Abstract
Nucleosome assembly protein 1 (Nap1) is widely conserved from yeasts to humans and facilitates nucleosome formation in vitro as a histone chaperone. Nap1 is generally localized in the cytoplasm, except that subcellular localization of Drosophila melanogaster Nap1 is dynamically regulated between the cytoplasm and nucleus during early development. The cytoplasmic localization of Nap1 is seemingly incompatible with the proposed role of Nap1 in nucleosome formation, which should occur in the nucleus. Here, we have examined the roles of a putative nuclear export signal (NES) sequence in yeast Nap1 (yNap1). yNap1 mutants lacking the NES-like sequence were localized predominantly in the nucleus. Deletion of NAP1 in cells harboring a single mitotic cyclin gene is known to cause mitotic delay and temperature-sensitive growth. A wild-type NAP1 complemented these phenotypes while nap1 mutant genes lacking the NES-like sequence or carboxy-terminal region did not. These and other results suggest that yNap1 is a nucleocytoplasmic shuttling protein and that its shuttling is important for yNap1 function during mitotic progression. This study also provides a possible explanation for Nap1's involvement in nucleosome assembly and/or remodeling in the nucleus.
Chromatin is one of the hallmarks of eukaryotes. The eukaryotic genome DNA is complexed with chromosomal proteins to form the chromatin structure in the nucleus. Nuclear reactions such as DNA replication, transcription, DNA repair, and recombination take place on the chromatin and are regulated by the disruption and assembly of chromatin, namely chromatin remodeling. Recently, a variety of chromatin remodeling factors has been identified, including histone modification enzymes and ATP-dependent factors (reviewed in references 1, 2, 22, and 42). The histone chaperone family is one of the chromatin remodeling factors that binds to core histones and facilitates assembly and remodeling of the chromatin in an ATP-independent manner. This family includes various kinds of proteins, such as nucleoplasmin (31) and N1/N2 (28) in Xenopus, nucleosome assembly protein 1 (designated Nap1 in this paper) and Nap2 (13, 48), template-activating factor Iα (TAF-Iα) and TAF-Iβ/SET (41), chromatin assembly factor 1 (CAF-1) (6), dNLP (17), Spt6 (5), DF31 (9), CIA (40), TAF-III/B23 (45), and so on.
Nap1 was first identified from HeLa cell extracts as a protein that stimulates nucleosome formation from DNA and purified core histones (14). Nap1 directly binds to core histones and transfers them onto naked DNA (15, 36, 54). Recent studies suggest that Nap1 is involved in transcriptional regulation through chromatin remodeling (32). For example, Nap1 remodels the chromatin-like structure of the adenovirus genome DNA and stimulates transcription as well as replication (24). Nap1 acts synergistically with ATP-dependent chromatin remodeling factors such as NURF and ACF to remodel the chromatin structure (18, 19, 23, 43). Furthermore, Nap1 and Nap2 are found to interact with p300, a histone acetyltransferase, and to stimulate transcription from p21 and E2F promoters (20, 49). Considering the roles of Nap1 in nucleosome formation and/or transcription, Nap1 should exist in the nucleus. However, it is widely accepted that Nap1 is localized in the cytoplasm but not in the nucleus (26, 35; unpublished observations). One exceptional observation is that Drosophila melanogaster Nap1 is present in nuclei limitedly during some stages of development (16). A possible explanation for this contradiction is that Nap1 is transiently present in nuclei and regulatedly exported from nuclei, although the static subcellular localization of Nap1 is cytoplasmic. Here, we have addressed this issue and further tried to understand the physiological meaning of nuclear localization and nucleocytoplasmic shuttling of Nap1.
In the yeast Saccharomyces cerevisiae, deletion of NAP1 causes no phenotypes or very weak phenotypes, if any (26, 50). In yeasts, there are four cyclin B homologs, Clb1, Clb2, Clb3, and Clb4. Nap1 physically and genetically interacts with Clb2 (25, 26). Deletion of NAP1 in a Clb2-dependent strain, in which CLB1, CLB3, and CLB4 are disrupted, shows a temperature-sensitive growth phenotype and an elongated bud phenotype (25, 26). It is suggested that the elongated bud phenotype is caused by a prolonged mitotic delay because disappearance of Clb2 at the end of mitosis is delayed in the ΔCLB1 ΔCLB3 ΔCLB4 ΔNAP1 strain (25). Different kinds of factors and genes are known to be involved in the elongated bud phenotype. A cdc28-1N mutation shows the elongated bud phenotype (52); a clb2ts mutant, in which CLB1, CLB3, and CLB4 are disrupted, shows the elongated bud phenotype at nonpermissive temperatures (10); and overexpression of Swe1 kinase, which inactivates Cdc28 by phosphorylation, also causes the elongated bud phenotype (4, 34). These observations suggest that the compromised activity of Cdc28-Clb kinase causes the elongated bud phenotype and that a target protein(s) of this kinase is responsible for the elongated bud phenotype. Gin4 is a protein kinase that is localized in the bud neck and activated during mitosis by phosphorylation. Mutations in this gene also cause the elongated bud phenotype (3). The mitosis-specific phosphorylation of Gin4 requires Nap1 and Clb2 (3). Gin4 phosphorylation is downstream of Clb2 because an ectopic expression of destruction box-deleted Clb2 causes phosphorylation of Gin4 in interphases (3). It is interesting that Gin4 physically and genetically interacts with Nap1 (3). Thus, Nap1 may play an important role during mitotic progression from Cdc28-Clb regulation to cytokinesis, although an exact in vivo function(s) of yeast Nap1 (yNap1) remains unclear. Identification of interactors of Nap1 further emphasizes this point. Nap1 interactors identified by biochemical methods and two-hybrid screening (8) includes Clb2 (26), Nbp1 (50), Gin4 (3), CK2 (33), Hta1, Kcc4, Yrb1, Jip1 (53), Yap6, and Kap95 (21). Note that Yrb1 and Kap95 are closely related to the nuclear import mechanism (30). Recently, Mosammaparast et al. reported that Kap114 is involved in the nuclear import of Nap1 (38).
In this study, we show that yNap1 is a nucleocytoplasmic shuttling protein. A region of yNap1 involved in histone binding was found to be important for nuclear import of yNap1, and nuclear export of yNap1 was dependent on a nuclear export signal (NES)-like sequence. To elucidate the role of the Nap1 nucleocytoplasmic shuttling, we used a yNAP1-disrupted strain in complementation assays and showed that nucleocytoplasmic shuttling, but not cytoplasmic localization of yNap1, is important for mitotic progression. Our findings solve the contradiction in Nap1 localization and show the possibility that Nap1 acts as a histone chaperone in vivo. In addition, these data provide a cue to its physiological function during mitotic progression.
MATERIALS AND METHODS
Strains and culture conditions.
Except where noted, all yeast strains were grown at 25°C in yeast extract-peptone-dextrose (YPD) medium. For protein expression induced by galactose, single colonies were picked up and first inoculated into synthetic defined (SD) medium containing 2% glucose. Cells were grown at 25°C until the optical density at 600 nm (OD600) reached 0.8. Then cells were collected by centrifugation at 800 × g and washed twice with phosphate-buffered saline (PBS). The collected cells were grown in synthetic galactose (SG) medium containing 2% galactose for 6 to 8 h, allowing the induced expression of proteins.
Vector constructions.
To construct expression vectors of yNap1 derivatives in yeast cells, promoter and terminator fragments of the yNAP1 gene were amplified by PCR with primers 5′-GTGGCGGCCGCCGGGCACTCCCTTCTTTCTTCC-3′ and 5′-GGAGGATCCACTAGTTCTAGAGATCTTGCGCTTTGCTCTTGGTCC-3′ for the promoter and 5′-ATCTCTAGAACTAGTGGATCCTCCCAACGCACTTCGCAAGAGTG-3′ and 5′-CCCCTCGAGTAACTCGTCCAACTTGAAGGCCCC-3′ for the terminator, with the yeast genomic DNA as a template. Overlap extension was performed by PCR with primers 5′-GTGGCGGCCGCCGGGCACTCCCTTCTTTCTTCC-3′ and 5′-CCCCTCGAGTAACTCGTCCAACTTGAAGGCCCC-3′, with promoter and terminator PCR products as templates. PCR products were digested with NotI and XhoI and cloned into NotI- and XhoI-digested pRS316 plasmid, generating pRS316-yNap1pt, which contains a unique BamHI site between the promoter and the terminator fragments. pBluescript-Flag was constructed from BamHI- and XhoI-digested pBluescript SK(+) (Stratagene) and a DNA fragment made by annealing two synthetic oligonucleotides, 5′-GATCCGCCGCCACCATGGACTACAAGGATGACGACGACAAGCATATGC-3′ and 5′-TGCAGCATATGCTTGTCGTCGTCATCCTTGTAGTCCATGGTGGCGGCG-3′. yNAP1 cDNA fragments were amplified by PCR with primers 5′-AGCGCAAGCATATGTCAGACCCTATCAGAACGAAAC-3′ and 5′-GCGGGATCCTTATGACTGCTTGCATTCAGGAG-3′. The amplified DNA fragment was phosphorylated with T4 polynucleotide kinase (Toyobo), blunted with Klenow fragment, and digested with NdeI. Then the fragment was cloned into NdeI- and EcoRV-digested pBluescript-Flag (pBS-Flag-yNap1). For yNap1ΔNES, DNA fragments corresponding to the N- and C-terminal segments were prepared by PCR with primers 5′-AGCGCAAGCATATGTCAGACCCTATCAGAACGAAAC-3′ and 5′-CAACTTCGAATAGCTCGCTTTGACCCCCCACATACCCGCTGT-3′ and primers 5′-CAAAGCGAGCTATTCGAAGTTG-3′ and 5′-GCGGGATCCTTATGACTGCTTGCATTCAGGAG-3′, respectively, with the yNap1 cDNA as a template. The two segments were ligated by overlap extension PCR with primers 5′-AGCGCAAGCATATGTCAGACCCTATCAGAACGAAAC-3′ and 5′-GCGGGATCCTTATGACTGCTTGCATTCAGGAG-3′. Amplified DNA fragments were phosphorylated with T4 polynucleotide kinase, blunted with Klenow fragment, and digested with NdeI. The fragment was cloned into NdeI- and EcoRV-digested pBS-Flag and designated pBS-Flag-yNap1ΔNES. For yNap1 (L99A L102A), DNA fragments corresponding to the N- and C-terminal segments were prepared by PCR with primers 5′-AGCGCAAGCATATGTCAGACCCTATCAGAACGAAAC-3′ and 5′-CGCTTTGTGCAGTCTTAGCGCTCAGCAGC-3′ and primers 5′-GCTGAGCGCTAAGACTGCACAAAGCGAGC-3′ and 5′-GCGGGATCCTTATGACTGCTTGCATTCAGGAG-3′, respectively, with the yNap1 cDNA as a template. These two segments were ligated by overlap extension PCR with the primers 5′-AGCGCAAGCATATGTCAGACCCTATCAGAACGAAAC-3′ and 5′-GCGGGATCCTTATGACTGCTTGCATTCAGGAG-3′. Amplified DNA fragments were phosphorylated with T4 polynucleotide kinase, blunted with Klenow fragment, and digested with NdeI. The fragment was cloned into NdeI- and EcoRV-digested pBS-Flag and designated pBS-Flag-yNap1 (L99A L102A). Flag-yNap1 wild-type (WT) and mutant fragments, except for ΔC1 mutants, were excised from pBS-Flag-yNap1 derivatives by digestion with BamHI and cloned into the BamHI site of pRS316-yNap1pt or pYES2. Flag-yNap1ΔC1 and yNap1ΔNESΔC1 were excised from pBS-Flag-yNap1 or pBS-Flag-yNap1ΔNES by digestion with BamHI and XhoI and ligated with the plasmid pYES2, which was digested with BamHI and XhoI. To construct yNap1 expression vectors for HeLa cells, Flag-yNap1 fragments excised from pBS-Flag-yNap1 plasmids with BamHI were cloned into BamHI-digested pcDNA3 (Invitrogen). To construct Flag-yNap1ΔC1 and Flag-yNap1ΔNESΔC1 expression vectors, plasmids pBS-Flag-yNap1 and pBS-Flag-yNap1ΔNES were digested with BamHI and XhoI, blunted with Klenow, and cloned into the EcoRV site of pcDNA3. To construct enhance green fluorescent protein (EGFP)-Nap1 expression vectors, Nap1 coding sequences were cut out with NdeI and BamHI from the pBS-Flag-yNap1 plasmids and cloned into the BglII site of pEGFP-C1 (Clontech) by blunt-end ligation. To construct the glutathione S-transferase (GST)-fused recombinant yNap1 protein expression vector, yNap1 cDNA fragments digested with NdeI and BamHI and then blunted with Klenow fragment were cloned into pGEX-5X-3 (Amersham Biosciences), which had been digested with SalI and filled with Klenow fragment. To construct pYES2-Clb2-EGFP, two DNA fragments were made by PCR as follows. The Clb2 fragment was obtained by PCR with oligonucleotides 5′-CGCTAGCGAATTCCGCCACCATGTCCAACCCAATAGAAAACACA-3′ and 5′-TGGTGGCGACCGGTGGATCTTCATGCAAGGTCATTATATCATAGCC-3′ as primers and with CLB2 cDNA as a template. The EGFP fragment was obtained by PCR with oligonucleotides 5′-GATCCACCGGTCGCCACCATGG-3′ and 5′-CGGATCCTCGAGTTACTTGTACAGCTCGTCCATGCC-3′ as primers and with pEGFP-N1 (Clontech) as a template. Then overlap extension PCR was performed with oligonucleotides 5′-CGCTAGCGAATTCCGCCACCATGTCCAACCCAATAGAAAACACA-3′ and 5′-CGGATCCTCGAGTTACTTGTACAGCTCGTCCATGCC-3′ as primers and with the two DNA fragments as templates. The overlap extension products were gel purified, digested with EcoRI and BamHI, and cloned into EcoRI- and BamHI-digested pYES2 to generate pYES2-Clb2-EGFP. To construct yCP, a low-copy-number plasmid carrying the NAP1 gene, the coding region of NAP1 was cloned between SmaI and SphI sites in a TRP1/CEN4-based yeast plasmid, YCp19, so that the region between the SmaI and SphI sites in URA3 was replaced by NAP1. To construct yEP, a high-copy-number plasmid carrying the NAP1 gene, the coding region of NAP1 was cloned between the SmaI and SphI sites in a TRP1, URA3, 2-based plasmid, YEp24.
Preparation of recombinant proteins.
To prepare recombinant GST-yNap1, Escherichia coli BL21(DE3) cells were transformed with pGEX-5X-3 derivatives and grown in Lurai-Bertani medium containing 100 μg of ampicillin/ml. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a concentration of 1 mM when the OD600 reached 0.3, and cells were further incubated for 3 h. The cells were harvested by centrifugation and washed with PBS. Recombinant proteins were purified by using glutathione Sepharose beads according to the manufacturer's instructions (Amersham Biosciences).
Examination of morphology and temperature sensitivity.
Single colonies of DK168 and its transformants were grown at 25°C in YPD for 12 h. Cells were plated onto slide glasses, covered with coverslips, and subjected to examination under a microscope (Axioscope; Zeiss). To examine the temperature sensitivity of each strain, single colonies were inoculated into 2 ml of YPD medium and incubated at 25°C for 24 h. Cell concentrations were adjusted to ODs of 0.1, 0.01, and 0.001 with YPD, and 2 μl of each dilution was spotted onto SD plates lacking His, Trp, Leu, and Ura and incubated at either 25 or 37°C for 1 or 2 days. To examine the growth rate of yNap1-overexpressing strains, a single colony of each strain was streaked onto SD and SG plates lacking His, Trp, Leu, and Ura and incubated at 25°C for 2 or 3 days.
Construction of NAP1-deleted strains.
The NAP1 gene was disrupted by insertion of LEU2. To this end, a LEU2 gene sandwiched with upstream and downstream flanking regions of a nap1 gene was amplified by PCR from the genomic DNA of DK168, whose NAP1 is disrupted by LEU2. The Δnap1::LEU2 DNA was transformed into PSY1785 and PSY1784 strains carrying WT KAP114 and disrupted KAP114 alleles, respectively (37). The disruption of the NAP1 gene was confirmed by PCR.
Western blotting analysis of yeast cell lysates.
Exponentially growing yeast cells were harvested by centrifugation and washed twice with PBS. Cells were suspended in a cell lysis buffer containing 62.5 mM Tris-HCl (pH 6.8), 10% glycerol, 145 mM β-mercaptoethanol, and 0.25% sodium dodecyl sulfate (SDS). Glass beads were added, and cells were lysed by vigorously shaking and boiling at 98°C for 5 min. The lysates were cleared by centrifugation at 15,000 × g for 5 min, and their protein concentrations were determined by using the protein assay kit (Bio-Rad). Aliquots (equivalent to 10 μg of protein) were subjected to SDS-7.5% polyacrylamide gel electrophoresis (PAGE) followed by Western blotting with the antibodies indicated in each figure legend.
Immunoprecipitation assay.
Yeast cell extracts were prepared from exponentially growing cells cultured in SG (without Ura) for 6 to 8 h. The cell cultures were harvested by centrifugation and frozen in liquid nitrogen. The cells were sonicated in lysis buffer (50 mM HEPES-KOH [pH 7.6], 50 mM KCl, 0.5 mM EGTA, 1 mM MgCl2, 10% glycerol, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 2 μg of pepstatin A/ml, 1 mM phenylmethylsulfonyl fluoride) in the presence of glass beads. The cell lysates were obtained from sonicated lysates by centrifugation. The protein concentration was determined by the Bradford method. The soluble yeast cell extracts (200 μg of protein) and anti-Flag antibody-conjugated agarose beads (Sigma) were mixed and incubated at 4°C for 30 min. Then the beads were washed four times with the lysis buffer, and bound proteins were separated by SDS-7.5% PAGE followed by immunoblotting with anti-Flag or anti-Clb2 (Santa Cruz) antibodies.
GST pull-down assays.
To prepare resins bound by GST or GST fusion proteins, glutathione-Sepharose beads (Amersham Bioscience) were mixed with E. coli lysates containing GST or GST fusion proteins, washed three times with a binding buffer containing 500 mM NaCl, 50 mM Tris (pH 7.9), 1 mM EDTA, and 1% Triton X-100, and washed three times with a buffer containing 50 mM KCl, 50 mM HEPES-KOH (pH 7.6), 1 mM EGTA, 1 mM MgCl2, and 0.01% Tween 20. To prepare [35S]methionine-labeled yNap1 proteins, in vitro transcription-translation reactions were performed according to the manufacturer's instructions (Invitrogen). As a template for transcription with T7 RNA polymerase, pYES2-Clb2-EGFP was digested with NcoI, which removes the EGFP coding region downstream of Clb2. In vitro-translated Clb2 was incubated with the resins bound by GST or GST-Nap1 at 4°C for 1 h. Beads were washed 3 times with binding buffer and eluted with an SDS sample buffer. Bound materials were separated by SDS-10% PAGE and visualized by Coomassie brilliant blue (CBB) staining and autoradiography.
Supercoiling assay.
Supercoiling assays were performed essentially as described previously (11). Recombinant yNap1 and its derivatives were prepared as GST fusion proteins.
Mammalian cell culture and transfection experiments.
HeLa cells were grown in minimal essential medium supplemented with 10% fetal calf serum. For the subcellular localization study, HeLa cells were split and grown on coverslips placed in 6-cm-diameter tissue culture dishes for 16 h before transfection. Cells in each dish were transfected with 10 μg of mammalian expression vectors by the standard calcium phosphate method (27). At 6 h posttransfection, the medium was changed and cells were further incubated for 18 h. Cells were washed twice with PBS and fixed for 10 min at room temperature with 4% formaldehyde that was freshly prepared from paraformaldehyde. Coverslips were washed with 0.5% Triton X-100 in PBS for 10 min and washed three times with PBS. To visualize Flag-tagged yNap1 proteins, fixed cells were serially incubated with 1% nonfat dried milk in PBS for 30 min, with 10 μg of anti-Flag M2 antibody (Sigma)/ml in 1% nonfat dry milk in PBS for 1 h, with rhodamine-conjugated anti-mouse immunoglobulin G (Cappel) for 1 h, and with 1 μg of 4′,6′-diamino-2-phenylindole dihydrochloride (DAPI)/ml in PBS for 10 min. The cells were then observed under a fluorescence microscopy (Axioscope). To measure the fluorescent intensity in the nucleus and cytoplasm, EGFP-Nap1, EGFP-Nap1ΔNES, EGFP-Nap1ΔC1, and EGFP-Nap1ΔNESΔC1 were expressed in HeLa cells by conventional calcium phosphate transfection and fixed with 4% paraformaldehyde for 10 min and DNA was stained with DAPI. Transfected cells were analyzed by the laser scanning cell cytometer LSC2 (Olympus).
Indirect immunostaining of yeast cells.
Indirect immunostaining of yeast cells with anti-Flag antibody was performed as described previously (51). DNA was visualized by DAPI staining.
RESULTS
Identification of NES in the yNAP1 sequence.
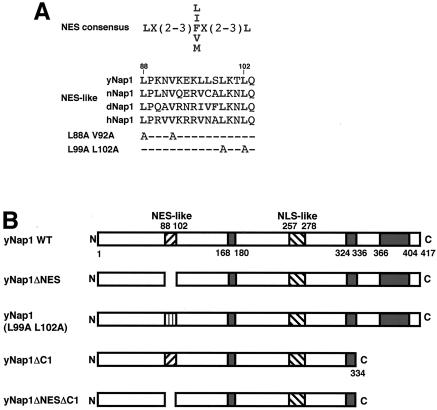
Nap1 has been identified as a protein that facilitates formation of nucleosomes from DNA and histones in vitro. Therefore, Nap1 should be present in the nucleus where nucleosome formation occurs. However, it is reported that the subcellular localization of Nap1 in mammalian and yeast cells is restricted to the cytoplasm (26, 35). To get insight into a physiological function(s) of Nap1 as a histone chaperone, we initiated this study by carefully reexamining the subcellular localization of Nap1. In a previous study (16), a putative NES-like sequence was pointed out in Drosophila Nap1, leading to a hypothesis that Nap1 may be imported into and exported from the nucleus. To study this point further, we looked for NES-like sequences in Nap1 homologs of other species. We found NES-like sequences in Nap1's in humans, nematodes, and yeasts (Fig. 1A). To examine whether the putative NES is functionally active, we constructed expression vectors for Flag-tagged yNap1ΔNES mutant protein, which lacks the region between amino acid positions 88 to 102 encompassing the NES-like sequence, and two kinds of amino acid substitution mutant proteins in the NES-like sequence, Flag-yNap1 (L88A V92A) and Flag-yNap1(L99A L102A) (Fig. 1). We tested the subcellular localization of these yNap1 mutant proteins in mammalian (Fig. 2) and yeast (see Fig. 4) cells. Flag-tagged proteins were visualized by using anti-Flag antibody combined with a fluorescein-conjugated secondary antibody, whereas nuclei were visualized by staining DNA with DAPI. WT Flag-yNap1 is predominantly localized in the cytoplasm (Fig. 2a and e). In contrast, Flag-yNap1ΔNES and Flag-yNap1 (L99A L102A) are predominantly localized in the nucleus (Fig. 2b, c, d, f, g, and h). We could not see expression of Flag-yNap1 (L88A V92A) for unknown reasons (data not shown). These results show that the NES-like sequence is functional and responsible for the export of yNap1 to the cytoplasm. Therefore, it is quite possible that the cytoplasmic localization of yNap1 is not static and yNap1 is dynamically shuttled between the nucleus and the cytoplasm.
FIG. 1.
(A) NES-like sequences of Nap1. The NES consensus sequence is taken from reference 29. The NES-like sequences of yNap1, nematode Nap1 (nNap1), Drosophila Nap1 (dNap1), and human Nap1 (hNap1) are aligned. Amino acid substitution mutants in yNap1 proteins used in this study are also indicated (L88A V92A and L99A L102A). Numbers indicate amino acid positions in the yNap1 protein. (B) yNap1 mutant proteins used this study are schematically summarized. An NES-like sequence (88 to 102), NLS-like sequence (257 to 278), and acidic regions (168 to 180, 324 to 336, and 404 to 417) are indicated.
FIG. 2.
Subcellular localization of Flag-yNap1 derivatives in mammalian cells. HeLa cells grown on coverslips were transfected with expression plasmids for Flag-yNap1 (a and e), Flag-yNap1ΔNES (b, c, f, and g), Flag-yNap1 (L99A L102A) (d and h), Flag-yNap1ΔC1 (i and k), or Flag-yNap1ΔNESΔC1 (j and l). For cells expressing Flag-yNap1ΔNES, two independent optical fields are shown. Cells were fixed, stained with DAPI (a to d, i, and j) or anti-Flag antibody (e to h, k, and l), and then examined under a fluorescence microscope and analyzed by a decombolution system (Zeiss).
FIG. 4.
Subcellular localization of yNap1 derivatives in yeast cells. (A) Exponentially growing cells expressing WT Flag-yNap1, Flag-yNap1ΔNES, or Flag-yNap1 (L99A L102A) were fixed, and DNA and Flag-tagged proteins were stained with DAPI and anti-Flag antibody, respectively. (B) Amounts of Flag fusion proteins in transformed cells. DK168 cells were transformed with pRS316 (lane 1), pRS316-Flag-yNap1 (lane 2), pRS316-Flag-yNap1ΔNES (lane 3), or pRS316-Flag-yNap1 (L99A L102A) (lane 4). Exponentially growing transformants were lysed, and lysates were subjected to Western blot analysis with anti-Flag antibody. DK168 cells were separately transformed with pYES2 (lanes 5 and 6), pYES2-Flag-yNap1 (lanes 7 and 8), or pYES2-Flag-yNap1ΔNES (lanes 9 and 10). Lysates prepared from exponentially growing transformants in either SD (lanes 5, 7, and 9) or SG (lanes 6, 8, and 10) were subjected to Western blot analysis.
Subcellular localization of yNap1 that loses the histone-binding activity.
An interesting question is raised as to how Nap1 is imported into the nucleus. Several lines of the evidence suggest that histones could be involved in the nuclear import of Nap1. First, Nap1 interacts with H2A/H2B dimers in the cytoplasm (7, 16). Second, the N-terminal regions of histones act as nuclear localization signals (NLS) (39). Third, ectopically expressed NAP-2, one of the Nap1 family proteins, is translocated from the cytoplasm to the nucleus at the S phase, when histones are most actively synthesized in the cytoplasm and imported into the nucleus (47). Fourth, histone H2A/H2B forms a ternary complex with Kap114 and Nap1, and recently, it was shown that Kap114 is involved in the nuclear import of Nap1 (38). We therefore examined a possible relationship between histone binding and nuclear import of yNap1. We constructed vectors for the expression of the yNap1 mutant proteins yNap1ΔC1 and yNap1ΔNESΔC1, both of which lack the carboxy-terminal region of WT yNap1 and yNap1ΔNES (Fig. 1B), respectively. The deleted region contains a highly acidic stretch and a small region homologous to TAF-I (24). As previously reported, yNap1ΔC1 has no stimulatory activity in nucleosome formation in vitro (11). Since histone binding is one of the most basic activities for histone chaperones and the binding ability is likely to depend on the acidity of histone chaperones, it is important to examine whether yNap1ΔC1 binds to histones. To this end, we performed GST pull-down assays with GST-Nap1 proteins and core histones purified from HeLa cells. As shown in Fig. 3, WT yNap1 and yNap1ΔNES bind to core histones (Fig. 3, lanes 7 and 8). The histone-binding activity of yNap1ΔNES is approximately 80% of that of WT yNap1. It is clear that the amount of core histones binding to yNap1ΔC1 is less than that of WT yNap1 (Fig. 3, lanes 7 and 9). The ability of yNap1ΔC1 to bind to H2A/H2B is almost completely lost. Therefore, it is concluded that the C-terminal region is involved in H2A/H2B binding.
FIG. 3.
Interactions between yNap1 derivatives and core histones. GST pull-down assays were performed with purified core histones and GST-tagged recombinant yNap1 proteins. Lysates prepared from E. coli cells expressing GST (lanes 2 and 6), WT GST-yNap1 (lanes 3 and 7), GST-yNap1ΔNES (lanes 4 and 8), and GST-yNAP-1ΔC1 (lanes 5 and 9) were incubated with (lanes 6 to 9) or without (lanes 2 to 5) 2 μg of core histones purified from HeLa cells as described previously (44) and then mixed with glutathione-Sepharose beads. Beads were washed with a buffer containing 150 mM NaCl and 0.5% NP-40 (lanes 2 to 9). Core histone (lane 1, 0.4 μg) and the bound proteins (lanes 2 to 9) eluted by an SDS-PAGE loading buffer were separated by SDS-15% PAGE and visualized by CYPRO Orange staining. The positions of each core histone component are indicated. The amounts of the bound histones were measured by National Institutes of Health imaging.
We next examined the involvement of the C-terminal region of yNap1 in its nuclear import. Flag-yNap1ΔNES is clearly localized in the nucleus (Fig. 2b, c, f, and g). It is assumed that the subcellular localization could be changed by truncation of the C-terminal region from Flag-yNAP1ΔNES. The ectopically expressed Flag-yNap1ΔC1 and Flag-yNap1ΔNESΔC1 are localized in the cytoplasm in HeLa cells (Fig. 2i, j, k, and l). Therefore, it is likely that the C-terminal region that is important for H2A/H2B binding is involved in the nuclear import of Nap1. To more precisely examine the subcellular localization of yNap1, we employed a novel microscope system, termed laser scanning cytometer (LSC2; Olympus) and measured the fluorescent intensity of the nucleus and of the peripheral area of the nucleus. To minimize the background staining, we used EGFP-tagged yNap1's for transient expression in HeLa cells. As shown in Table 1, a majority of cells (89.1%) expressing EGFP-yNap1 showed the dominant EGFP signals in the cytoplasm. In 10.1% of the cells, EGFP signals were dominant in the nucleus. In sharp contrast, among cells expressing EGFP-yNap1ΔC1, we could not observe a cell in which the nuclear fluorescence is brighter than the cytoplasmic fluorescence. Thus, it is concluded that WT yNap1 is dominantly localized in the nucleus and that the C-terminal H2A/H2B binding region of yNap1 is important for the nuclear localization of yNap1. EGFP-yNap1ΔNES is a nuclear dominant in 89.2% of total cells expressing EGFP-yNap1ΔNES. This confirms that the NES-like sequence is involved in exclusion of yNap1 from the nucleus. Among cells expressing EGFP-yNap1ΔNESΔC1, we could not see the cell that is dominantly stained in the nucleus (data not shown). Again, the C-terminal region is likely to be essential for the nuclear localization of yNap1.
TABLE 1.
Quantitative analysis of cells expressing nuclear and cytoplasmic Nap1 proteinsa
| Protein | % of cells in nucleus | % of cells in cytoplasm | Total cell no. |
|---|---|---|---|
| WT yNap1 | 10.2 | 89.1 | 356 |
| yNap1ΔC | 0 | 100 | 212 |
| yNap1ΔNES | 89.2 | 10 | 120 |
EGFP-yNap1 and its derivatives were transiently expressed in HeLa cells. Cells were fixed with 4% paraformaldehyde, stained with DAPI, and observed under a laser scanning cytometer (LSC2; Olympus). The integral intensity value of fluorescence in the nucleus and that in the nuclear periphery area were measured, and the former value was plotted versus the latter value. Cells were sorted into nucleus- or cytoplasm-staining dominant populations by checking each area of each cell on the graph. The number of nucleus-staining dominant cells and that of cytoplasm-staining dominant cells are indicated as percentages of the total scanned cell number. This table does not include the results of the experiment with yNap1ΔNESΔC1, since the scanned cell number was not sufficient to correctly evaluate the result. However, as described in the text, we could not detect nucleus-staining dominant cells among cells expressing yNap1ΔNESΔC1 as far as we searched in the sample.
Nucleocytoplasmic shuttling of yNap1 is involved in mitotic progression.
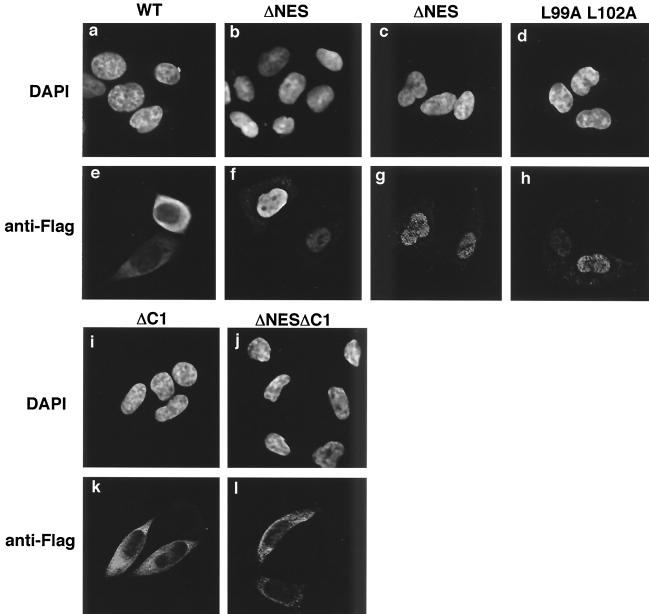
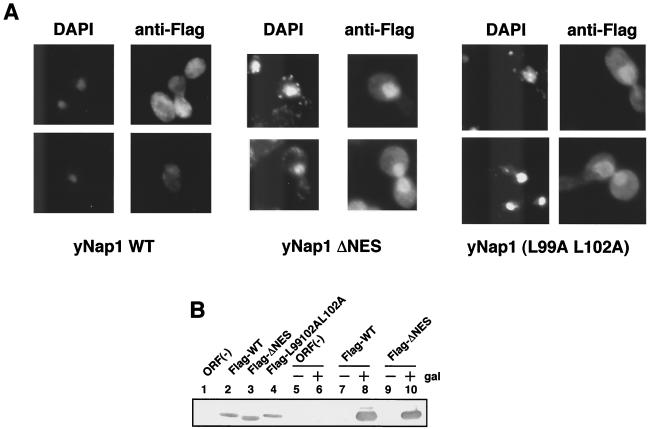
We tried to examine the physiological role of nucleocytoplasmic transport of yNap1 by genetic complementation assays with the DK168 strain (ΔNAP1 ΔCLB1 ΔCLB3 ΔCLB4). DK168 shows a temperature-sensitive growth and an elongated bud phenotype below the permissive temperature (25, 26). Before carrying out complementation assays, we first examined the subcellular localization of Flag-yNap1 mutant proteins in yeast DK168 cells (Fig. 4A). Flag-yNap1 is dominantly localized in the cytoplasm, whereas Flag-yNap1ΔNES and Flag-yNap1 (L99A L102A) are localized dominantly in the nucleus (Fig. 4A). Therefore, it is confirmed that the mechanism of nuclear import and export of yNap1 in yeast cells are similar to that in mammalian cells.
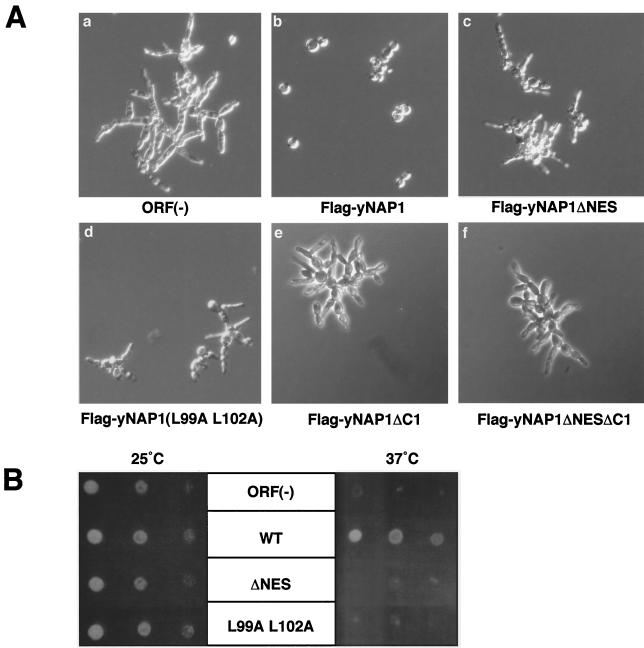
It is suggested that the elongated bud phenotype of DK168 is caused by the prolonged mitotic delay followed by the inability of Gin4 kinase phosphorylation (3). Next, we performed complementation experiments with various yNap1 derivatives by using the DK168 strain. WT Flag-yNap1 complements the elongated bud phenotype (Fig. 5A, b) and the temperature-sensitive (ts) phenotype (Fig. 5B). In sharp contrast, Flag-yNap1ΔNES and Flag-yNap1 (L99A L102A) cannot complement the elongated bud phenotype (Fig. 5A, c and d) and the ts phenotype (Fig. 5B). The artificial mutations sometimes affect protein stability so that it is possible that yNap1ΔNES and yNap1 (L99A L102A) could not complement the phenotypes due to its instability and low abundance. Western blot analyses of lysates with anti-Flag antibody have revealed that Flag-yNap1, Flag-yNap1ΔNES, and Flag-yNap1 (L99A L102A) driven by the native promoter are equally expressed (Fig. 4B, lanes 2 to 4). Therefore, cytoplasmic localization and/or nucleocytoplasmic shuttling of yNap1 is required for the putative mitotic function of yNap1. To distinguish these possibilities, we performed further complementation experiments with Flag-yNap1ΔC1 and Flag-yNap1ΔNESΔC1, which are predominantly localized in the cytoplasm (Fig. 2k and l). Flag-yNap1ΔC1 and Flag-yNap1ΔNESΔC1 do not complement the elongated bud phenotype (Fig. 5A, e and f). As these mutant proteins are localized in the cytoplasm and the C-terminal region is important for the nuclear import of Nap1, it is likely that nucleocytoplasmic shuttling, but not cytoplasmic localization, of yNap1 is important for the mitotic progression.
FIG. 5.
Complementation of a NAP1 disruptant with yNAP1 derivatives. The morphological change (A) and ts phenotype (B) of a NAP1-disrupted strain, DK168, were examined when DK168 was transformed with an empty vector [ORF(−)] (a) or expression vectors for Flag-yNap1 WT (WT) (b), Flag-yNap1ΔNES (ΔNES) (c), Flag-yNap1(L99A L102A) (L99A L102A) (d), Flag-yNap1ΔC1 (e), or Flag-yNap1ΔNESΔC1 (f). (A) The morphologies of exponentially growing transformants were observed under a phase microscope. (B) The ts phenotypes during growth were examined by incubation of serially diluted cell suspension spots at permissive (25°C) and nonpermissive (37°C) temperatures.
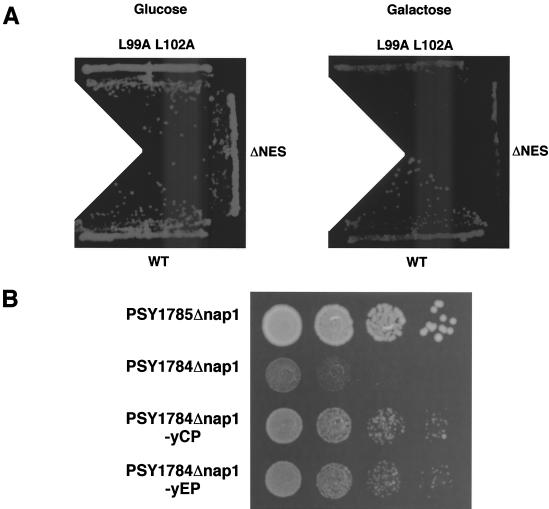
Overexpressed NES-deficient yNap1 mutants inhibit normal growth in the Clb2-dependent strain.
Nucleocytoplasmic shuttling could be divided into two steps, that is, nuclear import and export. To elucidate the molecular mechanisms of the role of Nap1, it is necessary to clarify whether either one of the two steps or both steps are required for the mitotic function of Nap1. As NES-deficient Nap1's do not complement the mitotic function (Fig. 5), it is likely that the nuclear export of Nap1 is essential for its function. However, it is possible that the loss of the NES function reduces the level of nuclear import of Nap1, possibly due to the decrease of recycling of Nap1 for repeated nuclear import processes by the loss of its nuclear export. To test this, we performed overexpression experiments of NES-deficient yNap1's. Since a part of yNap1ΔNES is localized in the cytoplasm, it is possible that overexpression of yNap1ΔNES may result in the increase of the cytoplasmic portion of yNap1ΔNES. Thus, the increased amount of NES-deficient yNap1 may overcome the reduced nuclear import. Overexpression of yNap1 and its derivatives was performed by using a galactose-inducible multicopy plasmid, pYES2 (Fig. 6A). The expression level of each galactose-inducible derivative was examined in comparison with that derived from a single-copy plasmid containing the native promoter by Western blot analysis with anti-Flag antibody (Fig. 4B, lanes 5 to 10, and data not shown). It is shown that the expression level in the galactose-inducible derivatives tested here was at least five times or more than that derived from the native promoter. Under these conditions, the growth rate of each strain was observed. Overexpression of yNap1ΔNES and yNap1 (L99A L102A) could not complement the ts and elongated bud phenotypes (data not shown). To be more precise, the slight growth inhibition by overexpression of yNap1ΔNES or yNap1 (L99A L102A) was observed even at the permissive temperature (Fig. 6A). yNap1ΔNES appears to inhibit cell growth more potently than yNap1 (L99A L102A). Possibly, yNap1 (L99A L102A), but not yNap1ΔNES, may retain a weak NES function, although they both show similar subcellular localizations (Fig. 2 and 4) and both are unable to complement the WT function (Fig. 5). These results show that the increased nuclear import of NES-deficient Nap1 is still insufficient for mitotic function of Nap1. Furthermore, the nuclear accumulations of NES-deficient Nap1's have a trans dominant-negative effect for mitotic progression. Therefore, it is strongly suggested that the nuclear export of Nap1 is essential for the mitotic function of Nap1.
FIG. 6.
Genetic analyses of NAP1. (A) Effect of overexpression of NES-deficient yNap1 in a Clb2-dependent strain. DK168 cells were transformed with pYES2-Flag-yNap1 (WT), pYES2-Flag-yNap1ΔNES (ΔNES), or pYES2-Flag-yNap1 (L99A L102A) (L99A L102A). Single colonies on an SD (without Ura) plate were isolated, and well-grown cultures were streaked onto SD (without Ura) and SG (without Ura) plates and incubated at 25°C for 2 and 3 days, respectively. (B) Genetic interaction between NAP1 and KAP114. Yeasts (PSY1785Δnap1 = KAP114 nap1::LEU2; PSY1784Δnap1 = kap114::HIS3 nap1::LEU2; PSY1784Δnap1-yCP = kap114::HIS3 nap1::LEU2 carrying a low copy number of NAP1; PSY1784Δnap1-yEP = kap114::HIS3 nap1::LEU2 carrying a high copy number of NAP1) fully grown in liquid media were diluted to OD600s of 0.1 with distilled water. Then 1/10 serially diluted cultures were prepared from the 0.1-OD600 cell suspensions in distilled water, and 5 μl of each dilution was grown on YPD agar plates for 2 days at 30°C.
Genetic interaction between NAP1 and KAP114.
We further examined the effect of the balance between nuclear import and export on the Nap1 function. As Flag-yNap1ΔC1 did not complement the mitotic function of Nap1 and the C-terminal region was shown essential for nuclear import of Nap1, it is possible that not only nuclear export but also nuclear import is important for the Nap1 function. For instance, it is possible that the loss of nuclear import causes reduced nuclear export, since nuclear import is the primary step for nuclear export. To know the effect of nuclear import of Nap1 on the Nap1 function, we examined a genetic interaction between NAP1 and KAP114, which is involved in the nuclear import of Nap1 (38) and other proteins, including TATA-binding protein and histone H2A/2B (37, 46). It has been reported that disruption of either NAP1 or KAP114 does not show any cell growth defect (25, 26, 37, 46). The NAP1 and KAP114 double-gene-disrupted strain PSY1784Δnap1 shows slower growth than the KAP114-positive strain PSY1785Δnap1 (Fig. 6B). The decreased growth rate caused by NAP1 gene disruption was partially complemented by the NAP1 gene with both low- and high-copy-number plasmids. These results indicate the genetic interaction between these two genes. Disruption of KAP114 shows the reduced level of nuclear import of some proteins, such as histone H2A/2B and TATA-binding protein (37, 46), so that the genetic interaction between NAP1 and KAP114 suggested that Nap1 has some functions in nuclear import which is important for the cell growth.
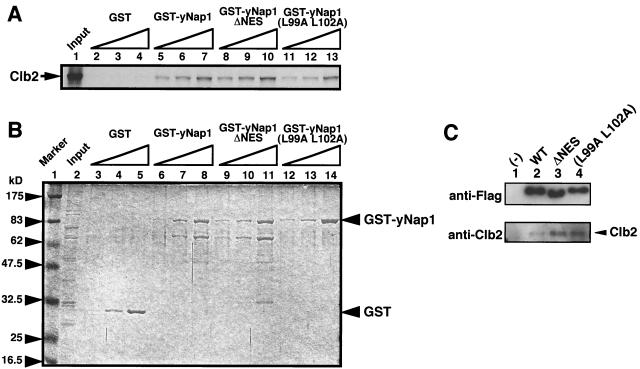
Interaction of NES-deficient yNap1 proteins with Clb2.
Clbs play key roles in mitotic progression (10). It has been noted that the physical and genetic interactions between Clb2 and yNap1 have been indicated (25, 26). As clb2ts mutant shows the elongated bud phenotype at nonpermissive temperatures, it is likely that Nap1 regulates some Clb2 function through a direct interaction. Thus, we examined the Clb2-binding ability of yNap1 mutant proteins by GST pull-down assays. Clb2 was prepared by in vitro transcription-translation reactions in the presence of [35S]methionine. GST-fused WT yNap1, but not GST alone, interacts with Clb2 in a dose-dependent manner (Fig. 7A, lanes 2 to 7), which is in good agreement with the previous result (3). yNap1ΔNES and yNap1 (L99A L102A) also interact with Clb2 as efficiently as WT yNap1 (Fig. 7, lanes 8 to 13). Therefore, the mutation in the NES sequence does not affect the interaction ability of Nap1 to Clb2.
FIG. 7.
Interaction between yNap1 and Clb2. (A) Fifty nanograms (lanes 2, 5, and 8), 150 ng (lanes 3, 6, and 9), or 500 ng (lanes 4, 7, and 10) of GST (lanes 2 to 4), GST-yNap1 (lanes 5 to 7), GST-yNap1ΔNES (lanes 8 to 10), or GST-yNap1 (L99A L102A) (lanes 11 to 13) was mixed with glutathione beads and incubated with in vitro-translated [35S]methionine-labeled Clb2 (lane 1, input). Bound proteins were separated by SDS-10% PAGE and visualized by autoradiography. (B) The amount of loaded GST proteins used in panel A is shown by CBB R-250 staining. (C) Yeast cell lysates prepared from DK168::pYES2 (lane 1), DK168::pYES-Flag-yNap1 (lane 2), DK168::pYES-Flag-yNap1ΔNES (lane 3), or DK168::pYES-Flag-yNap1 (L99A L102A) (lane 4) were precipitated with anti-Flag antibody-conjugated agarose beads, and bound proteins were separated by SDS-7.5% PAGE and stained with anti-Flag and anti-Clb2 antibodies.
Next, we examined the in vivo Clb2 interacting abilities of WT yNap1, yNap1ΔNES, and yNap1 (L99A L102A) by coimmunoprecipitation assays (Fig. 7C). Flag-tagged WT yNap1, yNap1ΔNES, and yNap1 (L99A L102A) proteins were expressed in DK168 strains by using the Gal1 promoter. Flag-tagged WT yNap1, yNap1ΔNES, and yNap1 (L99A L102A) proteins were precipitated by anti-Flag antibody (Fig. 7, upper panel). Clb2 was coprecipitated with WT Flag-yNap1, Flag-yNap1ΔNES and Flag-yNap1 (L99A L102A) (Fig. 7, lower panel). Therefore, the interaction between Nap1 and Clb2 is not dependent on either the NES-like sequence or the subcellular localization.
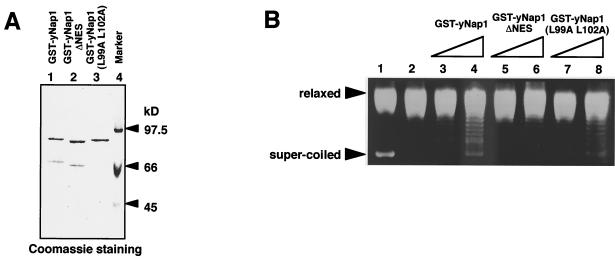
Nucleosome assembly activity of yNap1 mutant proteins.
A question is raised whether the mitotic function of yNap1 is correlated with its nucleosome assembly activity. We next examined the nucleosome assembly activity of NES-mutated yNap1 proteins (Fig. 8B) by using purified recombinant GST-yNap1 proteins (Fig. 8A). In the supercoiling assay, a protein containing the nucleosome assembly activity facilitates nucleosome formation with core histones and a relaxed circular plasmid DNA in the presence of topoisomerase I. Removal of proteins after the reaction gives a supercoiled DNA. A relaxed DNA (Fig. 8B, lane 9) generated by treatment of a supercoiled DNA with E. coli topoisomerase I (Fig. 8B, lanes 1 and 2) was mixed with core histones that had been preincubated with yNap1 proteins. The supercoiled DNA was formed by GST-yNap1 in a dose-dependent manner (Fig. 8B, lanes 3 and 4). GST-yNap1 (L99A L102A) has the same level of nucleosome assembly activity as GST-yNap1 (Fig. 8B, lanes 3, 4, 7, and 8). Therefore, the substitution mutations in the NES region of yNap1 do not affect the nucleosome assembly activity in vitro. GST-yNap1ΔNES has less, but significant, activity than WT GST-yNap1 (Fig. 8B, lanes 3, 4, 5, and 6). Densitometric scanning revealed that the activity of GST-yNap1ΔNES is approximately 50% of that of WT GST-yNap1 (Fig. 8B, lanes 4 and 5). The low nucleosome assembly activity of yNap1ΔNES could be due to its low histone-binding ability, as shown in Fig. 2. We could not find any strict correlation between the mitotic complementation ability and the nucleosome assembly activity.
FIG. 8.
Nucleosome assembly activities of yNap1 mutants. (A) Purified GST-fused yNap1 proteins used for nucleosome assembly assays. Four hundred nanograms of each protein (lane 1, GST-yNap1; lane 2, GST-yNap1ΔNES; lane 3, GST-yNap1 [L99A L102A]) was separated by SDS-7.5% PAGE and stained with CBB R-250. (B) Supercoiling assays. Nucleosome assembly reactions (lanes 2 to 8) were carried out with relaxed circular DNA and purified core histones in the absence (lane 2) or presence of GST-yNap1 (lanes 3 and 4), GST-yNap1ΔNES (lanes 5 and 6), or GST-yNap1 (L99A, L102A) (lanes 7 and 8). After the reaction, DNA in each reaction mixture was deproteinized, separated by 1% agarose gel electrophoresis, and visualized by staining with Cyber gold (Molecular Probes). A mixture of fully supercoiled DNA and relaxed DNA is also shown in lane 1.
DISCUSSION
Our analyses described in this paper started with a question of whether Nap1 is stably localized in the cytoplasm because the nucleosome assembly activity should be a nuclear function. By examining the subcellular localization and biochemical activities of mutant Nap1 proteins, we could propose a novel regulatory role of Nap1 in mitotic progression through its nucleocytoplasmic shuttling. We have shown that yNap1 is localized in the nucleus when the NES-like sequence is deleted or mutated (Fig. 2 and 4A). The histone binding was found to be important for nuclear import of yNap1 (Fig. 2 and 3). Since NES-deficient Nap1's had no complementing activity for mitotic function of Nap1 (Fig. 5), it is suggested that nucleocytoplasmic shuttling of Nap1 is required for its function. Overexpression of NES-deficient Nap1's did not complement the mitotic function of Nap1 (Fig. 6). Therefore, it is concluded that nuclear import of Nap1 is insufficient and the nuclear export is essential for Nap1 function.
Nucleocytoplasmic shuttling of Nap1.
As yNap1 is predominantly localized in the cytoplasm (26) (Fig. 4A) and Gin4, which is one of the Nap1 interactors which plays a critical role in mitosis, is localized in the bud neck, it is reasonably assumed that Nap1 acts as part of the mitotic function in the cytoplasm. However, our results suggest that nucleocytoplasmic shuttling, but not stable cytoplasmic localization, of Nap1 is required for the mitotic function of Nap1. First, C-terminal deletion mutants (Flag-yNap1ΔC1 and Flag-yNap1ΔNESΔC1), which are not effectively imported into the nucleus, thereby staying in the cytoplasm, have no complementing activity (Fig. 5A). Second, overexpression of NES mutant proteins is still insufficient to complement the DK168 phenotypes (Fig. 6A). Ectopically expressed NES-deficient yNap1's are predominantly localized in the nucleus, but a significant level of cytoplasmic staining is also observed (Fig. 4A). Therefore, the cytoplasmic amount of the mutant yNap1 could be increased, as the total amount of mutant yNap1 proteins is increased by its overexpression. These observations strongly suggest that the constitutive cytoplasmic existence of Nap1 is not enough and that nucleocytoplasmic shuttling is required for the mitotic function of Nap1.
Nuclear import of Nap1.
As discussed above, the nuclear export of Nap1 is important for the mitotic function of Nap1. However, we could not rule out the possibility that nuclear import of Nap1 is also critical because nuclear import is the primary step for nuclear export. We addressed this point by using KAP114 mutants. Kap114 physically interacts with Nap1 and is involved in the nuclear import of TATA-binding protein (37, 46) and histone H2A/2B (38). Kap114 and Nap1 make a complex with H2A/2B (38). The nuclear import of Nap1 is shown to be dependent on its histone-binding domain (Fig. 2). Therefore, it is likely that Nap1 assists the Kap114 function in nuclear import of some proteins as a nuclear import partner. Indeed, a double gene disruption (kap114 and nap1) resulted in a decreased growth rate phenotype (Fig. 6B), although a single gene disruption of either one of these two genes did not give any growth defect. This synthetic phenotype between two genes suggests that Nap1 may be involved in nuclear import of some nuclear proteins.
Nuclear export of Nap1.
Experimental results strongly suggest that the nuclear export of Nap1 is essential for its mitotic function. Overexpression of NES-deficient Nap1s caused the cell growth inhibition (Fig. 6A). Such growth inhibition was not observed when they were expressed from the authentic yNAP1 promoter on a single-copy plasmid (Fig. 5B). A yNap1 itself may be inhibitory when stably localized in the nucleus. On this line, it is noted that NAP1 has the chromatin disruption activity (24) as well as the nucleosome assembly activity (36, 54). The cell cycle is likely to be inhibited at the mitotic phase because nuclei of these cells were localized at the bud neck (data not shown). Such inhibition was not observed in NAP1-positive strains, a WT YPH499 strain, and a DK166 strain (ΔCLB1 ΔCLB3 ΔCLB4) (data not shown). Under the same conditions, yNap1-NLS, which is a fusion yNap1 protein with simian virus 40 NLS at the C terminus of yNap1 and predominantly localized in nucleus, could complement the DK168 phenotypes and did not have any inhibitory effect on the cell growth (data not shown). Thus, the inhibitory effect is suppressed by endogenous Nap1. These observations suggest that nuclear yNap1 does not have a mitotic cell arrest ability, but an export-deficient Nap1 that accumulates in the nucleus provides the inhibitory effect on cell growth.
An alternative explanation for the growth inhibition would be that the yNap1 mutants in the nucleus may anchor a yNap1 interactor(s) that exerts its normal function transiently in the nucleus and is needed to be exported to the cytoplasm. The target of Nap1 during its nuclear export is an open question. Several lines of evidence suggest that Clb2 is one of the candidates. First, Clb2 is also a nucleocytoplasmic shuttling protein and is dominantly localized in the nucleus, in particular during the G2/M phase (12). Second, an NES mutation of Clb2 causes an elongated bud phenotype (12). Therefore, we examined the difference of Clb2 localization in the absence and presence of NAP1. We confirmed that Nap1 and NES-mutated Nap1s bind to Clb2 (Fig. 7). We tried to detect the difference of Clb2-EGFP localization in NAP1-positive and -negative strains. However, the difference is not so significant between two strains.
Although systematic analyses are needed, we would like to propose the term shuttling chaperone for Nap1. The shuttling chaperone may modulate nuclear import and/or export of proteins rather than itself. A variety of Nap1-interacting proteins are being identified, and their intracellular localization patterns are closely related to the regulation of mitotic progression. It is possible that these interacting proteins are also targets of the Nap1 shuttling chaperone activity. Thus, it is interesting and necessary to examine the involvement of Nap1 in the subcellular localization of these interacting proteins and to ask whether their aberrant localization eliminates the proper mitotic progression.
Regulation of Nap1 localization.
We have discussed the function of Nap1 associated with its nucleocytoplasmic shuttling. It is interesting how the shuttling is regulated. It has long been thought that Nap1 is localized in the cytoplasm in a variety of species. Since the NES-like sequence in yNap1 is well conserved among species, the conclusion from our analyses with yeast Nap1 is probably applicable for other species. It is reported that the fly Nap1 is temporally localized in the nucleus at some stages during development (17). We have shown that EGFP-Nap1 is dominantly detected in cytoplasm, whereas in 10.1% of cells expressing EGFP-yNap1, the signal is dominantly detected in nucleus (Table 1). These observations point out that Nap1 could be localized in the nucleus under a certain condition(s). It is possible that the rate of nuclear import and export is varied depending on cell conditions. Based on the histone-binding activity and nucleosome assembly activity of Nap1, it is assumed that Nap1 functions as a chaperone for histones during nuclear import and in the cytoplasm as well as at the deposition to DNA. Thus, the rate of the nuclear import of Nap1 might be increased at the S phase, since histones are most actively synthesized at the S phase. The modification may also be involved in the regulation of nuclear transport and nuclear function, if any, of Nap1. It is reported that a cytoplasmic fraction of Nap1, but not the nuclear fraction, is possibly phosphorylated by CK2 and that the histone-binding activity is stimulated by phosphorylation of Nap1 by CK2 (48).
The balance of nuclear import and export of Nap1 may be important for the nuclear function of Nap1. Nap1 could be involved in not only de novo assembly of nucleosomes but also structural change of chromatin, although we do not know an exact function of Nap1 in the nucleus. Nap1 family proteins are shown to be involved in transcription regulation (20, 24, 49). Our preliminary experiments with one-hybrid assays indicated that Nap1 stimulates the transcription from chromatin under a certain condition (data not shown). To consider the nuclear function of Nap1, it is worthwhile to note that the nuclear localizations of NES-mutated yNap1s show speckled patterns in the nuclei (Fig. 2g, h, and j). Determination of the nuclear domain may support understanding of the nuclear function of Nap1.
Acknowledgments
We thank Doug Kellogg for providing yeast strains and helpful discussion. kap114Δ strains were kindly provided by Steve Buratowski and Pamela A. Silver. We thank Yoshimasa Kiyomatsu (Olympus ProMarketing, Inc.) for permitting us to use the laser scanning cytometer and for technical support. We thank Yuki Yamaguchi for critical comments on the manuscript.
This work was supported by a grant-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by a grant from the Bioarchitect Research Project of RIKEN.
REFERENCES
- 1.Aalfs, J. D., and Kingston, R. E. 2000. What does ‘chromatin remodeling’ mean? Trends Biochem. Sci. 25:548-555. [DOI] [PubMed] [Google Scholar]
- 2.Adams, C. R., and R. T. Kamakaka. 1999. Chromatin assembly: biochemical identities and genetic redundancy. Curr. Opin. Genet. Dev. 9:185-190. [DOI] [PubMed] [Google Scholar]
- 3.Altman, R., and D. Kellogg. 1997. Control of mitotic events by Nap1 and the Gin4 kinase. J. Cell Biol. 138:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booher, R. N., R. J. Deshaies, and M. W. Kirschner. 1993. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 12:3417-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bortvin, A., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272:1473-1476. [DOI] [PubMed] [Google Scholar]
- 6.Bulger, M., T. Ito, R. T. Kamakaka, and J. T. Kadonaga. 1995. Assembly of regularly spaced nucleosome arrays by Drosophila chromatin assembly factor 1 and a 56-kDa histone-binding protein. Proc. Natl. Acad. Sci. USA 92:11726-11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, L., S. S. Loranger, C. Mizzen, S. G. Ernst, C. D. Allis, and A. T. Annunziato. 1997. Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry 36:469-480. [DOI] [PubMed] [Google Scholar]
- 8.Chien, C. T., P. L. Bartel, R. Sternglanz, and S. Fields. 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88:9578-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crevel, G., and S. Cotterill. 1995. DF 31, a sperm decondensation factor from Drosophila melanogaster: purification and characterization. EMBO J. 14:1711-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitch, I., C. Dahmann, U. Surana, A. Amon, K. Nasmyth, L. Goetsch, B. Byers, and B. Futcher. 1992. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 3:805-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujii-Nakata, T., Y. Ishimi, A. Okuda, and A. Kikuchi. 1992. Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J. Biol. Chem. 267:20980-20986. [PubMed] [Google Scholar]
- 12.Hood, J. K., W. W. Hwang, and P. A. Silver. 2001. The Saccharomyces cerevisiae cyclin Clb2p is targeted to multiple subcellular locations by cis- and trans-acting determinants. J. Cell Sci. 114:589-597. [DOI] [PubMed] [Google Scholar]
- 13.Ishimi, Y., J. Hirosumi, W. Sato, K. Sugasawa, S. Yokota, F. Hanaoka, and M. Yamada. 1984. Purification and initial characterization of a protein which facilitates assembly of nucleosome-like structure from mammalian cells. Eur. J. Biochem. 142:431-439. [DOI] [PubMed] [Google Scholar]
- 14.Ishimi, Y., M. Kojima, M. Yamada, and F. Hanaoka. 1987. Binding mode of nucleosome-assembly protein (AP-I) and histones. Eur. J. Biochem. 162:19-24. [DOI] [PubMed] [Google Scholar]
- 15.Ishimi, Y., and A. Kikuchi. 1991. Identification and molecular cloning of yeast homolog of nucleosome assembly protein I which facilitates nucleosome assembly in vitro. J. Biol. Chem. 266:7025-7029. [PubMed] [Google Scholar]
- 16.Ito, T., M. Bulger, R. Kobayashi, and J. T. Kadonaga. 1996. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 16:3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, T., J. K. Tyler, M. Bulger, R. Kobayashi, and J. T. Kadonaga. 1996. ATP-facilitated chromatin assembly with a nucleoplasmin-like protein from Drosophila melanogaster. J. Biol. Chem. 271:25041-25048. [DOI] [PubMed] [Google Scholar]
- 18.Ito, T., M. Bulger, M. J. Pazin, R. Kobayashi, and J. T. Kadonaga. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90:145-155. [DOI] [PubMed] [Google Scholar]
- 19.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, T., T. Ikehara, T. Nakagawa, W. L. Kraus, and M. Muramatsu. 2000. p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 14:1899-1907. [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, W., S. K. Nordeen, and J. T. Kadonaga. 2000. Transcriptional analysis of chromatin assembled with purified ACF and dNAP1 reveals that acetyl-CoA is required for preinitiation complex assembly. J. Biol. Chem. 275:39819-39822. [DOI] [PubMed] [Google Scholar]
- 24.Kawase, H., M. Okuwaki, M. Miyaji, R. Ohba, H. Handa, Y. Ishimi, T. Fujii-Nakata, A. Kikuchi, and K. Nagata. 1996. NAP-I is a functional homolog of TAF-I that is required for replication and transcription of the adenovirus genome in a chromatin-like structure. Genes Cells 1:1045-1056. [DOI] [PubMed] [Google Scholar]
- 25.Kellogg, D. R., and A. W. Murray. 1995. NAP1 acts with Clb2 to perform mitotic functions and to suppress polar bud growth in budding yeast. J. Cell Biol. 130:675-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellogg, D. R., A. Kikuchi, T. Fujii-Nakata, C. W. Turck, and A. W. Murray. 1995. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J. Cell Biol. 130:661-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kingston, R. E. 1997. Introduction of DNA into mammalian cells, p. 9.0.1-9.0.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 28.Kleinschmidt, J. A., and W. W. Franke. 1982. Soluble acidic complexes containing histones H3 and H4 in nuclei of Xenopus laevis oocytes. Cell 29:799-809. [DOI] [PubMed] [Google Scholar]
- 29.Kudo, N., H. Taoka, T. Toda, M. Yoshida, and S. Horinouchi. 1999. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J. Biol. Chem. 274:15151-15158. [DOI] [PubMed] [Google Scholar]
- 30.Künzler, M., T. Gerstberger, F. Stutz, F. R. Bischoff, and E. Hurt. 2000. Yeast Ran-binding protein 1 (Yrb1) shuttles between the nucleus and cytoplasm and is exported from the nucleus via a CRM1 (XPO1)-dependent pathway. Mol. Cell. Biol. 20:4295-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskey, R. A., B. M. Honda, A. D. Mills, and J. T. Finch. 1978. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275:416-420. [DOI] [PubMed] [Google Scholar]
- 32.LeRoy, G., A. Loyola, W. S. Lane, and D. Reinberg. 2000. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275:14787-14790. [DOI] [PubMed] [Google Scholar]
- 33.Li, M., D. Strand, A. Krehan, W. Pyerin, H. Heid, B. Neumann, and B. M. Mechler. 1999. Casein kinase 2 binds and phosphorylates the nucleosome assembly protein-1 (NAP1) in Drosophila melanogaster. J. Mol. Biol. 293:1067-1084. [DOI] [PubMed] [Google Scholar]
- 34.Longtine, M. S., C. L. Theesfeld, J. N. McMillan, E. Weaver, J. R. Pringle, and D. Lew. 2000. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4049-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marheineke, K., and T. Krude. 1998. Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. J. Biol. Chem. 273:15279-15286. [DOI] [PubMed] [Google Scholar]
- 36.McQuibban, G. A., C. N. Commisso-Cappelli, and P. N. Lewis. 1998. Assembly, remodeling, and histone binding capabilities of yeast nucleosome assembly protein 1. J. Biol. Chem. 273:6582-6590. [DOI] [PubMed] [Google Scholar]
- 37.Morehouse, H., R. M. Buratowski, P. A. Silver, and S. Buratowski. 1999. The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc. Natl. Acad. Sci. USA 96:12542-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosammaparast, N., C. S. Ewart, and L. F. Pemberton. 2002. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 21:6527-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosammaparast, N., K. R. Jackson, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and L. F. Pemberton. 2001. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J. Cell Biol. 153:251-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munakata, T., N. Adachi, N. Yokoyama, T. Kuzuhara, and M. Horikoshi. 2000. A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells 5:221-233. [DOI] [PubMed] [Google Scholar]
- 41.Nagata, K., H. Kawase, H. Handa, K. Yano, M. Yamasaki, Y. Ishimi, A. Okuda, A. Kikuchi, and K. Matsumoto. 1995. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc. Natl. Acad. Sci. USA 92:4279-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagata, K., F. Momose, and M. Okuwaki. 1999. Acidic molrcular chaperones: their involvement in viral genome replication and transcription. Recent Res. Dev. Virol. 1:559-597. [Google Scholar]
- 43.Nakagawa, T., M. Bulger, M. Muramatsu, and T. Ito. 2001. Multistep chromatin assembly on supercoiled plasmid DNA by nucleosome assembly protein-1 and ATP-utilizing chromatin assembly and remodeling factor. J. Biol. Chem. 276:27384-27391. [DOI] [PubMed] [Google Scholar]
- 44.Okuwaki, M., and K. Nagata. 1998. Template activating factor-I remodels the chromatin structure and stimulates transcription from the chromatin template. J. Biol. Chem. 273:34511-34518. [DOI] [PubMed] [Google Scholar]
- 45.Okuwaki, M., A. Iwamatsu, M. Tsujimoto, and K. Nagata. 2001. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol. 311:41-55. [DOI] [PubMed] [Google Scholar]
- 46.Pemberton, L. F., J. S. Rosenblum, and G. Blobel. 1999. Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J. Cell Biol. 145:1407-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez, P., D. Munroe, D. Prawitt, L. L. Chu, E. Bric, J. Kim, L. H. Reid, C. Davies, H. Nakagama, R. Loebbert, A. Winterpacht, M. J. Petruzzi, M. J. Higgins, N. Nowak, G. Evans, T. Shows, B. E. Weissman, B. Zabel, D. E. Housman, and J. Pelletier. 1997. Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics 44:253-265. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez, P., J. Pelletier, G. B. Price, and M. Zannis-Hadjopoulos. 2000. NAP-2: histone chaperone function and phosphorylation state through the cell cycle. J. Mol. Biol. 298:225-238. [DOI] [PubMed] [Google Scholar]
- 49.Shikama, N., H. M. Chan, M. Krstic-Demonacos, L. Smith, C. W. Lee, W. Cairns, and N. B. La Thangue. 2000. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell. Biol. 20:8933-8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu. Y., T. Akashi, A. Okuda, A. Kikuchi, and K. Fukui. 2000. NBP1 (Nap1 binding protein 1), an essential gene for G2/M transition of Saccharomyces cerevisiae, encodes a protein of distinct sub-nuclear localization. Gene 246:395-404. [DOI] [PubMed] [Google Scholar]
- 51.Spector, L. D. 1998. Cells: a laboratory manual, p. 106.3-106.7, Cold Spring Harbor Laboratory, Cold Spring Harbor, N. Y.
- 52.Surana, U., H. Robitsch, C. Price, T. Schuster, I. Fitch, A. B. Futcher, and K. Nasmyth. 1991. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell 65:145-161. [DOI] [PubMed] [Google Scholar]
- 53.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 54.Walter, P. P., T. A. Owen-Hughes, J. Cote, and J. L. Workman. 1995. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol. Cell. Biol. 15:6178-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]