Abstract
An AU-rich element (ARE) consisting of repeated canonical AUUUA motifs confers rapid degradation to many cytokine mRNAs when present in the 3′ untranslated region. Destabilization of mRNAs with AREs (ARE-mRNAs) is consistent with the interaction of ARE-binding proteins such as tristetraprolin and the four AUF1 isoforms. However, the association of the AUF1-mRNA interaction with decreased ARE-mRNA stability is correlative and has not been directly tested. We therefore determined whether overexpression of AUF1 isoforms promotes ARE-mRNA destabilization and whether AUF1 isoforms are limiting components for ARE-mRNA decay. We show that the p37 AUF1 isoform and, to a lesser extent, the p40 isoform possess ARE-mRNA-destabilizing activity when overexpressed. Surprisingly, overexpressed p37 AUF1 also destabilized reporter mRNAs containing a noncanonical but AU-rich 3′ untranslated region. Since overexpressed p37 AUF1 could interact in vivo with the AU-rich reporter mRNA, AUF1 may be involved in rapid turnover of mRNAs that lack canonical AREs. Moreover, overexpression of p37 AUF1 restored the ability of cells to rapidly degrade ARE-mRNAs when that ability was saturated and inhibited by overexpression of ARE-mRNAs. Finally, activation of ARE-mRNA decay often involves a translation-dependent step, which was eliminated by overexpression of p37 AUF1. These data indicate that the p37 AUF1 isoform and, to some extent, the p40 isoform are limiting factors that facilitate rapid decay of AU-rich mRNAs.
Most cytokine and many proto-oncogene mRNAs display short half-lives, which are conferred by an AU-rich element (ARE) in the 3′ untranslated region (UTR) that functions as a cytoplasmic destabilizing motif (reviewed in reference 20). As many as 8% of other human mRNA 3′ UTRs also contain AREs (2). The granulocyte-macrophage colony-stimulating factor (GM-CSF) ARE is prototypical; it consists of eight AUUUA pentamers, five of which are contiguous, which are sufficient to confer a short half-life to reporter mRNAs (40).
The mechanism for ARE-mediated mRNA decay is poorly understood. The ARE promotes mRNA deadenylation in vitro (16) and in vivo (42) and may stimulate mRNA-decapping activity (19). A number of proteins can bind various ARE sequences in vitro, including hnRNP proteins A1, A2, C, and L (21-23), but a role in controlling mRNA stability is unlikely (36). To date, three ARE-binding proteins have been shown to be involved in regulating rapid mRNA decay in vivo: HuR (33); tristetraprolin (TTP) (10); and the ARE- and poly(U)-binding and degradation factor, AUF1 (5), also known as hnRNP D (48). HuR stabilizes ARE-containing reporter mRNAs when ectopically overexpressed (15, 35), and antisense RNA knockdown of endogenous HuR expression increases the half-lives of certain ARE-containing mRNAs (ARE-mRNAs) (31, 38, 49). TTP is important for the destabilization of tumor necrosis factor and GM-CSF mRNAs, as shown in knockout mice (10, 45) and in tissue culture by ectopic-overexpression studies (27).
AUF1 consists of four isoforms of 37, 40, 42, and 45 kDa, generated by alternative splicing of a single mRNA transcript (48). The different isoforms possess different ARE binding affinities (25), with the core p37 isoform exhibiting the highest affinity for the c-fos ARE and p40 exhibiting the lowest (48). Several lines of evidence suggest a role for AUF1 in destabilization of ARE-mRNAs. (i) Purified p37 and p40 proteins accelerate c-myc mRNA turnover in an in vitro decay system (5). (ii) The affinity of p37 for ARE sequences in vitro correlates with mRNA-destabilizing activity in vivo (12, 44). (iii) Decay of ARE-mRNAs correlates with ubiquitination and degradation of AUF1 p37 by the 26S proteasome (28, 30). (iv) Examination of ARE function during hematopoietic differentiation showed that the p37 isoform had the highest destabilizing activity for reporter ARE-mRNA (32).
Given these reports, we sought to determine whether overexpression of any AUF1 isoforms promotes greater ARE-mRNA turnover. We found that ectopic overexpression of only the p37 AUF1 isoform specifically decreased the stability of AU-rich mRNAs. Surprisingly, ectopic overexpression of p37 destabilized a normally stable reporter mRNA containing a mutated ARE sequence that still retains significant AU richness in the 3′ UTR. These data indicate that mRNAs containing noncanonical AU-rich sequences are also potential targets for AUF1-mediated rapid degradation. Finally, we show that overexpression of p37 AUF1 relieves the translation dependence for targeted degradation of ARE-mRNAs and restores rapid degradation of ARE-mRNAs that accumulate due to their overexpression and saturation of the decay machinery. These results indicate that the abundance of p37 AUF1 is limiting for the control of short-lived-ARE-mRNA stability and is associated with translational activation of ARE-mRNA turnover.
MATERIALS AND METHODS
Plasmids.
Flag-AUF1 constructs were made by PCR amplification of each AUF1 isoform cDNA to generate a HindIII site upstream of the initiating AUG codon and an EcoRI site downstream of the termination codon. PCR products were purified by gel electrophoresis (Qiagen), digested with HindIII and EcoRI, and then ligated into pFlag-CMV-2 (Sigma) digested with the same enzymes. The resulting construct contains the Flag epitope fused in-frame to the N terminus of each AUF1 isoform. β-Galactosidase (β-Gal) reporter plasmids containing either the 62-bp ARE from the GM-CSF mRNA 3′ UTR or its stable GC mutant counterpart (40) have been previously described (28). The enhanced green fluorescent protein (GFP) expression plasmid was from Clontech. The DNA sequence of the 3′ UTR of the plasmid pEGFP-C3 (Clontech) was determined to validate the lack of significant AU richness. The 3′ UTR (nucleotides 1414 to 1550) is shown in Fig. 1A.
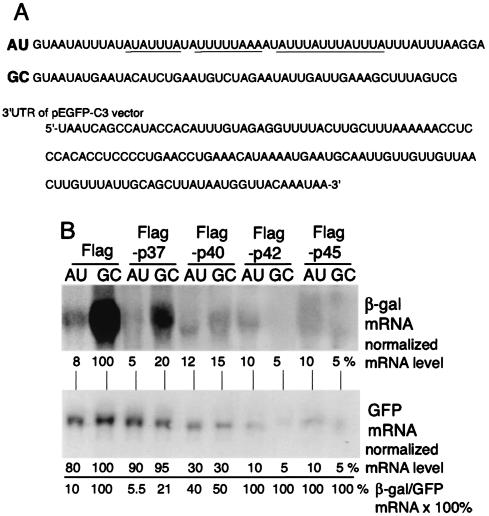
FIG. 1.
Steady-state reporter mRNA analysis with overexpression of Flag-AUF1 proteins. (A) ARE sequences cloned into the 3′ UTR of β-Gal reporter plasmids, depicted as RNA nucleotides with the canonical AUUUA motifs underlined. AU, wild-type ARE from the GM-CSF 3′ UTR; GC, AU sequence with G and C point mutations. Also shown is the 3′ UTR of the control plasmid pEGFP-C3. (B) CHO cells were cotransfected with 1.0 μg of either the Flag expression plasmid or each individual Flag-AUF1 isoform per 107 cells and plasmids encoding mRNAs containing the GM-CSF ARE (AU) or the GC mutant ARE (GC). Cells were transfected with 0.5 μg of a GFP expression vector that lacks a significant AU-rich 3′ UTR. At 24 h posttransfection, total RNA was isolated, and equal amounts were analyzed by Northern blot hybridization using probes prepared from the lacZ gene. A representative blot is shown. Autoradiograms were quantified by densitometry, and the ratio of the β-Gal/GFP mRNAs for the vector Flag sample (lane 1) was set to 100%. The ratio of normalized β-Gal to GFP mRNA indicates the percent abundance of β-Gal mRNA relative to that of the untreated vector sample.
Cell culture and treatments.
NIH 3T3, COS-1, HeLa, and 293T cells were maintained in Dulbecco's modification of Eagle's medium (DMEM) supplemented with 10% bovine calf serum and 50 μg of gentamicin/ml. Chinese hamster ovary (CHO) cells were grown in DMEM supplemented with 10% fetal bovine serum, nonessential amino acids, and 50 μg of gentamicin/ml. Cells were subcultured for 18 h prior to transfection with Lipofectamine Plus (Invitrogen) according to the manufacturer's directions. The cytotoxicity of transfected AUF1 protein expression was determined in three ways. Assay of lactate dehydrogenase (LDH) release into medium was routinely performed with an LDH detection kit (Invitrogen). In addition, apoptotic cell death was analyzed in some instances by MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay, and the absence of cytotoxicity was determined by measuring unimpaired cell proliferation.
RNA extraction, Northern blotting, half-life analysis, and quantification.
Actinomycin D (Sigma) was diluted in media to a final concentration of 5 μg/ml and added to cells for the times indicated in the figures. Cells were washed with 1× phosphate-buffered saline at 4°C, scraped from plates, and then collected in a microcentrifuge tube by a 30-s centrifugation at 5,000 × g and 4°C. Total RNA was extracted with 1 ml of Trizol reagent (Invitrogen) according to the manufacturer's directions. RNA was dissolved in formamide and separated by denaturing formaldehyde-agarose gel electrophoresis. RNA was transferred to GeneScreen Plus (NEN Lifescience) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), UV cross-linked to the membrane, hybridized to cDNA probes as indicated below, and labeled with [α-32P]CTP by using the Rediprime II kit (Amersham Bioscience) with PerfectHyb buffer (Sigma). Blots were washed in 2× SSC-1% sodium dodecyl sulfate (SDS), followed by 0.1× SSC-1% SDS, and then subjected to autoradiography. A GS800 digital densitometer (Bio-Rad) was used to quantify the signal from mRNAs with nonsaturated autoradiograms and Quantity One software (Bio-Rad). The β-Gal signal was normalized to the GFP signal to correct for transfection efficiency. Half-lives of mRNAs were calculated by plotting normalized β-Gal mRNA versus time; linear regression analysis was used to determine the slope of each decay curve.
Immunoblot analysis.
Anti-AUF1 antibodies were produced for these studies. Rabbits were immunized with glutathione S-transferase (GST)-p37 by following a standard protocol (Cocalico Biologicals, Inc.). Thrombin cleavage of the GST moiety produced a nearly full-length p37 protein, which was then coupled to Affigel-10 (Bio-Rad). Crude antiserum was passed over immobilized GST (Amersham-Pharmacia) and then affinity purified with p37 resin to produce an anti-AUF1 antibody. To produce cell lysates, cells were washed with ice-cold phosphate-buffered saline and then harvested with NP-40 lysis buffer (150 mM NaCl, 20 mM HEPES [pH 7.5], and 0.5% NP-40, supplemented with Complete protease inhibitor [Roche]). Then cells were disrupted by four passages through a 29-gauge needle, followed by incubation on ice for 20 min. This procedure disrupts nuclei and produces whole-cell lysates that contain soluble nuclear and cytoplasmic protein. Lysates were cleared by microcentrifugation at 4°C for 15 min. Protein concentrations were determined by the Bradford assay (Bio-Rad). Equal amounts of cell lysates were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to the membrane, and immunoblotting was performed using a primary antibody directed against AUF1, Flag, or tubulin. Immunoblots were developed by the enhanced chemiluminescence system.
AUF1-mRNA interaction analysis.
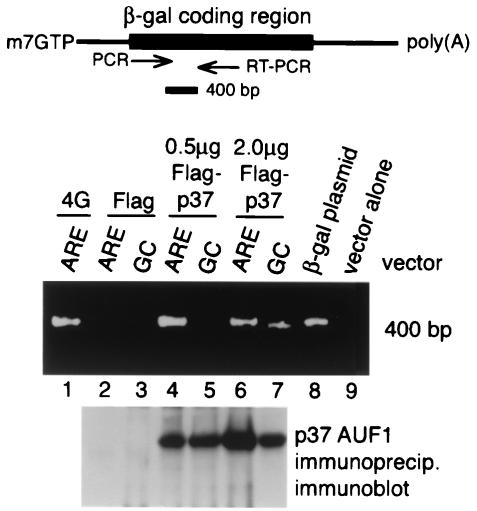
Ten-centimeter-diameter plates of 293 cells were transfected with 1.0 μg of the β-Gal ARE or GC reporter plasmid DNA and 0.5 or 2.0 μg of the Flag-p37 expression plasmid or 0.5 μg of the Flag control vector. At 48 h posttransfection, cell lysates were prepared as described above for immunoblot studies. For Flag-p37 immunoprecipitation, amounts of lysate were normalized for β-Gal mRNA levels and commercial Flag antibodies (Sigma) or eukaryotic initiation factor 4G (eIF4G) was immunoprecipitated with C-terminally directed antibodies prepared in the laboratory. Semiquantitative reverse transcription-PCR (RT-PCR) amplification of immunoprecipitated β-Gal mRNA was carried out with DNA oligonucleotide primers 5′-CCGCTGGGATCTGCCATTGTC-3′ and 5′-CGGTCAGCTGGAATTCCGCCG-3′. Following RT, 1% of the reaction product was used for 30 cycles of PCR. Products of RT-PCR were resolved by agarose gel electrophoresis, stained with ethidium bromide, and photographed.
Nucleotide sequence accession number.
The DNA sequence of the 3′ UTR of the plasmid pEGFP-C3 was assigned GenBank accession no. U57607.
RESULTS
Ectopic overexpression of p37 AUF1 selectively decreases the stability of AU-rich mRNAs.
The functional contribution of each AUF1 isoform to ARE-mRNA decay was examined by cotransfection of reporter plasmids encoding a β-Gal mRNA containing either the GM-CSF ARE or a mutant counterpart that contains G and C residues interspersed in the 3′ ARE (GC-mRNA) (Fig. 1A). Importantly, the G and C point mutations disrupt the canonical ARE sequence but some of the AU richness of this sequence is retained (Fig. 1A). The GC-mRNA typically has a long half-life (28, 40). CHO cells were cotransfected with β-Gal reporter plasmids and the Flag vector alone or with each Flag epitope-tagged AUF1 isoform. A GFP expression plasmid (pEGFP-C3) was included in each transfection to control for transfection efficiency. The GFP mRNA has several U-rich sequences but no ARE and only a small number of AU-rich sequences (Fig. 1A). Titration of each AUF1 isoform was conducted to avoid cytotoxicity. The absence of cell death was determined by a commercial assay for LDH, which is released into the medium by dying and dead cells; by measuring apoptotic death on the basis of MTT activity; and by measuring the proliferation of cells (data not shown). Total β-Gal mRNA levels were determined by Northern blot RNA hybridization analysis. ARE-mRNA reporters accumulated to a level approximately 10-fold lower than that of control GC-mRNA in the absence of cotransfection with an AUF1 isoform (Flag samples; Fig. 1B). Surprisingly, cotransfection of p37 AUF1 reduced the steady-state levels of both the ARE and GC β-Gal reporter mRNAs (to 5 and 20%, respectively, of the nontransfected control GC-mRNA level), but not that of the GFP mRNA, which lacks an AU-rich 3′ UTR (Fig. 1B). Importantly, the GC-mRNA retains some AU richness but does not contain the canonical GM-CSF ARE. Cotransfection of β-Gal reporters with the p40 AUF1 isoform reduced by 40 to 50% the abundance of ARE- and GC-mRNAs but also decreased the level of GFP reporter mRNA by 70% (Fig. 1A). These data suggest that overexpression of p37 AUF1 likely further destabilizes AU-rich mRNAs (Fig. 2 and 3). Cotransfection of ARE-mRNA or control GC-mRNA reporters with the p42 or p45 AUF1 isoform severely reduced β-Gal mRNAs (90 to 95%) but also decreased the levels of the GFP mRNA loading control similarly (Fig. 1B). Accordingly, GFP fluorescence was also reduced in cells cotransfected with the two larger AUF1 isoforms, but not the number of transfected cells (data not shown). These data suggest that the two larger AUF1 isoforms, when overexpressed, disrupt the RNA-processing events in which they are reported to participate (24, 46). These data also suggest that the p40 AUF1 isoform, when overexpressed, might disrupt RNA-processing events but also destabilize AU-rich mRNAs, a point addressed in Fig. 2 and 3. Transfection of smaller amounts of AUF1 expression plasmids for p40, p42, and p45 reduced the negative effect on mRNA abundance, with no evidence for selective reduction of AU-rich mRNAs (data not shown).
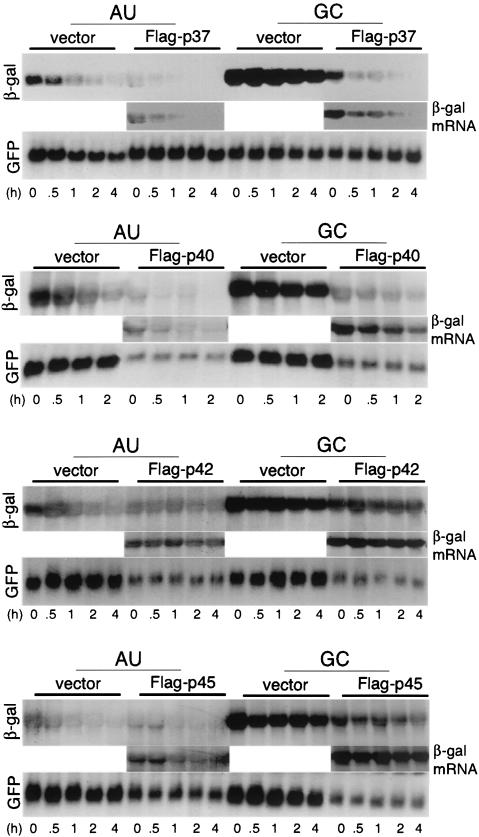
FIG. 2.
mRNA stability analysis with Flag-AUF1 proteins. CHO cells were transfected as described in Fig. 1 and then treated with 5 μg of actinomycin D/ml 24 h posttransfection for the indicated time periods prior to isolation of total RNA and Northern blot analysis. Representative blots are shown. Insets represent overexposures of β-Gal mRNA samples cotransfected with AUF1 proteins to permit better visualization of the low-abundance mRNA.
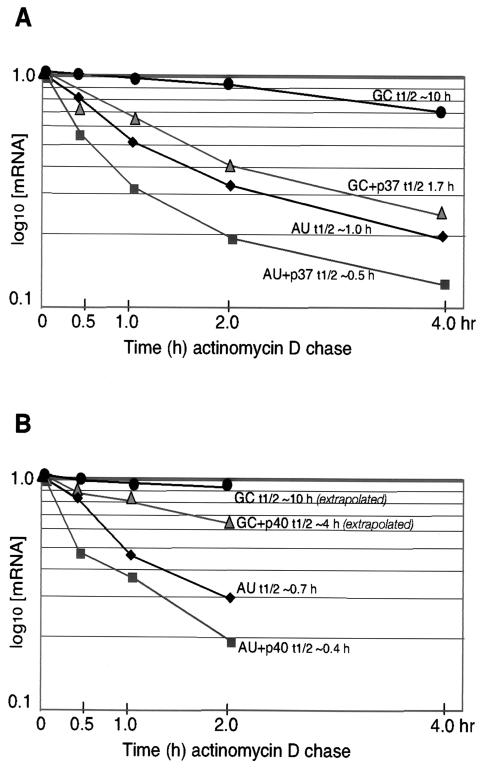
FIG. 3.
Analysis of the effect of p37 (A) and p40 (B) AUF1 supplementation on mRNA half-life. CHO cells were cotransfected with Flag-p37 or Flag-p40 and the vector alone and reporters containing the GM-CSF ARE (AU) or the GC mutant ARE (GC). The β-Gal mRNA level was normalized to the GFP mRNA level determined by densitometry of the results from Fig. 2 to correct for transfection efficiency. Normalized β-Gal mRNA decay curves were plotted as percentages of the initial mRNA concentration versus time of actinomycin D chase. Half-lives (t1/2) of GC-mRNA (with and without p40 AUF1) were estimated by extrapolation of decay curves.
We therefore conducted studies to determine whether ectopic overexpression of the different AUF1 isoforms alters the relative half-life of reporter mRNAs or suppresses transcription. CHO cells were cotransfected as described above and treated with actinomycin D to block new transcription, and mRNA abundance was determined by extraction of total RNA and Northern blot hybridization analysis. Autoradiograms were quantified by densitometry, and the relative rates of mRNA decay were determined (Fig. 3). The β-Gal reporter ARE-mRNA exhibited an average half-life of about 50 to 60 min (0.8 to 1.0 h) in CHO cells (Fig. 2 and 3). This is slightly more stable than it is in HeLa cells (half-life, 30 to 40 min) (28, 29). The control GC-mRNA displayed a long half-life of approximately 10 h in the absence of cotransfected AUF1 (Fig. 2 and 3). Ectopic overexpression of the p37 AUF1 isoform decreased the stability of the ARE-mRNA reporter compared to the vector control by ∼50%, which is consistent with the steady-state analysis of Fig. 1. Also as suggested by Fig. 1, overexpression of p37 AUF1 decreased by 80% the stability of the GC-mRNA which lacks the canonical ARE but retains significant AU richness in the 3′ UTR. Ectopic overexpression of Flag-p40 AUF1 was associated with an ∼50% decrease in the reporter ARE-RNA half-life and a slightly greater decrease in the half-life of the GC reporter mRNA, in addition to an overall decrease in the abundance of β-Gal and GFP reporter mRNAs (Fig. 2 and 3). The p37 AUF1 isoform and, to a lesser extent, the p40 isoform specifically destabilize mRNAs containing an ARE or an AU-rich 3′ UTR when overexpressed.
Ectopic overexpression of the p42 or p45 AUF1 isoforms severely reduced the abundance but not the half-lives of both β-Gal reporter mRNAs and the cotransfected GFP mRNA (Fig. 2). These data are consistent with the steady-state mRNA analysis of Fig. 1. Since total RNA was examined, the p42 and p45 AUF1 isoforms and, to some extent, the p40 isoform likely block transcription or other events in RNA processing when overexpressed. Consequently, the three larger AUF1 isoforms were not analyzed further.
Studies were carried out to determine whether overexpression of p37 AUF1 and decreased stability of the control GC-mRNA are associated with interaction with AUF1. It is possible that, with overexpression of p37 AUF1, its abundance increases sufficiently to promote interaction with the noncanonical AU-rich sequences in the control GC-mRNA, counteracting to some extent a decreased equilibrium constant for binding to noncanonical sequences. Alternatively, overexpression of p37 AUF1 could act indirectly, promoting the interaction of other AU-rich binding proteins that accelerate mRNA decay. To discriminate between these possibilities, 293 cells were transfected with 0.5 or 2.0 μg of a plasmid carrying β-Gal reporters containing an ARE or GC control sequence and plasmids expressing Flag-p37 AUF1 proteins or Flag alone. As shown below (see Fig. 6), Northern blot hybridization analysis demonstrated a 20-fold decrease in the abundance of β-Gal ARE-mRNA compared to the control GC-mRNA and a decrease in both ARE- and GC-mRNAs with ectopic overexpression of 2.0 μg of the Flag-p37 expression plasmid. Flag-p37 or the translation initiation factor was immunoprecipitated, and an RT-PCR amplification was carried out with β-Gal mRNA-specific primers. Products of PCR amplification were resolved by agarose gel electrophoresis (Fig. 4). Immunoprecipitation of eIF4G and RT-PCR products revealed a diagnostic 400-bp fragment specific for β-Gal mRNA which was not present in cells transfected with Flag-p37 but without the β-Gal reporter plasmid (lanes 1 and 2). In cells transfected with 0.5 μg of the Flag-p37 plasmid, immunoprecipitation of Flag and RT-PCR products revealed a specific β-Gal mRNA band only for the ARE-mRNA (lanes 4 and 5), not for the control GC-mRNA and not in cells transfected with Flag alone (lane 3). With a fourfold increase in the amount of transfected Flag-p37 DNA, which is associated with decreased ARE and GC-mRNA abundance (Fig. 6), immunoprecipitation of Flag-p37 and RT-PCR products revealed a somewhat-reduced level of products derived from the ARE-mRNA, possibly resulting from the decreased abundance (stability) of the mRNA. However, a low level of β-Gal GC-mRNA was also represented, consistent with increased binding to the noncanonical AU-rich sequence in this mRNA. Control studies failed to detect any evidence for interaction of Flag-p37 AUF1 with the GFP mRNA (data not shown). These results are consistent with the possibility that overexpression of p37 AUF1 likely promotes accelerated decay of noncanonical AU-rich mRNA by an increased ability to bind to the AU-rich sequences.
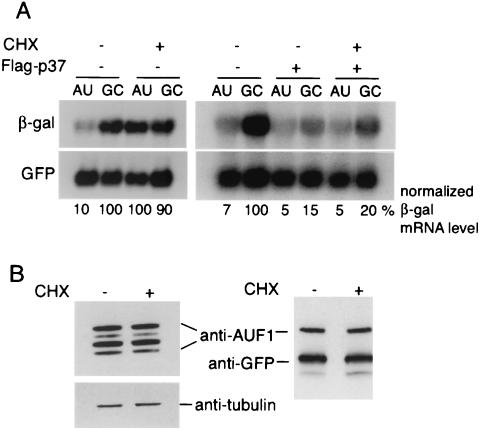
FIG. 6.
Flag-p37 expression inhibits cycloheximide-mediated stabilization of ARE-mRNA decay. (A) CHO cells were cotransfected with Flag-p37 AUF1 or vector alone, along with β-Gal reporter plasmid DNAs. At 24 h posttransfection, cells were treated with 100 μg of cycloheximide (CHX)/ml for 4 h prior to isolation of total RNA and Northern blot analysis. Autoradiograms were quantified by densitometry, β-Gal mRNA levels were normalized to the GFP control mRNA, and levels were expressed relative to that of the untreated and untransfected control GC-mRNA, which was set at 100%. (B) Untransfected (left) and Flag-p37-transfected (right) CHO cells were treated with CHX and then used to prepare whole-cell lysates. Equal amounts of total protein were resolved by SDS-PAGE and immunoblotted for endogenous AUF1 or Flag. The blot was stripped and reprobed with an antibody specific for tubulin or GFP to verify equal loading. Representative blots are shown.
FIG. 4.
Overexpressed p37 AUF1 is associated with AU-rich mRNAs in vivo. 293 cells were transfected with 1.0 μg of plasmids expressing β-Gal mRNAs and an ARE or GC reporter mRNA, 0.5 μg of a plasmid expressing Flag alone, or 0.5 or 2.0 μg of a plasmid expressing Flag-p37 AUF1. eIF4G and Flag were immunoprecipitated from cell lysates normalized to equal amounts of β-Gal mRNAs, and β-Gal mRNA-specific RT-PCR was carried out. The products of RT-PCR were resolved by agarose gel electrophoresis and visualized by staining with ethidium bromide.
Overexpression of p37 AUF1 relieves the saturation block in ARE-mRNA decay.
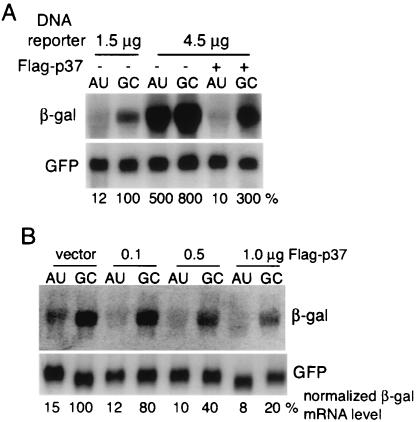
Since overexpression of p37 AUF1 could promote more-rapid degradation of ARE-mRNA and AU-rich mRNAs, we asked whether p37 represents a limiting factor in the decay machinery. It has been well established that the ability of the decay machinery to degrade ARE-mRNAs can be inhibited or saturated by overexpression of the ARE-mRNA (29, 43, 53). The level of ARE-mRNA reporter required to saturate the decay machinery was experimentally determined and varies in different cell lines. When CHO cells were transfected with a threefold-higher level of a plasmid encoding the β-Gal reporter ARE-mRNA (4.5 rather than 1.5 μg of DNA), the decay machinery was found to be near saturation (Fig. 5A). The steady-state level of β-Gal ARE-mRNA was elevated >10-fold at near-saturation conditions for decay (4.5 μg of plasmid), to a level roughly comparable to that of the GC-mRNA. Cells were also cotransfected with a GFP expression vector to normalize transfection efficiencies and to monitor alteration of non-AU-rich mRNAs. Ectopic expression of 1.0 μg of the Flag-p37 AUF1 plasmid decreased the steady-state level of β-Gal reporter ARE-mRNA to that found in the absence of reporter mRNA overexpression (Fig. 5A). Expression of Flag-p37 AUF1 also reduced by about one-half the abundance of the control GC-mRNA. The GFP control mRNA, which lacks significant AU richness in the 3′ UTR, was unaffected by overexpressing reporter mRNAs or p37 AUF1 (Fig. 5A). These data demonstrate that ectopic expression of p37 AUF1 relieves saturation of the ARE-mRNA decay machinery and likely restores the ability to degrade ARE-mRNAs. To determine whether p37 AUF1 is utilized in a dose-responsive manner to reduce ARE-mRNA levels, cells were transfected with subsaturating amounts of ARE-mRNA or β-Gal reporter GC-mRNA and increasing amounts of Flag-p37 plasmid (Fig. 5B). A dose-responsive reduction in the steady-state levels of both ARE- and reporter GC-mRNAs was evident, but not a reduction in that of the GFP control. Quantification of autoradiograms and determination of the specific reduction of β-Gal mRNA (normalized to the GFP mRNA control) indicated that increased transfection of the Flag-p37 AUF1 expression plasmid decreased mRNA more strongly than transfection of ARE-mRNA (50 and 80%, respectively), as observed in Fig. 2 and 3. Similar results were observed when GFP-p37 or p37 AUF1 lacking any epitope tag was transfected instead of Flag-p37 (data not shown). These data indicate that the level of p37 AUF1 is limiting in the control of AU-rich-mRNA turnover.
FIG. 5.
p37 AUF1 relieves saturation of the ARE-mRNA decay pathway. (A) CHO cells were cotransfected with either 1.0 μg of plasmid vector or the Flag-p37 plasmid and 1.5 or 4.5 μg of the β-Gal ARE or GC control reporter plasmids per 107 cells. Total RNA was isolated 24 h posttransfection, and Northern analysis was performed. (B) CHO cells were transfected as in panel A but with a fixed amount of the reporter plasmid (1.5 μg) and an increasing amount of Flag-p37 plasmid DNA (0.1, 0.5, and 1.0 μg), as shown. A representative blot is shown. Autoradiograms were quantified by densitometry, and data were normalized by transfection efficiency, as determined by GFP β-Gal control mRNA with the vector alone set at 100%, as described in the legend to Fig. 1.
Cycloheximide stabilization of ARE-mRNAs is overridden by overexpression of p37 AUF1.
AUF1 proteins complex with eIF4G and poly(A) binding protein, two factors involved in translation, and can be detected in polysomes (28, 36, 54). Cycloheximide, an inhibitor of ribosome elongation, inhibits protein synthesis and blocks degradation of ARE-mRNAs (reviewed in reference 20). The effect of cycloheximide is thought to involve some aspect of mRNA translation itself that is coupled to activation of ARE-mRNA degradation (1, 11, 39), although there may also be a requirement for continuous synthesis of a short-lived decay protein. Studies were carried out to determine whether overexpression of p37 AUF1 influences the requirement for protein synthesis in the degradation of ARE-mRNAs. Cells were cotransfected with vector DNA or Flag-p37 and β-Gal reporter plasmids (AU or GC). Cycloheximide was added to cells 24 h posttransfection at 100 μg/ml for 4 h, as previously described (6), and total RNA was extracted and examined by Northern blot hybridization analysis and autoradiography. Cycloheximide inhibited protein synthesis by > 95%, as shown by labeling cells with [35S]methionine and determining the specific activity of radiolabel incorporation into protein (data not shown). Treatment with cycloheximide increased the abundance of the ARE-mRNA reporter by approximately 10-fold, but not that of the GC mutant reporter, as expected (Fig. 6A, left). It is well documented that the cycloheximide effect is due to increased mRNA stability rather than increased transcription (1, 6, 9, 11, 37, 40). Surprisingly, cycloheximide did not stabilize ARE-mRNAs if Flag-p37 AUF1 was overexpressed. This was shown by overexpression of p37 AUF1, which decreased the abundance of the AU-rich and control GC-mRNAs by ∼50 and 80%, respectively, in the absence or presence of cycloheximide inhibition of protein synthesis (Fig. 6A, right). These data suggest that increased expression of p37 AUF1 uncouples the requirement for protein synthesis in ARE-mRNA decay. It was therefore determined whether the cycloheximide-mediated increase in ARE-mRNAs results from depletion of the endogenous p37 AUF1 protein or some other aspect of inhibition of protein synthesis. Whole-cell extracts were prepared from cycloheximide-treated and untreated cells, and endogenous or Flag-p37 AUF1 proteins were examined by immunoblot analysis. No changes in AUF1 protein levels were evident following incubation with cycloheximide for either the endogenous or transfected AUF1 protein (Fig. 6B). We previously determined by pulse-chase analysis that p37 AUF1 has an average half-life of 20 to 30 min (30). These results therefore suggest that p37 AUF1 degradation and ARE-mRNA decay are linked to protein synthesis in an unknown manner. These results further show that, when the p37 AUF1 protein is ectopically overexpressed, the requirement for protein synthesis in ARE-mRNA turnover is likely eliminated.
p37 AUF1 decreases ARE-mRNAs in various cell lines.
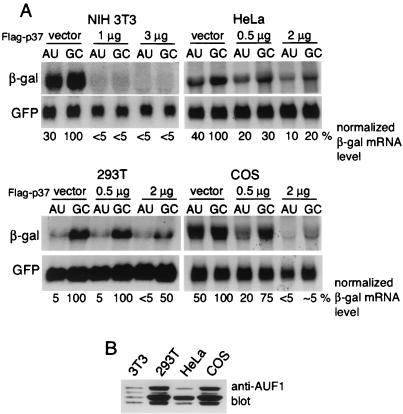
It has been reported that p37 AUF1 promotes degradation of ARE-mRNAs only in certain cell lines, and actually stabilizes ARE-mRNAs in NIH 3T3 cells (52). We therefore explored whether p37 AUF1 is limiting for ARE-mRNA decay in a variety of cell lines, including NIH 3T3 cells. Cells were cotransfected with the matched ARE or GC β-Gal reporter plasmids and Flag-p37 AUF1 at the indicated levels or with the vector alone (Fig. 7A). In the absence of ectopic expression of p37 AUF1, the ability of the ARE to promote mRNA destabilization varied among the cell lines, demonstrating the greatest effect in 293T cells compared to the control GC-mRNA (∼95% reduction in ARE-mRNA), moderate effects in NIH 3T3 and HeLa cells (60 to 70% reduction in ARE-mRNA), and the least effect in COS cells (50% reduction). In all cell types examined, however, ectopic overexpression of p37 AUF1 was associated with decreased abundance of both ARE- and GC-mRNAs, but not of the GFP control mRNA, which lacks any AU richness in its 3′ UTR (Fig. 7A). Different lines of NIH 3T3 cells all demonstrated a selective decrease in ARE-mRNA reporters and specifically decreased levels of AU-rich mRNAs with increased expression of p37 AUF1 (data not shown). None of the cell types, notably NIH 3T3 cells, exhibited increased levels of the reporter mRNAs when p37 was overexpressed, contrary to a recent report (52). We cannot explain the difference in results. The relative abundances of the different AUF1 isoforms were also examined. Whole-cell lysates were prepared from the cell lines, equal amounts of protein were resolved by SDS-PAGE, and endogenous AUF1 proteins were detected by immunoblotting (Fig. 7B). AUF 1 levels in different cell lines varied, consistent with previous reports (3, 26, 41). Although the level of p37 AUF1 relative to those of the other isoforms was most elevated in 293T cells, which also had the greatest uncomplemented ability to decrease ARE-mRNA levels, in all cases ectopic overexpression of AUF1 promoted greater reductions in ARE-mRNA abundance than in GC-mRNA abundance. However, the endogenous levels of p37 do not always parallel the inherent ability of cell lines to carry out rapid degradation of ARE-mRNAs, since COS cells had a high level of p37 but a weak ability to reduce ARE-mRNA levels. These results therefore suggest that p37 may be limiting for ARE-mRNA decay in some cell lines but that ectopic overexpression of the p37 AUF1 promotes greater decay of ARE-mRNAs than of GC-mRNAs in a variety of cell lines from different mammalian species.
FIG. 7.
Destabilization of ARE-mRNA by Flag-p37 in various cell lines. (A) The indicated cell lines were cotransfected with vector alone or the indicated amounts of Flag-p37, along with the β-Gal and GFP reporter plasmids. At 24 h posttransfection, total RNA was isolated and Northern blot analysis was performed. (B) Equal amounts of whole-cell lysates prepared from the various cell lines were resolved by SDS-PAGE and immunoblotted for endogenous or Flag-tagged AUF1 proteins. Representative blots from at least three independent experiments are shown. Autoradiograms were quantified by densitometry, β-Gal mRNA levels were normalized to the GFP control mRNA, and levels were expressed relative to that of the untreated and untransfected control GC-mRNA, which set at 100%.
DISCUSSION
Previous in vitro studies have provided valuable information regarding the function and properties of the AUF1 protein family. This includes identification of domains necessary for AUF1 dimerization (13), determination of binding affinities for ARE sequences (12, 48), and ordering of in vitro binding affinities of the four AUF1 proteins for the c-fos ARE (p37 > p42 > p45 ≫ p40) (48). However, in vitro studies are functionally limited, since the purified mammalian or bacterially expressed p37 AUF1 protein binds ARE sequences but does not intrinsically accelerate mRNA decay in vitro (50). In vitro binding affinities of AUF1 isoforms for ARE sequences generally correlate with in vivo destabilizing activity (with p37 the strongest), but exceptions exist (4). Moreover, in vitro systems also lack active translation, which is an important element in the activation of the ARE-mRNA decay process in vivo. For all of these reasons, we employed a cotransfection-and-overexpression assay to examine the functional role of each AUF1 isoform in vivo.
The finding that ectopic overexpression of the three larger AUF1 isoforms results in global reduction in mRNA synthesis to different extents was unanticipated (Fig. 1 and 2) but perhaps not unexpected. Several reports demonstrated that the three larger AUF1 proteins promote transcription of promoters for c-myc and thymidine kinase, complement receptor 2, and Epstein-Barr virus genes (7, 17, 46). Other genes and promoters have not been tested, so the extent of transcriptional participation of the different AUF1 isoforms is not known. There are some discrepancies regarding which isoforms possess transcriptional activity, although exon 2 found in the p40 and p45 genes is sufficient to drive transcription when fused to the DNA-binding domain of GAL4 (47). Moreover, p40 AUF1 was reported to bind TATA-binding protein and the p300 coactivator (47). It is likely that overexpression of p40, p42, and p45 AUF1s impairs transcription complex function or formation, resulting in the observed decrease in mRNA levels. Importantly, overexpression of the two larger isoforms had no effect on ARE-mRNA stability. These data do not suggest, however, that the two larger AUF1 isoforms fail to play a role in ARE-mRNA decay. Rather, these results indicate that the two larger AUF1 isoforms are not generally limiting for ARE-mRNA decay and also likely function in other aspects of mRNA metabolism.
Our functional analyses indicate that the p37 AUF1 isoform and, to a lesser extent, the p40 isoform when overexpressed in the context of all endogenous AUF1 isoforms exhibit destabilizing activity toward a reporter ARE-mRNA (Fig. 1 and 2). Indeed, titration of ectopically expressed p37 AUF1 demonstrated a clear dose-responsive decrease in AU-rich mRNA levels with increasing levels of p37 protein (Fig. 5B). No such profile was found for the two larger AUF1 isoforms (data not shown). This finding is in agreement with several reports specifically correlating p37 (and to some extent) p40 AUF1 protein levels with ARE-mRNA-destabilizing activity (8, 28, 32, 34, 54). The normally stable GC-mRNA was also destabilized by overexpression of p37 AUF1 (Fig. 3), initially a somewhat surprising result. Although the G and C point mutations disrupt the canonical GM-CSF ARE, the sequence still possesses an AU content of 70% (Fig. 1A). The reporter GC-mRNA is normally stable compared to the ARE-mRNA. It is therefore most likely that overexpression of p37 AUF1 raises its abundance sufficiently to overcome the reduced binding affinity for noncanonical AU-rich sequences. Support for this hypothesis was provided in Fig. 4, in which weak interaction of p37 AUF1 with the control GC-mRNA on p37 AUF1 overexpression was observed. In further support of this possibility, purified mammalian and recombinant p37 AUF1s retain the ability to bind c-fos and c-myc AREs that similarly possess AU-rich sequences but that lack the canonical AUUUA repeats (12, 54). As the stoichiometry of AUF1 complexes remains unknown, it is not possible to determine which isoform directly mediates the decreased abundance of mRNAs. Nevertheless, the mRNA-destabilizing effect of p37 AUF1 overexpression was observed in a variety of cell lines of different mammalian origins, indicating that it is often a limiting component in ARE-mRNA decay. It is important, however, that the three larger AUF1 isoforms might also participate in ARE-mRNA destabilization when expressed at typical endogenous levels.
A great many studies have shown that cycloheximide treatment blocks the rapid degradation of natural and reporter ARE-mRNAs (20). Rapid ARE-mRNA turnover appears to require both ongoing translation of the mRNA itself and the presence of a labile factor, based on several observations. Removal of all initiation codons from a reporter ARE-mRNA or insertion of a ribosome-blocking secondary structure at any position 5′ to the ARE results in stabilization of the mRNA (1, 11, 39). In addition, stabilization of GM-CSF mRNA by cycloheximide occurs within 15 min of treatment, arguing that ribosome movement is required for ARE-mRNA decay (37). On the other hand, only extracts from untreated cells, not those from cells treated with cycloheximide for 2 h, accelerate c-myc mRNA decay in vitro (6), suggesting the involvement of an unstable protein factor.
In this regard, one surprising finding of our work is that overexpression of p37 AUF1 overrides the stabilization of ARE-mRNAs mediated by cycloheximide inhibition of protein synthesis (Fig. 6A). Since cycloheximide treatment did not decrease AUF1 protein levels, we suggest that ongoing translation of ARE-mRNA itself may be a prerequisite for both p37 and ARE-mRNA turnover. ARE-mRNAs are stabilized when sequestered from polysomes (51), suggesting that essential components of the degradation machinery are located on, or associated with, polysomes. Moreover, AUF1 proteins are found on translationally active polysomes (28, 36), raising the possibility that the AUF1 interaction with the ARE is altered by translation of the ARE-mRNA. A precedent for this is the Y14 shuttling RNA binding protein, which is found in the exon junction complex on translationally inactive mRNAs and which is removed with translation (14). It is therefore possible that AUF1 is similarly displaced by the act of mRNA translation, potentially from an inactive mRNP site (possibly the cap initiation complex [28]) to an active site for mRNA decay (possibly the ARE). Alternatively, mRNA translation might result in the displacement of HuR, a likely inhibitor of ARE-mRNA decay (18, 31, 49), from the ARE, allowing binding by p37 AUF1. How HuR and AUF1 mitigate each other's functions remains a key question. AUF1 destabilizing activity seems to predominate, as most ARE-mRNAs are labile and stabilized only with various stimuli, which the results presented here indicate best coincide with the p37 AUF1 isoform.
Acknowledgments
We thank Gary Brewer (UMDNJ) for cDNA clones of human AUF1.
This work was supported by National Institutes of Health grant GM60428 (R.J.S.). B.S. was supported by NIH training grant 5T32CA09161. C.H. was supported by Canadian NSERC fellowship 253767.
REFERENCES
- 1.Aharon, T., and R. J. Schneider. 1993. Selective destabilization of short-lived mRNAs with the granulocyte-macrophage colony-stimulating factor AU-rich 3′ noncoding region is mediated by a cotranslational mechanism. Mol. Cell. Biol. 13:1971-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakheet, T., M. Frevel, B. R. Williams, W. Greer, and K. S. Khabar. 2001. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29:246-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaxall, B. C., L. D. Dwyer-Nield, A. K. Bauer, T. J. Bohlmeyer, A. M. Malkinson, and J. D. Port. 2000. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol. Carcinog. 28:76-83. [PubMed] [Google Scholar]
- 4.Blaxall, B. C., A. Pende, S. C. Wu, and J. D. Port. 2002. Correlation between intrinsic mRNA stability and the affinity of AUF1 (hnRNP D) and HuR for A+U-rich mRNAs. Mol. Cell. Biochem. 232:1-11. [DOI] [PubMed] [Google Scholar]
- 5.Brewer, G. 1991. An A+U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 11:2460-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brewer, G., and J. Ross. 1989. Regulation of c-myc mRNA degradation in vitro by a labile destabilizer with an essential nucleic acid component. Mol. Cell. Biol. 9:1996-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brys, A., and N. Maizels. 1994. LR1 regulates c-myc transcription in B-cell lymphomas. Proc. Natl. Acad. Sci. USA 91:4915-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buzby, J. S., G. Brewer, and D. J. Nugent. 1999. Developmental regulation of RNA transcript destabilization by A + U-rich elements is AUF1-dependent. J. Biol. Chem. 274:33973-33978. [DOI] [PubMed] [Google Scholar]
- 9.Buzby, J. S., S. M. Lee, P. V. Van Winkle, C. T. DeMaria, G. Brewer, and M. S. Cairo. 1996. Increased granulocyte-macrophage colony stimulating factor mRNA instability in cord versus adult mononuclear cells is translation-dependent and associated with increased levels of A+U rich element binding factor. Blood 88:2889-2897. [PubMed] [Google Scholar]
- 10.Carballo, E., W. S. Lai, and P. J. Blackshear. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95:1891-1899. [PubMed] [Google Scholar]
- 11.Curatola, A. M., M. S. Nadal, and R. J. Schneider. 1995. Rapid mRNA degradation of AU-rich element (ARE) mRNAs is blocked by secondary structure at any position 5′ to the ARE. Mol. Cell. Biol. 15:6331-6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeMaria, C. T., and G. Brewer. 1996. AUF1 binding affinity to A+U rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 271:12179-12184. [DOI] [PubMed] [Google Scholar]
- 13.DeMaria, C. T., Y. Sun, L. Long, B. J. Wagner, and G. Brewer. 1997. Structural determinants in AUF1 required for high affinity binding to A+U-rich elements. J. Biol. Chem. 272:27635-27643. [DOI] [PubMed] [Google Scholar]
- 14.Dostie, J., and G. Dreyfuss. 2002. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 12:1060-1067. [DOI] [PubMed] [Google Scholar]
- 15.Fan, X. C., and J. A. Steitz. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17:3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ford, L. P., J. Watson, J. D. Keene, and J. Wilusz. 1999. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 13:188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuentes-Panana, E. M., R. Peng, G. Brewer, J. Tan, and P. D. Ling. 2000. Regulation of the Epstein-Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway. J. Virol. 74:8166-8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallouzi, I. E., C. M. Brennan, M. G. Stenberg, M. S. Swanson, A. Eversole, N. Maizels, and J. A. Steitz. 2000. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA 97:3073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, M., C. J. Wilusz, S. W. Peltz, and J. Wilusz. 2001. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 20:1134-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guhaniyogi, J., and G. Brewer. 2001. Regulation of mRNA stability in mammalian cells. Gene 265:11-23. [DOI] [PubMed] [Google Scholar]
- 21.Hamilton, B. J., E. Nagy, J. S. Malter, B. A. Arrick, and W. F. C. Rigby. 1993. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J. Biol. Chem. 268:8881-8887. [PubMed] [Google Scholar]
- 22.Hamilton, B. J., R. C. Nichols, H. Tsukamoto, R. J. Boado, W. M. Pardridge, and W. F. Rigby. 1999. hnRNP A2 and hnRNP L bind the 3′UTR of glucose transporter 1 mRNA and exist as a complex in vivo. Biochem. Biophys. Res. Commun. 261:646-651. [DOI] [PubMed] [Google Scholar]
- 23.Henics, T., A. Sanfridson, B. J. Hamilton, E. Nagy, and W. F. Rigby. 1994. Enhanced stability of interleukin-2 mRNA in MLA 144 cells. Possible role of cytoplasmic AU-rich sequence-binding proteins. J. Biol. Chem. 269:5377-5383. [PubMed] [Google Scholar]
- 24.Ishikawa, F., M. J. Matunis, G. Dreyfuss, and T. R. Cech. 1993. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol. 13:4301-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kajita, Y., J.-I. Nakayama, M. Aizawa, and F. Ishikawa. 1995. The UUAG-specific RNA binding protein, heterologous nuclear ribonucleoprotein D0. J. Biol. Chem. 270:22167-22175. [DOI] [PubMed] [Google Scholar]
- 26.Lafon, I., F. Carballes, G. Brewer, M. Poiret, and D. Morello. 1998. Developmental expression of AUF1 and HuR, two c-myc mRNA binding proteins. Oncogene 16:3413-3421. [DOI] [PubMed] [Google Scholar]
- 27.Lai, W. S., E. Carballo, J. M. Thorn, E. A. Kennington, and P. J. Blackshear. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J. Biol. Chem. 275:17827-17837. [DOI] [PubMed] [Google Scholar]
- 28.Laroia, G., R. Cuesta, and R. J. Schneider. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499-502. [DOI] [PubMed] [Google Scholar]
- 29.Laroia, G., B. Sarkar, and R. J. Schneider. 2002. Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc. Natl. Acad. Sci. USA 99:1842-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laroia, G., and R. J. Schneider. 2002. Alternate exon insertion controls selective ubiquitination and degradation of different AUF1 protein isoforms. Nucleic Acids Res. 30:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy, N. S., S. Chung, H. Furneaux, and A. P. Levy. 1998. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 273:6417-6423. [DOI] [PubMed] [Google Scholar]
- 32.Loflin, P., C. Y. Chen, and A. B. Shyu. 1999. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13:1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma, W.-J., S. Cheng, C. Campbell, A. Wright, and H. Furneaux. 1996. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271:8144-8151. [DOI] [PubMed] [Google Scholar]
- 34.Pende, A., K. D. Tremmel, C. T. DeMaria, B. C. Blaxall, W. A. Minobe, J. A. Sherman, J. D. Bisognano, M. R. Bristow, G. Brewer, and J. D. Port. 1996. Regulation of the mRNA-binding protein AUF1 by activation of the β-adrenergic receptor signal transduction pathway. J. Biol. Chem. 271:8493-8501. [DOI] [PubMed] [Google Scholar]
- 35.Peng, S. S., C. Y. Chen, N. Xu, and A. B. Shyu. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17:3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pioli, P. A., B. J. Hamilton, J. E. Connolly, G. Brewer, and W. F. Rigby. 2002. Lactate dehydrogenase is an AU-rich element-binding protein that directly interacts with AUF1. J. Biol. Chem. 277:35738-35745. [DOI] [PubMed] [Google Scholar]
- 37.Rajagopalan, L. E., and J. S. Malter. 1996. Turnover and translation of in vitro synthesized messenger RNAs in transfected, normal cells. J. Biol. Chem. 271:19871-19876. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez-Pascual, F., M. Hausding, I. Ihrig-Biedert, H. Furneaux, A. P. Levy, U. Forstermann, and H. Kleinert. 2000. Complex contribution of the 3′-untranslated region to the expressional regulation of the human inducible nitric-oxide synthase gene. Involvement of the RNA-binding protein HuR. J. Biol. Chem. 275:26040-26049. [DOI] [PubMed] [Google Scholar]
- 39.Savant-Bhonsale, S., and D. W. Cleveland. 1992. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a >20S degradation complex. Genes Dev. 6:1927-1939. [DOI] [PubMed] [Google Scholar]
- 40.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-667. [DOI] [PubMed] [Google Scholar]
- 41.Sheflin, L. G., and S. W. Spaulding. 2000. Testosterone and dihydrotestosterone regulate AUF1 isoforms in a tissue-specific fashion in the mouse. Am. J. Physiol. 278:E50-E57. [DOI] [PubMed] [Google Scholar]
- 42.Shyu, A.-B., J. G. Belasco, and M. E. Greenberg. 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in mRNA decay. Genes Dev. 5:221-231. [DOI] [PubMed] [Google Scholar]
- 43.Shyu, A. B., M. E. Greenberg, and J. G. Belasco. 1989. The c-fos transcript is targeted for rapid mRNA decay by two distinct mRNA degradation pathways. Genes Dev. 3:60-72. [DOI] [PubMed] [Google Scholar]
- 44.Sirenko, O. I., A. K. Lofquist, C. T. DeMaria, J. S. Morris, G. Brewer, and S. Haskill. 1997. Adhesion-dependent regulation of an A+U-rich element-binding activity associated with AUF1. Mol. Cell. Biol. 17:3898-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, G. A., E. Carballo, D. M. Lee, W. S. Lai, M. J. Thompson, D. D. Patel, D. I. Schenkman, G. S. Gilkeson, H. E. Broxmeyer, B. F. Haynes, and P. J. Blackshear. 1996. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4:445-454. [DOI] [PubMed] [Google Scholar]
- 46.Tolnay, M., L. A. Vereshchagina, and G. C. Tsokos. 1999. Heterogeneous nuclear ribonucleoprotein D0B is a sequence-specific DNA-binding protein. Biochem. J. 338:417-425. [PMC free article] [PubMed] [Google Scholar]
- 47.Tolnay, M., L. A. Vereshchagina, and G. C. Tsokos. 2002. NF-κB regulates the expression of the human complement receptor 2 gene. J. Immunol. 169:6236-6243. [DOI] [PubMed] [Google Scholar]
- 48.Wagner, B., C. T. DeMaria, Y. Sun, G. M. Wilson, and G. Brewer. 1998. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four isoforms. Genomics 48:195-202. [DOI] [PubMed] [Google Scholar]
- 49.Wang, W., H. Furneaux, H. Cheng, M. C. Caldwell, D. Hutter, Y. Liu, N. Holbrook, and M. Gorospe. 2000. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20:760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson, G. M., and G. Brewer. 1999. The search for trans-acting factors controlling messenger RNA decay. Prog. Nucleic Acid Res. Mol. Biol. 62:257-291. [DOI] [PubMed] [Google Scholar]
- 51.Winstall, E., M. Gamache, and V. Raymond. 1995. Rapid mRNA degradation mediated by the c-fos 3′ AU-rich element and that mediated by the granulocyte-macrophage colony-stimulating factor 3′ AU-rich element occur through similar polysome-associated mechanisms. Mol. Cell. Biol. 15:3796-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, N., C. Y. Chen, and A. B. Shyu. 2001. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 21:6960-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, N., P. Loflin, C. Y. Chen, and A. B. Shyu. 1998. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 26:558-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, W., B. J. Wagner, K. Ehrenman, A. W. Schaefer, C. T. DeMaria, D. Crater, K. DeHaven, L. Long, and G. Brewer. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 13:7652-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]