse oligonucleotide(s) (source), sequence(s) |
|---|---|---|
| Promoter | ||
| S | P107, CCAGATCTGCGCCAGCAGAGCCAGC | GL primer 2 (Promega), CTTTATGTTTTTGGCGTCTTCC |
| T | P110, GGGGAGCTCGCTCGGTCCGCTCGC | GL primer 2 |
| U | P111, TCCGAGCTCCGCGCCGGAGAGAGCTGC | GL primer 2 |
| V | P105, AGAGATCTGAGACGCAGCCAGCCAG | GL primer 2 |
| W | P109, CCAGGAGCTCGGCGCCACGCCGCCGAGC | GL primer 2 |
| X | RV primer 3 (Promega), CTAGCAAAATAGGCTGTCCC | P206, CGAGATCTCGCACTCAGGGACTCCGG |
| Y | RV primer 3 | P207, GTGGCCTCGAGCTCGGCGGCGTGGCGCC |
| Z | RV primer 3 | P208, GCGTGCTCGAGGCTGGCTGGCTGGCTGC |
| AA | P110 | P206 |
| BRE-1 m1 | P520, TCGAGATCTGAGACGCTGCCTGCCTGCCTGCCGGCGCCACG | GL primer 2 |
| BRE-1 m2 | P518, TCGAGATCTGAGACGCAGCCAGCCAGCCAGCTAACGCCACGC | GL primer 2 |
| BRE-1 m3 | RV primer 3 | P608, CAGATCTCGAGTCCTTAGTAATACGCTGGGGGCTCG |
| Intron | ||
| Q | GAGCTCGGAGGGGGAGACTCTGAATT | GL primer 2 on clone P |
| I-BRE (R) | CTGTTTGTCTTAGCTGTGGG | GL primer 2 on clone P |
| I-BRE M1 | I502, GGTGATTGCCTTAGCGTTTAGCTGTC; RV primer 3 | GL primer 2; I602, GACAGCTAAACGCTAAGGCAATCACC |
| I-BRE M2 | I503, GCTGTCAGACGGGCAAAAGAGGCC; RV primer 3 | GL primer 2; I603, GGCCTCTTTTGCCCGTCTGACAGC |
| I-BRE M3 | I512, GCTGTCAAGTAAACAAAAGAGGCC; RV primer 3 | GL primer 2; I612, GGCCTCTTTTGTTTACTTGACAGC |
| I-BRE M4 | I511 TGTCAGATAACACAAAGAGGCCGCC; RV primer 3 | GL primer 2; I611, CGGCCTCTTTGTGTTATCTGACAG |
| GATA-5 | ||
| Flag-GATA-5 | G5-101, GCGGTACCATGGACTACAAGGACGACGATGACAAGGGATCCTACCAA AGCTTGGCGTTAGC | G5-201, AACTCCTCCAAGAAGTCAGG |
| 1-312 | G5-101 | G5-202, GCCCTCGAGTCATCCTGAGGAGCCCTTGATTTTG |
| 1-249 | G5-101 | G5-204, GCCCTCGAGTCAGCATAGGCCGGATCTTCGG |
| 1-195 | G5-101 | G5-205, GCCCTCGAGTCACTCCCGGCCCTCACCAGG |
| Δ49-91 | G5-120, ATGCTGCCGTACCTGCCTTCGACCTTCCCGTTCGCTCACAG; G5-101 | G5-202; G5-212, CGAAGGCAGGTACGGCAGCATG |
| Δ92-139 | T7; G5-106, GCCGGATCCACAACCTACCCAGCATACATG | G5-606, CATGTATGCTGGGTAGGTTGTGGTGGCTCCCGGTGGGTG; G5-202 |
| Δ140-195 | G5-505, TGCGTCAACTGTGGAGCC; T7 | G5-202; G5-605, GGCTCCACAGTTGACGCAGGGGTACGAGGCGCCCATG |
| Δ196-220 | G5-503, CCTGGTGAGGGCCGGGAGGGCCTCTATCACAAGATG; T7 | G5-202; G5-205 |
| Δ196-249 | G5-121, CCTGGTGAGGGCCGGGAGTGCTCCAACTGCCATACTGCCAC; G5-101 | G5-202; G5-205 |
| Δ196-274 | G5-504, CCTGGTGAGGGCCGGGAGGGCCTCTACATGAAGCTC; T7 | G5-202; G5-205 |
| C253G | G5-501, TGCTCCAACGGCCATACTGC; G5-101 | G5-202; G5-601, AGTATGGCCGTTGGAGCAG |
| GATA-5/GATA-2 zinc finger chimeras | G5-101; G5-107, GCCGGGAGTGTGTCAACTGTG; G5-108, GTCTGCAACGCCT GTGGCCTCTAC | G5-209, GGCCCCACAGTTGACACACTC; G5-208, GTAGAGGCCACAGGCGTTGC; G5-202 |
| GATA-5 1-91/GATA-2 170-480 | G5-101; G2-102, GCACACCCACCGGGAGCCACCCTTTTCGGCTTCCCACCCACG | G5-213, GGTGGCTCCCGGTGGGTGTGC; G2-202, GGGCAGCCGCCGGCACATAG |
| GATA-5 1-48/GATA-2 170-480 | G5-101; G2-103, ATGCTGCCGTACCTGCCTTCGCTTTTCGGCTTCCCACCCACG | G5-212; G2-202 |
siRNA.
Sequences for siRNA are AAGAUGAAUGGCGUCAACCGG for GATA-5, AAGCGCGUGCCUUCAUCACGG for GATA-6, AAUCAGGGGUUUUCUUCCCCU for GATA-1, and AAACAACAUAGGGUGACUAGA for the scrambled siRNA. RNAs were transfected by the calcium phosphate precipitation method at a concentration of 120 nM.
RESULTS
Identification of BMP-specific response elements.
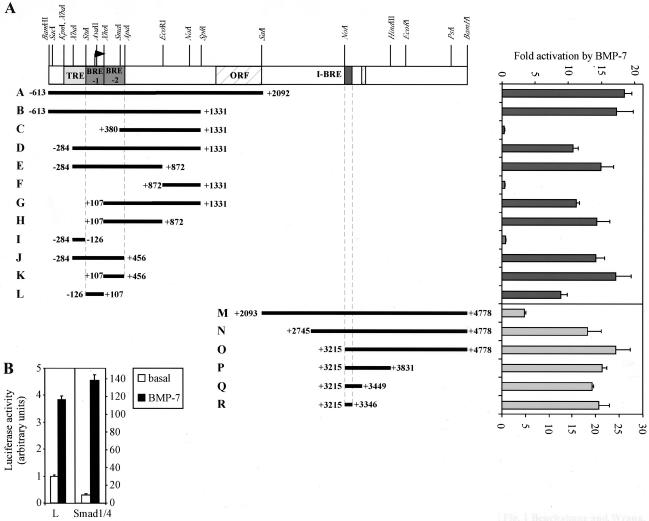
Smad7 is an antagonistic Smad shown to inhibit TGF-β/activin and BMP signaling and is upregulated by TGF-β, activin, and BMPs as part of an inhibitory feedback loop (3, 34, 58). To study the molecular mechanism of BMP-dependent regulation of this inhibitory feedback, we cloned the Smad7 promoter region by screening a Lambda Dash 129Sv female mouse kidney genomic DNA library with the +1204 to +1498 region of the Smad7 5′ untranslated region, obtained from a cDNA in the expressed sequence tag database. From this screening, we isolated a 5.4-kbp BamHI fragment, spanning base pairs −613 to +4778 of the Smad7 gene (according to reference 57). The BMP regulation of fragments of this region was then analyzed in a heterologous reporter assay. The initial results identified two fragments, A and M, which were responsive to BMP-7 in HepG2 cells (Fig. 1). To test whether increasing Smad concentration would alter the responsiveness of these fragments, we cotransfected Smad1 and Smad4 along with either fragment A or fragment M. Interestingly, increasing Smad1 and Smad4 expression resulted in a strong enhancement in BMP-dependent induction of fragment A, which corresponds to the promoter region, but had no effect on the BRE contained within fragment M (data not shown). Thus, to facilitate mapping of the BRE in the promoter, we tested numerous fragments for BMP-dependent induction in the presence of Smad1 and Smad4 expression. This analysis led to the identification of two BREs (BRE-1 and BRE-2) upstream of the Smad7 coding sequence. In the absence of increased Smad1 expression, BRE-1 was induced fourfold by BMP-7 while expression of exogenous Smads strongly enhanced the activity of the element (Fig. 1B). BRE-2 showed a similar response to Smad1 as did BRE-1. We also analyzed fragment M and identified a single BRE located within the first intron of the gene (I-BRE). Together, these studies show that Smad7 transcription is induced by BMPs through multiple cis-acting sequences.
FIG. 1.
Mapping of specific BRE regulating the Smad7 gene. (A) A schematic representation of a 5.4-kbp genomic fragment cloned from a lambda phage genomic DNA library is presented. The Smad7 coding region is hatched (ORF). The TGF-β responsive element (TRE) and specific BREs (BRE-1, BRE-2, and I-BRE) are indicated. Various restriction fragments derived from the 5.4-kbp genomic clone were subcloned into the pGL3 vector, upstream of the E1b promoter and luciferase reporter gene, and are shown as black bars with the corresponding clone letter. Diagrammed constructs were transiently transfected into HepG2 cells and incubated overnight in the absence or presence of 5 nM BMP-7. For constructs A to L, 0.03 μg of Smad1 and Smad4 cDNA/well were transfected into the cells. The relative luciferase activities of cell lysates were determined. The induction (n-fold) by BMP-7 is indicated. (B) The BRE-1 fragment was transiently transfected into HepG2 cells with or without Smad1 and Smad4 cDNA. Cells were treated with BMP-7, as indicated above, and the luciferase activities of cell lysates were determined.
Three elements in BRE-1 cooperate to mediate BMP responsiveness.
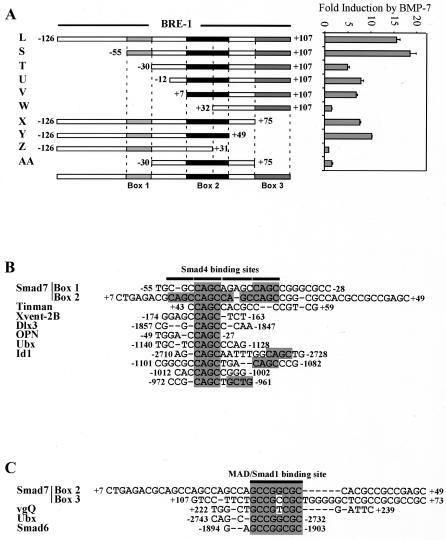
To investigate the mechanism of BMP-dependent induction of the Smad7 promoter, we first focused on BRE-1. BRE-2 was not further investigated, as it was a larger element than the first BRE and showed similar responses to Smad addition. To identify the minimal DNA sequence responsible for BMP inducibility, 5′ and 3′ deletions of BRE-1 were assessed for BMP-dependent regulation (Fig. 2A). Deletion of the −55 to −30 region from the 5′ end of BRE-1 resulted in a threefold reduction in reporter activity (Fig. 2A, construct T). Further 5′ deletion of the region between +7 and +32 completely eliminated the BMP response (Fig. 2A, construct W). Similarly, deletion from the 3′ end, spanning base pairs +75 to +107, reduced the response by approximately twofold, and additional deletions of the +31 to +49 region completely eliminated the response (Fig. 2A, constructs X and Z, respectively). Similar results were obtained by cloning these fragments into a TATA-less reporter (data not shown), indicating that these elements also function in the context of the native Smad7 promoter, which is contained within BRE-1 (57). Thus, three regions in BRE-1 are important for BMP-dependent induction, and we have termed these box 1 (−55 to −30), box 2 (+7 to +49), and box 3 (+75 to +107). While the presence of either boxes 1 and 2 or boxes 2 and 3 is sufficient for BMP responsiveness, all three boxes cooperate for maximal activation by BMP-7. Furthermore, none of the three boxes alone can be activated by BMP-7.
FIG. 2.
Mapping of a minimal BRE in Smad7 BRE-1 and identification of putative Smad binding sites. (A) Various fragments derived from the 233-bp BRE-1 were amplified by PCR and subcloned into the pGL3 vector, upstream of the E1b promoter and luciferase reporter gene. Diagrammed constructs were transiently transfected into HepG2 cells, with 0.03 μg of Smad1 and Smad4 cDNA/well, incubated overnight in the absence or presence of 5 nM BMP-7. The relative luciferase activities of cell lysates were determined. The induction (n-fold) by BMP-7 is presented. Three important regions of BRE-1 were delineated and termed box 1, box 2, and box 3. (B) Box 1 and box 2 of BRE-1 are aligned with fragments of the Dpp response elements of the tinman and Ubx genes, and BREs of the Xvent-2B, Dlx3, OPN, and Id1 promoters. Putative Smad4 binding sites (CAGC boxes) are highlighted. (C) Box 2 and box 3 of Smad7 BRE-1 are aligned to a region of the vestigial quadrant enhancer (vgQ) known to bind Mad and regions of the Ubx (72) and Smad6 promoters containing Smad1 binding sites. The MAD/Smad1 binding site sequence is highlighted.
BREs have been identified in many genes, including the Xvent-2B, Smad6, Id1, Dlx3, OPN, and Tlx2 genes of vertebrates and the tinman, Ubx, and vestigial genes of Drosophila melanogaster (27, 31, 33, 41, 43, 50, 64, 67, 72, 75, 84). To determine whether boxes 1, 2, and 3 display any similarity to these elements, we aligned them to each other and to that of these known BREs. Boxes 1 and 2 are highly homologous and contain multiple repeats of a 5′-CAGC sequence that is similar to that of the Smad4 binding site, 5′-CAGA, identified by Dennler et al. (16) (Fig. 2B). Interestingly, this repeat is present in a decapentaplegic (Dpp) response element that was mapped to the 3′ flanking region, tin-D, of the Drosophila tinman gene as well as in a Dpp response element in the Ubx promoter (72, 84). We also found a similar sequence in the BREs of Xvent-2B, Dlx3, OPN, and Id1 (27, 31, 43, 64).
BMP-regulated Smad1 binds DNA directly at the consensus-binding site 5′-GCCGNCGC (41, 44). Analysis of box 2 and box 3 showed that both contain these Smad1 binding elements (Fig. 2C), which are identical to those found in the vestigial quadrant enhancer and in the Ubx and Smad6 promoters (33, 41, 72). These results indicate that a BRE-1-like response element may be present in multiple BMP target genes. We confirmed that Smad1 and Smad4 can bind to boxes 2 and 3 by electrophoretic mobility shift assays (EMSA), by using bacterially produced glutathione S-transferase fusion proteins and small probes covering parts of the boxes. We found that Smad1 bound to the Smad1 elements of boxes 2 and 3 but did not bind to the CAGC repeats in box 2, whereas Smad4 bound to the CAGC repeats and the Smad1 sites of boxes 2 and 3 (data not shown). These results indicate that BRE-1 contains multiple copies of both Smad1 and Smad4 binding elements.
Endogenous Smad1 binds BRE-1 in ChIP assays.
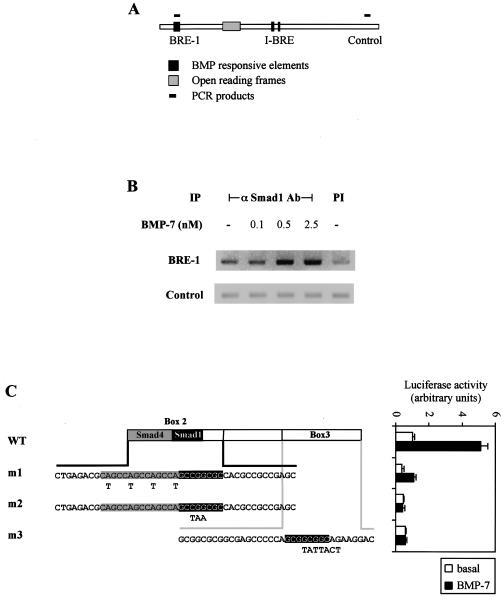
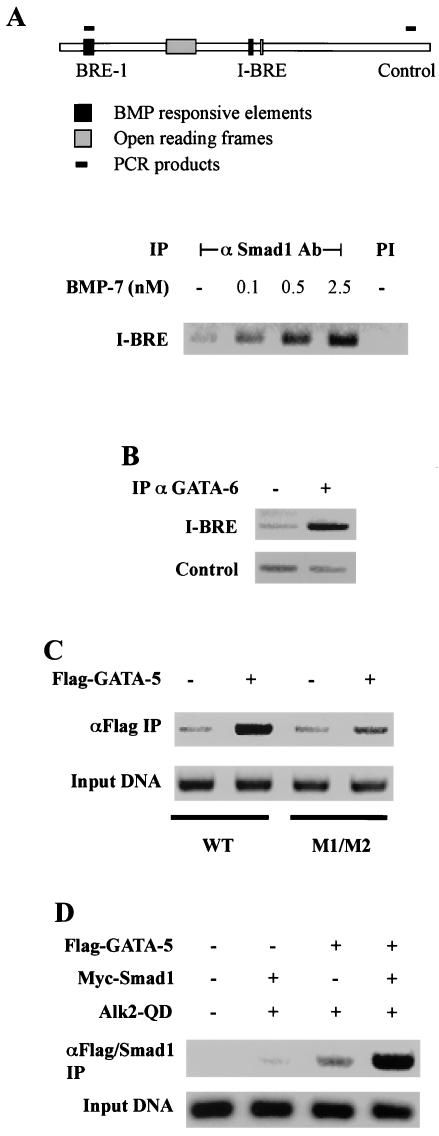
Having identified potential Smad1 binding sites in BRE-1, we wanted to verify that Smads bound to BRE-1 in a physiologically relevant setting. We thus performed a ChIP assay, which has emerged as a key tool for defining interactions of factors with specific chromosomal sites in living cells. ChIP thereby provides a snapshot of native chromatin structure and the endogenous factors bound to regulatory elements in different functional states (37). For this, ChIP assays were carried out in HepG2 cells that were treated for 1 h with increasing amounts of BMP-7. Endogenous protein-DNA complexes were then covalently cross-linked with formaldehyde, and after cell lysis, the chromatin was sheared by sonication. Endogenous Smad1-DNA complexes were then immunoprecipitated with an anti-Smad1 antibody, and the DNA purified from this coprecipitation was analyzed by PCR with two pairs of PCR primers. One pair amplified BRE-1 while the second amplified a control region of the Smad7 gene (Fig. 3A). The control PCR served to verify that DNA shearing was sufficient to ensure that I-BRE and BRE-1 were on separate DNA fragments. In untreated cell lysates, we observed close to background levels of BRE-1 amplified in Smad1 immunoprecipitates. However, in cells stimulated with 0.5 and 2.5 nM BMP-7, we observed amplification of BRE-1 DNA (Fig. 3B). In contrast, PCR analysis of the control region revealed no signals over background levels. These results demonstrate that BMP-7 induces binding in vivo of endogenous Smad1 to the native BRE-1 in the Smad7 gene.
FIG. 3.
Smad1 binds to BRE-1 in living cells. (A) Schematic representation of PCR products amplified following ChIP assays. (B) ChIP of Smad1 bound to BRE-1. HepG2 cells were treated for 1 h with increasing concentrations of BMP-7. Protein-DNA complexes were cross-linked by formaldehyde treatment, cells were lysed, and DNA was sheared by sonication. Cell lysates were subjected to immunoprecipitation with an anti-Smad1 antibody. DNA recovered from the immunoprecipitation was amplified by PCR. PI, preimmune; −, absent. (C) Diagrammed clones derived from BRE-1 were transiently transfected in HepG2 cells with 0.03 μg of Smad1 and Smad4 cDNA/well. Cells were incubated overnight in the absence (white bars) or presence (black bars) of 5 nM BMP-7. The relative luciferase activities of cell lysates were determined. Mutations created in box 2 or box 3 are indicated in bold letters under the appropriate sequences. The putative Smad4 and Smad1 binding sites are highlighted in gray and black, respectively. WT, wild type.
Since BRE-1 is a bona fide BMP-dependent response element, we next tested the requirement for the Smad binding sites by creating point mutations in boxes 2 and 3 of the BRE. Mutations were created in a reporter construct containing only boxes 2 and 3 (corresponding to fragment V in Fig. 2), since the presence of box 1 partially compensated for either mutations in box 2 (data not shown) or loss of box 3 (Fig. 2A). Mutation of the Smad1 binding sites in box 2 or 3 completely abolished BMP responsiveness while mutations in the CAGC repeat strongly reduced the activity of the BRE (Fig. 3C). We confirmed by EMSA with bacterially expressed Smad proteins that mutation of the CAGC repeat in box 2 blocked the binding of Smad4 while mutation of the Smad1 site in box 2 blocked Smad1 binding (data not shown). Together, these results show that BMP induces Smad1 binding to the BRE-1 in vivo and that efficient induction of BRE-1 requires adjacent Smad1 and Smad4 binding sites located in two or more boxes. Of note, substitution of the sequences between the Smad-binding boxes with unrelated DNA did not affect the activity of BRE-1 (data not shown), suggesting that Smad1/Smad4 binding to BRE-1 may be sufficient to activate the promoter in the absence of a DNA binding partner.
The I-BRE is a GATA- and Smad-dependent element.
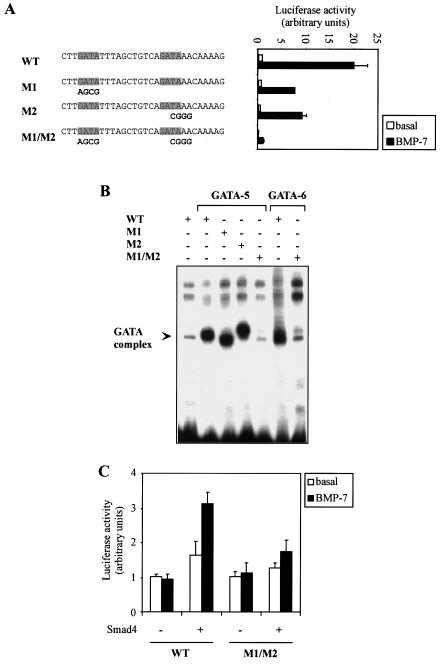
Next, we evaluated the BRE contained in intron 1. Unlike BRE-1, I-BRE contains no 5′-GCCGNCGC Smad1 binding motifs but contained three GC-rich regions that bound Smad4 in vitro (data not shown). This suggested to us that BMP-dependent induction of I-BRE might require cooperation with a DNA binding cofactor. Sequence analysis of I-BRE revealed the presence of two consensus GATA factor binding sites separated by 11 bp. To investigate whether these potential binding sites are important for regulation of I-BRE, we created point mutations in the GATA sites and evaluated I-BRE activity. Mutation of either GATA binding site alone reduced I-BRE's activity by half, while mutation of both binding sites reduced it by more than 10-fold (Fig. 4A). To verify that GATA proteins bind to these GATA sites, EMSAs with labeled I-BRE were performed on nuclear extracts of 293T cells transfected with GATA-5 or GATA-6. In control transfectants, there was little binding to I-BRE. However, in cells expressing GATA-5 or -6, we observed a strong band shift (Fig. 4B). Mutation of the two GATA sites in I-BRE eliminated complex formation, thus confirming that GATA-5 and -6 bind to the GATA sites of I-BRE.
FIG. 4.
I-BRE is a GATA- and Smad-dependent element. (A) Diagrammed clones derived from I-BRE were transiently transfected in HepG2 cells. Cells were incubated overnight in the absence (white bars) or presence (black bars) of 5 nM BMP-7. The relative luciferase activities of cell lysates were determined. Mutations created in I-BRE are indicated in bold letters under the appropriate sequences. The GATA binding sites are indicated. (B) I-BRE was 32P-labeled and incubated with lysates of 293T cells transfected with GATA-5 and -6 cDNA. Mutations in the I-BRE probe (M1, M2, and M1/M2) correspond to mutations in panel A. +, present; −, absent. (C) Wild-type (WT) and M1/M2 mutant I-BRE (as shown in panel A) were transfected in MEF Smad4−/− cells in the presence or absence of exogenous Smad4 cDNA. Cells were incubated overnight in the absence (white bars) or presence (black bars) of BMP-7. The relative luciferase activities of cell lysates were determined.
We next evaluated the requirement for Smad4. For this, the activity of the I-BRE was assessed in a MEF cell line deficient for Smad4 (70), a key partner required for Smad-dependent transcriptional activation. We did not examine BMP-specific R-Smads, since there is considerable redundancy in the pathway (reviewed in reference 35). The I-BRE was unresponsive in Smad4−/− MEFs, but BMP-dependent induction was restored by ectopic Smad4 expression (Fig. 4C). Interestingly, when both GATA sites were mutated, the element was unresponsive in either the presence or the absence of Smad4. Expression of GATA-6 in these cells was confirmed by RT-PCR (data not shown). These results demonstrate that BMP regulation of the I-BRE is dependent on both the Smad pathway and GATA factors.
The I-BRE binds GATA and Smad proteins in vivo.
Our analysis of the I-BRE suggested that the element is a Smad and GATA target. To confirm this, we performed ChIP assays to evaluate the binding of endogenous Smad1 and GATA factors to the I-BRE in HepG2 cells. As for the BRE-1 assay, two pairs of primers were utilized. One pair amplified the I-BRE while the second amplified a control region of the Smad7 gene (Fig. 5A). No I-BRE was amplified from samples subjected to a preimmune immunoprecipitation. However, in BMP-7-treated cells, we observed dose-dependent binding of endogenous Smad1 to the I-BRE. We next tested whether endogenous GATA-6 could bind the I-BRE in living cells by using ChIP assays. Consistent with our EMSA analysis, we observed endogenous GATA-6 bound to the I-BRE in HepG2 cells (Fig. 5B), and this binding was unaffected by BMP treatment (data not shown). Thus, GATA factors constitutively occupy the element, while Smad1 binding to the I-BRE is induced by BMP signaling. We did not investigate binding of endogenous Smad4 by ChIP. However, Smad4 is a well described partner of all R-Smads, and we showed that Smad4 is required to activate the I-BRE and binds the element in vitro. This strongly suggests that Smad4 is a component of this DNA binding complex.
FIG. 5.
I-BRE binds Smad and GATA proteins in living cells. (A) ChIP of Smad1 bound to I-BRE. HepG2 cells were treated for 1 h with increasing concentrations of BMP-7. Protein-DNA complexes were cross-linked by formaldehyde treatment, cells were lysed, and DNA was sheared by sonication. Cell lysates were subjected to immunoprecipitation (IP) with an anti-Smad1 antibody. DNA recovered from the immunoprecipitation was amplified by PCR. PCR products amplified are diagrammed. PI, preimmune. PCR of the control region is presented in Fig. 3B. (B) ChIP of GATA-6 bound to I-BRE. HepG2 cells were treated as described for panel A, and cell lysates were subjected to immunoprecipitation with the goat C-20 anti-GATA-6 (αGATA-6) antibody (Santa Cruz) or with an irrelevant goat antibody (−) (C) 293T cells were transfected with cDNA for Flag-GATA-5 and the wild-type or mutated (M1/M2 as in Fig. 4A) I-BRE reporter construct. Protein-DNA complexes were cross-linked by formaldehyde treatment, the DNA was sheared by sonication, and cell lysates were subjected to immunoprecipitation with an anti-Flag (αFlag) antibody. The DNA recovered from the immunoprecipitation was amplified by PCR with primers specific to I-BRE. (D) 293T cells were transfected with the I-BRE reporter construct and cDNA for Flag-GATA-5, Myc-Smad1, and Alk2-QD. Cells were treated as described for panel C. Lysates were subjected to immunoprecipitation with an anti-Flag antibody followed by elution with Flag peptide and immunoprecipitation of the eluate with an anti-Smad1 antibody. I-BRE recovered from the double immunoprecipitation was amplified by PCR. +, present; −, absent.
To confirm that the observed association between GATA and I-BRE is dependent on the GATA binding sites, we performed DNA immunoprecipitation experiments using exogenous I-BRE constructs in which the GATA sites were mutated. This was done by transfecting 293T cells with wild-type or mutated I-BRE containing reporter constructs and cDNA encoding Flag-GATA-5. Protein-DNA complexes were cross-linked by formaldehyde treatment of cells and cell lysates subjected to immunoprecipitation with an anti-Flag antibody. Coprecipitation of I-BRE bound to GATA-5 was then assessed by PCR. Amplification of anti-Flag immunoprecipitates clearly showed that I-BRE was coprecipitated in GATA-5-expressing cells (Fig. 5C), whereas there was little interaction with the mutated I-BRE, confirming that GATA-5 interacts with the I-BRE via the GATA sites.
Smad1 and GATA proteins simultaneously occupy the I-BRE.
To verify that they can cooccupy the same DNA element in living cells, we next performed sequential DNA immunoprecipitation of Flag-GATA-5 followed by Smad1. For this, 293T cells were transfected with the I-BRE-containing reporter construct as well as cDNA encoding Flag-GATA-5, Myc-Smad1, and activated BMP-7 type I receptor (Alk2-QD). Protein-DNA complexes were cross-linked by formaldehyde treatment of cells, and cell lysates were subjected to immunoprecipitation with an anti-Flag antibody. Protein-DNA complexes were then eluted from the immunoprecipitate, under nondenaturing conditions with Flag-peptide, and the eluates were subjected to immunoprecipitation with an anti-Smad1 antibody. Levels of immunoprecipitated I-BRE were then assessed by PCR. In the absence of GATA-5 expression, only trace levels of I-BRE were detected in the PCR. However, when Smad1 and GATA-5 were coexpressed in the presence of BMP signaling, we detected a strong I-BRE signal (Fig. 5D). Interestingly, we also detected I-BRE coprecipitating in the absence of ectopic Smad1 expression. This is likely due to binding of endogenous Smad1 to the I-BRE. These results thus demonstrate that GATA factors and Smad1 assemble on the I-BRE simultaneously in response to BMP signaling and may therefore cooperate to activate Smad7 transcription.
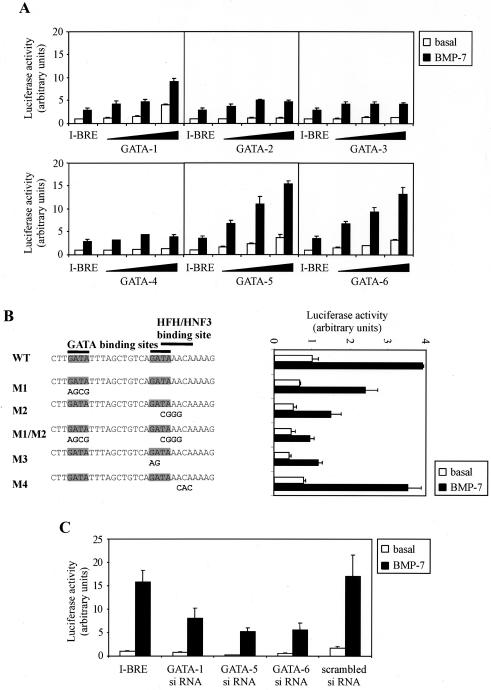
BMP-dependent induction of the I-BRE is mediated by GATA-1, -5, and -6.
Since GATA proteins bind to the I-BRE, we wanted to identify whether distinct GATA factors differentially regulate Smad7 transcription. To facilitate this, we looked for a cell line in which the I-BRE was minimally responsive due to a lack of GATA expression. GATA family members include GATA-1, -2, and -3, which are key regulators of hematopoietic cell lineages. GATA-2 and -3 are also important in the neural system, whereas GATA-4, -5, and -6 are important in mesoderm- and endoderm-derived tissues (7, 54, 59, 86). Consistent with reports on GATA expression patterns, RT-PCR analysis confirmed that GATA-1, -5, and -6 are not expressed in Neuro-2A cells but are expressed in HepG2 cells, which are endoderm derived (data not shown). Therefore, we analyzed I-BRE responsiveness in the mouse neuroblastoma cell line, Neuro-2A, and found I-BRE was only weakly induced by BMP-7 (Fig. 6A) but that this activity was enhanced by exogenous GATA-1, -5, and -6, whereas GATA-2, -3, and -4 had minimal effect. Furthermore, mutation of the GATA sites blocked activation observed upon exogenous expression of GATA-5 (Fig. 6B). We also noted that the second GATA site overlaps with a binding site for a protein of the hepatocyte nuclear factor 3/forkhead family (62, 63, 65). However, mutations in the second GATA site that also affected the forkhead site have the same effect as mutations that leave the forkhead site intact, and mutations in the forkhead site alone did not affect signaling. This suggests that binding of a forkhead DNA binding protein to the I-BRE is not critical for BMP-dependent regulation in Neuro-2A cells.
FIG. 6.
Exogenous expression of GATA-1, -5, and -6 rescue the I-BRE in a GATA-deficient cell line. (A) The I-BRE luciferase reporter construct was transiently transfected in Neuro-2A cells with increasing concentrations of GATA-1, -2, -3, -4, -5, or -6 cDNA. Cells were incubated overnight in the absence (white bars) or presence (black bars) of 5 nM BMP-7. The relative luciferase activities of cell lysates were determined. (B) I-BRE constructs, with wild-type (WT) sequence or GATA and HFH/HNF3 site mutations, were transiently transfected in Neuro-2A cells with GATA-5 cDNA. Cells were incubated overnight in the absence (white bars) or presence (black bars) of 5 nM BMP-7. The relative luciferase activities of cell lysates were determined. Mutations created in I-BRE are indicated in bold letters under the appropriate sequences, and the GATA binding sites are highlighted. (C) HepG2 cells were transiently transfected with the I-BRE reporter construct and siRNA against GATA-1, -5, and -6 as well as a scrambled siRNA. Cells were incubated overnight in the absence (white bars) or presence (black bars) of 5 nM BMP-7. The relative luciferase activities of cell lysates were determined at 72 h posttransfection.
To confirm that GATA-1, -5, and -6 expression in HepG2 cells mediates the strong BMP-dependent activation of the I-BRE, we disrupted GATA expression by RNA interference. We first assessed whether 21-bp siRNA fragments specific to GATA-5 or GATA-6 sequences disrupted the expression of their target proteins. 293T cells were used to evaluate this, as they contain endogenous levels of both GATA-5 and GATA-6 and are transfected at high efficiencies with the siRNA. GATA-5 and GATA-6 siRNA disrupted endogenous GATA-5 and GATA-6 expression, respectively (data not shown). We next tested the effect of the siRNAs on I-BRE activity in HepG2 cells and observed that transfection of siRNAs against GATA-5 or GATA-6 reduced the activity of the I-BRE reporter construct, whereas a scrambled siRNA had no effect (Fig. 6C). Since HepG2 cells also express GATA-1, we also targeted GATA-1 with siRNA and observed a decrease in BMP responsiveness. The GATA siRNAs reduced the basal level of I-BRE activity, which may reflect autocrine BMP signaling or the ability of GATA factors to activate the I-BRE to a certain extent on their own. However, maximal activation of the element clearly requires input from both the BMP pathway and GATA factors. We also attempted to knock down all three GATA factors but observed that simultaneous transfection of all three siRNAs blocked the inhibitory effect observed with individual siRNAs on both GATA expression and I-BRE activity (data not shown). Altogether, these results indicate that BMP-dependent induction of the I-BRE is dependent on Smads and GATA-1, -5, and -6.
Identification of domains in GATA-5 required for I-BRE activation.
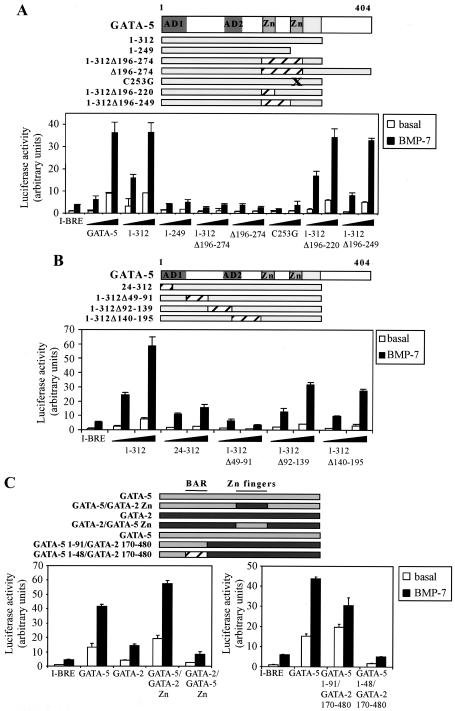
We next addressed which regions of the GATA proteins are critical for GATA to cooperate with Smad signaling to activate the I-BRE. We first evaluated the ability of GATA-5 proteins mutated in the C-terminal half to activate the I-BRE in Neuro-2A cells (Fig. 7A). Deletion of amino acids 313 to 404 of the protein did not affect transcription. As expected, all mutants that lacked the DNA binding C-terminal zinc finger were unable to activate transcription, while deletion of the N-terminal zinc finger and adjacent basic region had no effect. We next evaluated the requirement for additional determinants in the N-terminal half of the protein by analyzing a series of deletion mutants of GATA-5 (Fig. 7B). Studies with GATA-4 previously identified two activation domains (AD1 and AD2) important for activity on the cTnC promoter, which are conserved in GATA-5 and -6 (56). Consistent with these previous studies, deletions within AD1 significantly reduced GATA-dependent activation of the I-BRE (Fig. 7B). However, internal deletions that leave AD1 intact revealed removal of amino acids 49 to 91, just downstream of AD1, eliminated BMP-dependent activation. We refer to this 42-amino-acid stretch as the BMP activation region (BAR). In contrast, deletions of amino acids 92 to 139 and 140 to 195, which interrupt the second activation domain, only slightly reduced activation of the I-BRE by GATA-5. These results show that the DNA binding zinc finger of GATA-5 together with a 42-amino-acid region in the N terminus of GATA-5 mediates a critical GATA-dependent activation of the I-BRE in response to BMP.
FIG. 7.
Mapping of regions in GATA-5 required for I-BRE activation. (A and B) The I-BRE was transiently transfected in Neuro-2A cells with deletion mutants of the GATA-5 cDNA. Cells were incubated overnight in the absence (white bars) or presence (black bars) of BMP-7. The relative luciferase activities of cell lysates were determined. The zinc fingers (Zn) and activation domains (AD) of GATA-5 are indicated. (C) The I-BRE was transiently transfected in Neuro-2A cells with GATA-5, GATA-2, and chimeras of the two proteins. Cells were incubated overnight in the absence (white bars) or presence (black bars) of BMP-7. The relative luciferase activities of cell lysates were determined. The zinc fingers (Zn) and BAR of GATA-5 are indicated.
To assess whether the N-terminal region of GATA-5 is sufficient to mediate BMP-dependent regulation of the I-BRE, we created chimeras between GATA-2 and GATA-5 proteins (Fig. 7C). Both proteins can bind the I-BRE in EMSAs (Fig. 4B and data not shown), and GATA-5, in which the zinc finger region was replaced with that from GATA-2, still mediated strong activation of I-BRE, whereas the GATA-2 chimera that contained the GATA-5 zinc fingers but is missing the BAR, was unable to activate the element. In contrast, swapping the amino-terminal 92 amino acids of GATA-2 with the AD1 and BAR from GATA-5 restored activation of I-BRE, and deleting the BAR from this construct abolished activation. Of note, deletion of a region corresponding to the BAR in GATA-6 also reduced activation of the I-BRE (data not shown). Altogether, these data indicate that the BAR mediates GATA-5- and -6-specific activation of the Smad7 I-BRE.
Many DNA binding proteins that function in mediating Smad transcriptional responses interact directly with Smads. Furthermore, GATA-3 has been reported to interact with Smad3 and mediate TGF-β-dependent induction of interleukin-10 (8). Therefore, we also evaluated whether GATA and Smad1 and 4 physically interact. Although association of GATA-5 and the Smad1 MH2 domain was detected in vitro (data not shown), we were unable to observe interactions in mammalian cells. Moreover, Smad interaction did not require the BAR, and mutations in GATA-5 that abolished the in vitro interaction with Smad1 still mediated strong activation of the I-BRE. From these data, we conclude that physical interaction between GATA-5 and Smad1 is not required for BMP-dependent regulation of the I-BRE. Nevertheless, Smads, GATA factors, and the GATA sites cooperate to mediate BMP-dependent activation of the I-BRE. Therefore, although physical interaction is not required, these components clearly synergize functionally at this element.
I-BRE is active at lower BMP concentrations than BRE-1.
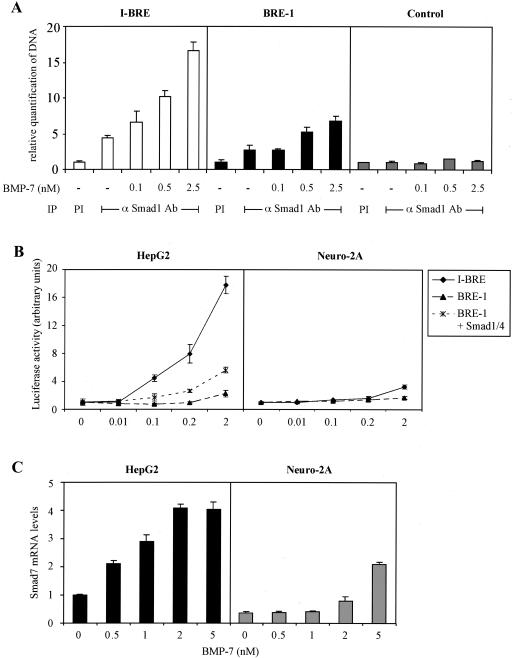
TGF-β family members are known to act as morphogens, emanating from a localized source and diffusing across tissues to produce a concentration gradient. This induces different transcriptional responses in target cells depending on the concentration of the ligand. Having demonstrated that BRE-1 and I-BRE are BMP-dependent enhancers, we wanted to assess their relative contribution to transcription of the Smad7 gene in response to different BMP concentrations. For this, we performed ChIP assays in HepG2 cells that express endogenous GATA factors and quantified Smad1 occupancy of the BRE-1 and I-BRE upon increasing levels of BMP stimulation by using real-time PCR. Quantitative ChIP revealed some binding of Smad1 to the I-BRE in the absence of BMP stimulation. However, even at the lowest doses of BMP, Smad1 occupancy increased 1.5-fold, and this was further enhanced to 3.6-fold at the highest doses of BMP tested (Fig. 8A). In contrast, the BRE-1 showed little or no occupancy in the absence of BMP stimulation and only displayed Smad1 binding at the highest BMP doses tested.
FIG. 8.
BRE-1 and I-BRE respond to different concentrations of BMP-7. (A) ChIP of Smad1 bound to I-BRE and BRE-1. HepG2 cells were treated for 1 h with increasing concentrations of BMP-7. Protein-DNA complexes were cross-linked by formaldehyde treatment, cells were lysed, and DNA was sheared by sonication. Cell lysates were subjected to immunoprecipitation (IP) with an anti-Smad1 antibody (αSmad1 Ab). DNA recovered from the immunoprecipitation was quantified by real-time PCR. Diagrammed representations of PCR products are presented in Fig. 3A and 4A. PI, preimmune; −, absent. (B) The BRE-1 and I-BRE luciferase reporter constructs were transiently transfected in HepG2 and Neuro-2A cells in the presence or absence of 0.03 μg of Smad1 and Smad4 cDNA/well. Cells were incubated overnight with increasing concentrations of BMP-7. The relative luciferase activities of cell lysates were determined. (C) Smad7 expression is induced by different concentrations of BMP-7 in HepG2 and Neuro-2A cells. RNA was extracted from HepG2 (black bars) and Neuro-2A (gray bars) cells treated for 1 h with increasing concentrations of BMP-7. The RNA was reverse transcribed, and cDNA for Smad7 was quantified by real-time PCR. Levels of hypoxanthine phosphoribosyltransferase cDNA were used to normalize the results.
The ChIP analysis showed that Smad1 bound more robustly to I-BRE than to BRE-1 following BMP treatment, implying that I-BRE is more active than BRE-1 and is induced at lower BMP concentrations. To confirm this, we transfected the I-BRE and BRE-1 reporter constructs in HepG2 cells that were then treated overnight with increasing amounts of BMP-7. Luciferase assays performed with lysates of these cells revealed that I-BRE displayed robust activation at a 0.1 nM concentration of BMP-7, whereas BRE-1 required 2 nM BMP treatment and was only weakly activated compared to I-BRE (Fig. 8B, left panel). As previously shown, BRE-1 activity is enhanced by exogenous Smad1/4 expression. Since increased Smad protein levels may facilitate Smad binding to BRE-1, we next tested whether ectopic expression of Smad1 and Smad4 would reduce BMP levels required for BRE-1 activation. Accordingly, increased Smad1/4 expression induced more robust activation of BRE-1 and at lower BMP concentrations. Next, we tested the signaling activity of BRE-1 and I-BRE in Neuro-2A, which lack endogenous GATA factors (Fig. 8B, right panel). In these cells, the I-BRE displayed a low level of activity that was similar to BRE-1. The lower induction of the I-BRE in Neuro-2A cells was not due to a general reduction in BMP responsiveness, as BMP-dependent phosphorylation of Smad1 was more robust than in HepG2 cells, and the tlx-2 promoter, another BMP-responsive element (75) that does not employ GATA factors, was strongly induced by BMP-7 (data not shown).
To confirm that these differences translate into different levels of responsiveness of the endogenous Smad7 gene, we analyzed Smad7 mRNA levels in Neuro-2A and HepG2 cells following 1 h of stimulation with increasing BMP concentrations. At a 0.5 nM concentration of BMP-7, we noted an induction of Smad7 in HepG2, which peaked at a 2 nM concentration of BMP-7 (Fig. 8C, left panel). In contrast, the Smad7 gene was relatively insensitive to BMP-7 induction in Neuro-2A cells and was only induced significantly by the highest dose of ligand tested (Fig. 8C, right panel). These results, together with our analysis of individual elements by reporter assays and ChIP suggest that in cells in which GATA-5 or -6 is expressed, Smad7 expression is robustly stimulated by BMPs acting through Smad1/Smad4/GATA-dependent regulation of the I-BRE element, whereas in cells deficient in GATA factors, Smad7 expression is regulated by the weaker Smad1/4-dependent BRE-1 element.
DISCUSSION
The TGF-β/activin and BMP signaling pathways are negatively regulated by antagonistic Smads (26, 32, 58). The inhibitory Smads, which include Smad6 and Smad7, form a stable interaction with type I receptors, thus preventing the phosphorylation of receptor-regulated Smads. I-Smads also mediate the interaction between receptors and E3-ubiquitin ligases, Smurf1 and Smurf2, leading to the degradation of receptors (19, 23, 38). The mRNA levels of Smad6 and Smad7 have been shown to be upregulated by TGF-β, activin, and BMP (3, 34, 58, 74), suggesting that the I-Smads function in an inhibitory feedback loop.
The general model of Smad transcriptional regulation suggests that Smads cooperate with specific transcription factors to enhance their affinity for DNA and to increase their specificity (reviewed in references 4, 35, 52, and 53). This model, however, is based primarily on TGF-β signaling studies, where genes are regulated via Smad2/4 or Smad3/4 complexes cooperating with DNA binding partners. Although all endogenous Smad2 and Smad3 elements characterized to date likely involve DNA binding partners, model elements containing concatemerized Smad3/4 binding sites do respond to TGF-β (36). No native promoter has yet been found that possesses these types of TGF-β responsive elements. However, in BRE-1 of the Smad7 promoter, we identified multiple Smad1 and Smad4 binding sites in close proximity, and mutagenesis of individual sites revealed that they all cooperate in regulation of the element. Mutation of any other sequence in BRE-1 had little effect on induction, suggesting the possibility that the BRE is a naturally occurring cluster of Smad binding sites that is regulated directly by Smad1/4 complexes. Consistent with this, the BRE was highly sensitive to the Smad concentration in the cell and required high doses of BMP stimulation for maximal activation. The ability of Smad1 and Smad4 to bind to multiple independent sequences in this element may thus confer sufficient affinity for the Smad complex to occupy the element independent of a DNA partner.
I-BRE is a Smad1/Smad4/GATA-dependent enhancer.
Regulation of the Smad7 I-BRE, on the other hand, appears to follow the more classical model of Smad transcriptional regulation. Activation of the element requires Smad4, and it contains GC-rich Smad4-binding sequences but no Smad1 binding sites. However, the element contains two binding sites for transcription factors of the GATA family, and when the GATA sites were mutated, BMP activation was reduced. Furthermore, in GATA-deficient cells, the I-BRE was minimally responsive. Therefore, GATA factors cooperate with Smads of the BMP signaling pathway to regulate BMP-dependent induction of the I-BRE.
The GATA family of transcription factors is comprised of six members in vertebrates that can be subdivided into two groups. GATA-1, -2, and -3 are predominantly expressed in hematopoietic and neuronal cell lineages while GATA-4, -5, and -6 are involved in visceral and parietal endoderm formation and are expressed in overlapping patterns in mesoderm- and endoderm-derived tissues such as heart, liver, lung, gut, bladder, testes, and ovary (reviewed in reference 54). We tested the effect of all GATA factors on I-BRE by using Neuro-2A cells, which do not express GATA-1, -5, and -6, and found a strong activation by these three GATA factors but not by GATA-2, -3, and -4. Furthermore, we demonstrated, by ChIP, that GATA-6 and Smad1 can bind the native element and, by DNA immunoprecipitation, that GATA and Smad1 coassemble on I-BRE in living cells. This suggests that Smad1/4 regulates the I-BRE in cooperation with GATA-1, -5, or -6 protein. This is reminiscent of how BMP signaling cooperates with the zinc finger protein OAZ to induce the Xvent-2B promoter (25) and how Smad2 and Smad3 signaling control a number of TGF-β-regulated elements (4, 17, 52). However, in marked contrast to these previous studies, GATA-dependent activation of the I-BRE was dependent on the BAR region near the amino terminus of GATA-5 and GATA-6 that does not appear to be required for physical interaction with Smads. It is currently unclear how the BAR mediates GATA specificity in regulation of the I-BRE. This region may bind another required factor or may be subject to posttranslational modification. For instance, GATA factors interact with numerous proteins including Sp1, TTF1, dHAND, Runx1, HNF-1, SRF, and MEF2 (6, 14, 20, 40, 48, 55, 79) and are phosphorylated at several sites in their N termini (11, 13, 39, 42, 46, 78). We are currently investigating how the BAR functions to specifically mediate I-BRE induction in response to BMP.
Recent studies have shown that GATA-3 interacts, through its N-terminal region, with the MH1 domain of Smad3, activating interleukin promoters in response to TGF-β (8). We found that the I-BRE was not responsive to TGF-β, even in cells expressing GATA-3, further supporting the notion that interaction between GATA and Smads is not sufficient to mediate I-BRE activation but that additional determinants in the N terminus of the protein are required. Nevertheless, these results argue that GATA factors may function generally as Smad partners to regulate distinct GATA regulatory elements in a variety of target genes. Consistent with this, a Smad responsive element recently identified in the Nkx2-5 promoter contains paired GATA binding sites similar to that found in I-BRE (47). Although the role of GATA factors in regulating the Nkx2-5 element has not been explored, the findings indicate that this might represent another GATA-Smad target element in the BMP pathway. Interestingly, Marty et al. (51) found that mutation of a GATA binding site reduced the activity of the Dpp responsive labial enhancer, lab550, suggesting that cooperation between GATA- and BMP-regulated Smads is evolutionarily conserved.
BRE-1 and I-BRE interpret BMP gradients.
Ligands of the TGF-β family, including activin, Dpp, and the zebra fish Nodal-related Squint, have been shown to act as morphogens (12, 61), and cells can sense as low as a threefold change in ligand concentration (18). Dyson and Gurdon (18) have shown that increasing the occupancy of activin receptors at the cell surface causes cells to switch gene expression from Xbra to Xgsc. A similar change in Smad2 concentration also leads to changes in gene transcription (69), and in the Drosophila wing, Dpp induces Spalt expression near its source and Omb expression further from it (60) while in zebra fish, different concentrations of Nodal-related factors, Cyclops and Squint, induce different downstream genes (21, 77). How morphogen gradients are interpreted at promoters has been unclear.
Using ChIP assays, we quantified binding of Smad1 to endogenous I-BRE and BRE-1 in HepG2 cells, a cell line expressing GATA factors. Although Smad1 bound to both BREs, binding was weaker on BRE-1 and required much higher BMP concentrations than I-BRE, which was occupied at higher levels and was induced by lower BMP concentrations. These data suggest a model in which Smad7 expression is induced along a BMP concentration gradient through different enhancer elements. Thus, the high affinity I-BRE functions through the cooperative activity of a Smad1/Smad4/GATA complex, in a cell type-specific manner, to mediate induction of Smad7 expression at low BMP concentrations. On the other hand, the low affinity BRE-1 can function in multiple cell types to regulate Smad7 expression, but only at high BMP concentrations, through direct binding of multiple Smad1/4 complexes. Accordingly, Smad7 expression was stimulated by BMPs to much higher levels and at much lower BMP concentrations in GATA-expressing cells. Furthermore, heterologous reporter assays showed that BRE-1 is induced weakly in multiple cell types, whereas I-BRE displays strong responses but only in GATA-expressing cells. Thus, we propose that these response elements can function in different cellular contexts to interpret the full range of BMP concentrations in which Smad7 must be turned on.
Our results show that expression of Smad7 is induced by BMP-7 through different responsive elements. We demonstrate binding of endogenous Smads to the endogenous elements, and show that GATA factors cooperate with Smad1/4. Furthermore, we show that two Smad7 BREs, BRE-1 and I-BRE, function in different contexts, the first being a low-affinity BRE active at high BMP concentrations and the second being a high-affinity BRE active at low BMP concentrations in GATA-5- and -6-expressing cells. These results may thus shed light on mechanisms used to read out BMP morphogen gradients in vertebrates.
Acknowledgments
cDNA encoding GATA-1 was a generous gift from L. Wall, cDNA for GATA-2 was from R. J. Rottier, and cDNA for GATA-4, -5, and -6 was from E. Morrisey.
The work was supported by grants to J.L.W. from the CIHR. H.B. was supported by a studentship from NSERC and the Heart and Stroke foundation of Canada.
REFERENCES
- 1.Abdollah, S., M. Macías-Silva, T. Tsukazaki, H. Hayashi, L. Attisano, and J. L. Wrana. 1997. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J. Biol. Chem. 272:27678-27685. [DOI] [PubMed] [Google Scholar]
- 2.Affolter, M., T. Marty, M. A. Vigano, and A. Jazwinska. 2001. Nuclear interpretation of Dpp signaling in Drosophila. EMBO J. 20:3298-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afrakhte, M., A. Moren, S. Jossan, S. Itoh, K. Sampath, B. Westermark, C. H. Heldin, N. E. Heldin, and P. ten Dijke. 1998. Induction of inhibitory Smad6 and Smad7 mRNA by TGF-beta family members. Biochem. Biophys. Res. Commun. 249:505-511. [DOI] [PubMed] [Google Scholar]
- 4.Attisano, L., and J. L. Wrana. 2000. Smads as transcriptional co-modulators. Curr. Opin. Cell Biol. 12:235-243. [DOI] [PubMed] [Google Scholar]
- 5.Bai, S., X. Shi, X. Yang, and X. Cao. 2000. Smad6 as a transcriptional corepressor. J. Biol. Chem. 275:8267-8270. [DOI] [PubMed] [Google Scholar]
- 6.Belaguli, N. S., J. L. Sepulveda, V. Nigam, F. Charron, M. Nemer, and R. J. Schwartz. 2000. Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators. Mol. Cell. Biol. 20:7550-7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell, E., A. Lumsden, and A. Graham. 1999. Expression of GATA-2 in the developing avian rhombencephalon. Mech. Dev. 84:173-176. [DOI] [PubMed] [Google Scholar]
- 8.Blokzijl, A., P. ten Dijke, and C. F. Ibanez. 2002. Physical and functional interaction between GATA-3 and Smad3 allows TGF-beta regulation of GATA target genes. Curr. Biol. 12:35-45. [DOI] [PubMed] [Google Scholar]
- 9.Boyd, K. E., J. Wells, J. Gutman, S. M. Bartley, and P. J. Farnham. 1998. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc. Natl. Acad. Sci. USA 95:13887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodin, G., A. Ahgren, P. ten Dijke, C. H. Heldin, and R. Heuchel. 2000. Efficient TGF-beta induction of the Smad7 gene requires cooperation between AP-1, Sp1, and Smad proteins on the mouse Smad7 promoter. J. Biol. Chem. 275:29023-29030. [DOI] [PubMed] [Google Scholar]
- 11.Charron, F., G. Tsimiklis, M. Arcand, L. Robitaille, Q. Liang, J. D. Molkentin, S. Meloche, and M. Nemer. 2001. Tissue-specific GATA factors are transcriptional effectors of the small GTPase RhoA. Genes Dev. 15:2702-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, Y., and A. F. Schier. 2001. The zebrafish Nodal signal Squint functions as a morphogen. Nature 411:607-610. [DOI] [PubMed] [Google Scholar]
- 13.Crossley, M., and S. H. Orkin. 1994. Phosphorylation of the erythroid transcription factor GATA-1. J. Biol. Chem. 269:16589-16596. [PubMed] [Google Scholar]
- 14.Dai, Y. S., P. Cserjesi, B. E. Markham, and J. D. Molkentin. 2002. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J. Biol. Chem. 277:24390-24398. [DOI] [PubMed] [Google Scholar]
- 15.Denissova, N. G., C. Pouponnot, J. Long, D. He, and F. Liu. 2000. Transforming growth factor beta-inducible independent binding of SMAD to the Smad7 promoter. Proc. Natl. Acad. Sci. USA 97:6397-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dennler, S., S. Itoh, D. Vivien, P. ten Dijke, S. Huet, and J. M. Gauthier. 1998. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17:3091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derynck, R., Y. Zhang, and X. H. Feng. 1998. Smads: transcriptional activators of TGF-beta responses. Cell 95:737-740. [DOI] [PubMed] [Google Scholar]
- 18.Dyson, S., and J. B. Gurdon. 1998. The interpretation of position in a morphogen gradient as revealed by occupancy of activin receptors. Cell 93:557-568. [DOI] [PubMed] [Google Scholar]
- 19.Ebisawa, T., M. Fukuchi, G. Murakami, T. Chiba, K. Tanaka, T. Imamura, and K. Miyazono. 2001. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 276:12477-12480. [DOI] [PubMed] [Google Scholar]
- 20.Elagib, K. E., F. K. Racke, M. Mogass, R. Khetawat, L. L. Delehanty, and A. N. Goldfarb. 2003. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood 101:4333-4341. [DOI] [PubMed] [Google Scholar]
- 21.Gritsman, K., W. S. Talbot, and A. F. Schier. 2000. Nodal signaling patterns the organizer. Development 127:921-932. [DOI] [PubMed] [Google Scholar]
- 22.Gurdon, J. B., and P. Y. Bourillot. 2001. Morphogen gradient interpretation. Nature 413:797-803. [DOI] [PubMed] [Google Scholar]
- 23.Hanyu, A., Y. Ishidou, T. Ebisawa, T. Shimanuki, T. Imamura, and K. Miyazono. 2001. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-beta signaling. J. Cell Biol. 155:1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hata, A., G. Lagna, J. Massagué, and A. Hemmati-Brivanlou. 1998. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 12:186-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hata, A., J. Seoane, G. Lagna, E. Montalvo, A. Hemmati-Brivanlou, and J. Massague. 2000. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell 100:229-240. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi, H., S. Abdollah, Y. Qiu, J. Cai, Y. Y. Xu, B. W. Grinnell, M. A. Richardson, J. N. Topper, M. A. Gimbrone, Jr., J. L. Wrana, and D. Falb. 1997. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell 89:1165-1173. [DOI] [PubMed] [Google Scholar]
- 27.Henningfeld, K. A., S. Rastegar, G. Adler, and W. Knochel. 2000. Smad1 and Smad4 are components of the bone morphogenetic protein-4 (BMP-4)-induced transcription complex of the Xvent-2B promoter. J. Biol. Chem. 275:21827-21835. [DOI] [PubMed] [Google Scholar]
- 28.Hill, C. S. 2001. TGF-beta signalling pathways in early Xenopus development. Curr. Opin. Genet. Dev. 11:533-540. [DOI] [PubMed] [Google Scholar]
- 29.Hogan, B. L. 1996. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 10:1580-1594. [DOI] [PubMed] [Google Scholar]
- 30.Hua, X., Z. A. Miller, H. Benchabane, J. L. Wrana, and H. F. Lodish. 2000. Synergism between transcription factors TFE3 and Smad3 in transforming growth factor-beta-induced transcription of the Smad7 gene. J. Biol. Chem. 275:33205-33208. [DOI] [PubMed] [Google Scholar]
- 31.Hullinger, T. G., Q. Pan, H. L. Viswanathan, and M. J. Somerman. 2001. TGFbeta and BMP-2 activation of the OPN promoter: roles of smad- and hox-binding elements. Exp. Cell Res. 262:69-74. [DOI] [PubMed] [Google Scholar]
- 32.Imamura, T., M. Takase, A. Nishihara, E. Oeda, J. Hanai, M. Kawabata, and K. Miyazono. 1997. Smad6 inhibits signalling by the TGF-beta superfamily. Nature 389:622-626. [DOI] [PubMed] [Google Scholar]
- 33.Ishida, W., T. Hamamoto, K. Kusanagi, K. Yagi, M. Kawabata, K. Takehara, T. K. Sampath, M. Kato, and K. Miyazono. 2000. Smad6 is a Smad1/5-induced smad inhibitor. Characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. J. Biol. Chem. 275:6075-6079. [DOI] [PubMed] [Google Scholar]
- 34.Ishisaki, A., K. Yamato, A. Nakao, K. Nonaka, M. Ohguchi, P. ten Dijke, and T. Nishihara. 1998. Smad7 is an activin-inducible inhibitor of activin-induced growth arrest and apoptosis in mouse B cells. J. Biol. Chem. 273:24293-24296. [DOI] [PubMed] [Google Scholar]
- 35.Itoh, S., F. Itoh, M. J. Goumans, and P. Ten Dijke. 2000. Signaling of transforming growth factor-beta family members through Smad proteins. Eur. J. Biochem. 267:6954-6967. [DOI] [PubMed] [Google Scholar]
- 36.Johnson, K., H. Kirkpatrick, A. Comer, F. M. Hoffmann, and A. Laughon. 1999. Interaction of Smad complexes with tripartite DNA-binding sites. J. Biol. Chem. 274:20709-20716. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, K. D., and E. H. Bresnick. 2002. Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods 26:27-36. [DOI] [PubMed] [Google Scholar]
- 38.Kavsak, P., R. K. Rasmussen, C. G. Causing, S. Bonni, H. Zhu, G. H. Thomsen, and J. L. Wrana. 2000. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell 6:1365-1375. [DOI] [PubMed] [Google Scholar]
- 39.Kerkela, R., S. Pikkarainen, T. Majalahti-Palviainen, H. Tokola, and H. Ruskoaho. 2002. Distinct roles of mitogen-activated protein kinase pathways in GATA-4 transcription factor-mediated regulation of B-type natriuretic peptide gene. J. Biol. Chem. 277:13752-13760. [DOI] [PubMed] [Google Scholar]
- 40.Kiela, P. R., J. LeSueur, J. F. Collins, and F. K. Ghishan. 2003. Transcriptional regulation of the rat NHE3 gene. Functional interactions between GATA-5 and Sp family transcription factors. J. Biol. Chem. 278:5659-5668. [DOI] [PubMed] [Google Scholar]
- 41.Kim, J., K. Johnson, H. J. Chen, S. Carroll, and A. Laughon. 1997. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature 388:304-308. [DOI] [PubMed] [Google Scholar]
- 42.Kitta, K., R. M. Day, Y. Kim, I. Torregroza, T. Evans, and Y. J. Suzuki. 2003. Hepatocyte growth factor induces GATA-4 phosphorylation and cell survival in cardiac muscle cells. J. Biol. Chem. 278:4705-4712. [DOI] [PubMed] [Google Scholar]
- 43.Korchynskyi, O., and P. ten Dijke. 2002. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277:4883-4891. [DOI] [PubMed] [Google Scholar]
- 44.Kusanagi, K., H. Inoue, Y. Ishidou, H. K. Mishima, M. Kawabata, and K. Miyazono. 2000. Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol. Biol. Cell. 11:555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Labbe, E., C. Silvestri, P. A. Hoodless, J. L. Wrana, and L. Attisano. 1998. Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol. Cell 2:109-120. [DOI] [PubMed] [Google Scholar]
- 46.Liang, Q., R. J. Wiese, O. F. Bueno, Y. S. Dai, B. E. Markham, and J. D. Molkentin. 2001. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol. Cell. Biol. 21:7460-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lien, C. L., J. McAnally, J. A. Richardson, and E. N. Olson. 2002. Cardiac-specific activity of an nkx2-5 enhancer requires an evolutionarily conserved smad binding site. Dev. Biol. 244:257-266. [DOI] [PubMed] [Google Scholar]
- 48.Liu, C., S. W. Glasser, H. Wan, and J. A. Whitsett. 2002. GATA-6 and thyroid transcription factor-1 directly interact and regulate surfactant protein-C gene expression. J. Biol. Chem. 277:4519-4525. [DOI] [PubMed] [Google Scholar]
- 49.Liu, F., A. Hata, J. C. Baker, J. Doody, J. Cárcamo, R. M. Harland, and J. Massagué. 1996. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature 381:620-623. [DOI] [PubMed] [Google Scholar]
- 50.Lopez-Rovira, T., E. Chalaux, J. Massague, J. L. Rosa, and F. Ventura. 2002. Direct binding of Smad1 and Smad4 to two distinct motifs mediates bone morphogenetic protein-specific transcriptional activation of Id1 gene. J. Biol. Chem. 277:3176-3185. [DOI] [PubMed] [Google Scholar]
- 51.Marty, T., M. A. Vigano, C. Ribeiro, U. Nussbaumer, N. C. Grieder, and M. Affolter. 2001. A HOX complex, a repressor element and a 50 bp sequence confer regional specificity to a DPP-responsive enhancer. Development 128:2833-2845. [DOI] [PubMed] [Google Scholar]
- 52.Massague, J., and D. Wotton. 2000. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 19:1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyazono, K., P. ten Dijke, and C. H. Heldin. 2000. TGF-beta signaling by Smad proteins. Adv. Immunol. 75:115-157. [DOI] [PubMed] [Google Scholar]
- 54.Molkentin, J. D. 2000. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 275:38949-38952. [DOI] [PubMed] [Google Scholar]
- 55.Morin, S., F. Charron, L. Robitaille, and M. Nemer. 2000. GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J. 19:2046-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrisey, E. E., H. S. Ip, Z. Tang, and M. S. Parmacek. 1997. GATA-4 activates transcription via two novel domains that are conserved within the GATA-4/5/6 subfamily. J. Biol. Chem. 272:8515-8524. [DOI] [PubMed] [Google Scholar]
- 57.Nagarajan, R. P., J. Zhang, W. Li, and Y. Chen. 1999. Regulation of Smad7 promoter by direct association with Smad3 and Smad4. J. Biol. Chem. 274:33412-33418. [DOI] [PubMed] [Google Scholar]
- 58.Nakao, A., M. Afrakhte, A. Moren, T. Nakayama, J. L. Christian, R. Heuchel, S. Itoh, M. Kawabata, N. E. Heldin, C. H. Heldin, and P. ten Dijke. 1997. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature 389:631-635. [DOI] [PubMed] [Google Scholar]
- 59.Nardelli, J., D. Thiesson, Y. Fujiwara, F. Y. Tsai, and S. H. Orkin. 1999. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev. Biol. 210:305-321. [DOI] [PubMed] [Google Scholar]
- 60.Nellen, D., R. Burke, G. Struhl, and K. Basler. 1996. Direct and long-range action of a DPP morphogen gradient. Cell 85:357-368. [DOI] [PubMed] [Google Scholar]
- 61.Neumann, C., and S. Cohen. 1997. Morphogens and pattern formation. Bioessays 19:721-729. [DOI] [PubMed] [Google Scholar]
- 62.Overdier, D. G., A. Porcella, and R. H. Costa. 1994. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol. Cell. Biol. 14:2755-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Overdier, D. G., H. Ye, R. S. Peterson, D. E. Clevidence, and R. H. Costa. 1997. The winged helix transcriptional activator HFH-3 is expressed in the distal tubules of embryonic and adult mouse kidney. J. Biol. Chem. 272:13725-13730. [DOI] [PubMed] [Google Scholar]
- 64.Park, G. T., and M. I. Morasso. 2002. Bone morphogenetic protein-2 (BMP-2) transactivates Dlx3 through Smad1 and Smad4: alternative mode for Dlx3 induction in mouse keratinocytes. Nucleic Acids Res. 30:515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peterson, R. S., L. Lim, H. Ye, H. Zhou, D. G. Overdier, and R. H. Costa. 1997. The winged helix transcriptional activator HFH-8 is expressed in the mesoderm of the primitive streak stage of mouse embryos and its cellular derivatives. Mech. Dev. 69:53-69. [DOI] [PubMed] [Google Scholar]
- 66.Schuster, N., and K. Krieglstein. 2002. Mechanisms of TGF-beta-mediated apoptosis. Cell Tissue Res. 307:1-14. [DOI] [PubMed] [Google Scholar]
- 67.Shi, X., X. Yang, D. Chen, Z. Chang, and X. Cao. 1999. Smad1 interacts with homeobox DNA-binding proteins in bone morphogenetic protein signaling. J. Biol. Chem. 274:13711-13717. [DOI] [PubMed] [Google Scholar]
- 68.Shi, Y., Y. F. Wang, L. Jayaraman, H. Yang, J. Massagué, and N. P. Pavletich. 1998. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell 94:585-594. [DOI] [PubMed] [Google Scholar]
- 69.Shimizu, K., and J. B. Gurdon. 1999. A quantitative analysis of signal transduction from activin receptor to nucleus and its relevance to morphogen gradient interpretation. Proc. Natl. Acad. Sci. USA 96:6791-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sirard, C., S. Kim, C. Mirtsos, P. Tadich, P. A. Hoodless, A. Itie, R. Maxson, J. L. Wrana, and T. W. Mak. 2000. Targeted disruption in murine cells reveals variable requirement for Smad4 in transforming growth factor beta-related signaling. J. Biol. Chem. 275:2063-2070. [DOI] [PubMed] [Google Scholar]
- 71.Stopa, M., D. Anhuf, L. Terstegen, P. Gatsios, A. M. Gressner, and S. Dooley. 2000. Participation of Smad2, Smad3, and Smad4 in transforming growth factor beta (TGF-beta)-induced activation of Smad7. The TGF-beta response element of the promoter requires functional Smad binding element and E-box sequences for transcriptional regulation. J. Biol. Chem. 275:29308-29317. [DOI] [PubMed] [Google Scholar]
- 72.Szuts, D., S. Eresh, and M. Bienz. 1998. Functional intertwining of Dpp and EGFR signaling during Drosophila endoderm induction. Genes Dev. 12:2022-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tabata, T. 2001. Genetics of morphogen gradients. Nat. Rev. Genet. 2:620-630. [DOI] [PubMed] [Google Scholar]
- 74.Takase, M., T. Imamura, T. K. Sampath, K. Takeda, H. Ichijo, K. Miyazono, and M. Kawabata. 1998. Induction of Smad6 mRNA by bone morphogenetic proteins. Biochem. Biophys. Res. Commun. 244:26-29. [DOI] [PubMed] [Google Scholar]
- 75.Tang, S. J., P. A. Hoodless, Z. Lu, M. L. Breitman, R. R. McInnes, J. L. Wrana, and M. Buchwald. 1998. The Tlx-2 homeobox gene is a downstream target of BMP signalling and is required for mouse mesoderm development. Development 125:1877-1887. [DOI] [PubMed] [Google Scholar]
- 76.Teleman, A. A., M. Strigini, and S. M. Cohen. 2001. Shaping morphogen gradients. Cell 105:559-562. [DOI] [PubMed] [Google Scholar]
- 77.Thisse, B., C. V. Wright, and C. Thisse. 2000. Activin- and Nodal-related factors control antero-posterior patterning of the zebrafish embryo. Nature 403:425-428. [DOI] [PubMed] [Google Scholar]
- 78.Tremblay, J. J., and R. S. Viger. 2003. Transcription factor GATA-4 is activated by phosphorylation of serine 261 via the cAMP/protein kinase A signaling pathway in gonadal cells. J. Biol. Chem. 278:22128-22135. [DOI] [PubMed] [Google Scholar]
- 79.van Wering, H. M., I. L. Huibregtse, S. M. van der Zwan, M. S. de Bie, L. N. Dowling, F. Boudreau, E. H. Rings, R. J. Grand, and S. D. Krasinski. 2002. Physical interaction between GATA-5 and hepatocyte nuclear factor-1alpha results in synergistic activation of the human lactase-phlorizin hydrolase promoter. J. Biol. Chem. 277:27659-27667. [DOI] [PubMed] [Google Scholar]
- 80.von Gersdorff, G., K. Susztak, F. Rezvani, M. Bitzer, D. Liang, and E. P. Bottinger. 2000. Smad3 and Smad4 mediate transcriptional activation of the human Smad7 promoter by transforming growth factor beta. J. Biol. Chem. 275:11320-11326. [DOI] [PubMed] [Google Scholar]
- 81.Whitman, M. 1998. Smads and early developmental signaling by the TGFbeta superfamily. Genes Dev. 12:2445-2462. [DOI] [PubMed] [Google Scholar]
- 82.Wotton, D., and J. Massague. 2001. Smad transcriptional corepressors in TGF beta family signaling. Curr. Top. Microbiol. Immunol. 254:145-164. [PubMed] [Google Scholar]
- 83.Wrana, J. L., and L. Attisano. 2000. The Smad pathway. Cytokine Growth Factor Rev. 11:5-13. [DOI] [PubMed] [Google Scholar]
- 84.Xu, X., Z. Yin, J. B. Hudson, E. L. Ferguson, and M. Frasch. 1998. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev. 12:2354-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zawel, L., J. L. Dai, P. Buckhaults, S. Zhou, K. W. Kinzler, B. Vogelstein, and S. E. Kern. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell 1:611-617. [DOI] [PubMed] [Google Scholar]
- 86.Zhou, Y., M. Yamamoto, and J. D. Engel. 2000. GATA2 is required for the generation of V2 interneurons. Development 127:3829-3838. [DOI] [PubMed] [Google Scholar]