Abstract
The c-myb proto-oncogene encodes two alternatively spliced mRNAs, which in turn code for proteins of 75 kDa and 89 kDa. It is at present unclear whether the two isoforms of c-Myb perform identical functions or whether they mediate different biological effects. To assess their role in apoptotic death of hematopoietic cells, we expressed the two isoforms of c-Myb in the murine myeloid cell lines 32Dcl3 and FDCP1. Our results show that while ectopic overexpression of p75 c-Myb results in the acceleration of cell death, similar overexpression of p89 c-Myb results in the protection of cells from apoptotic death. An analysis of gene expression changes with mouse cDNA expression arrays revealed that while p75 c-Myb blocked the expression of glutathione S-transferase μ mRNA, p89 c-Myb greatly enhanced the expression of this gene. These results were further confirmed by Northern blot analysis. Ectopic overexpression of the glutathione S-transferase μ gene in 32Dcl3 cells resulted in protection of cells from interleukin-3 withdrawal-induced cell death similar to that seen with the ectopic overexpression of p89 c-Myb. These results suggest that the two isoforms of c-Myb differentially regulate apoptotic death of myeloid cells through differential regulation of glutathione S-transferase μ gene expression.
Programmed cell death is an essential phenomenon that regulates normal development and homeostasis. Apoptotic machinery is normally suppressed or activated by signals from the extracellular environment as well as intracellular sensors that monitor DNA damage. The absence of external survival signals or irreparable DNA damage are some of the important events that appear to trigger apoptosis in lower organisms such as Caenorhabditis elegans (reviewed in references 4 and 15). While preserving these apoptotic responses, higher organisms such as mammals have evolved a distinctive mechanism that enables the organism to instruct certain cell populations to enter apoptotic pathways at different stages of development.
Accumulating evidence suggests that in higher organisms, apoptosis is regulated by two major pathways, one that originates at the membrane and another that involves mitochondria (reviewed in references 3, 25, 48, and 62). The apoptotic pathways that originate at the membrane involve death receptors such as Fas, TNF-R1, DR-3, DR-4, and DR-5 (3, 48). These death receptors are activated by their cognate ligands, resulting in the recruitment and activation of caspases (3, 48), and this process does not appear to require de novo transcription and translation (48). The apoptotic pathways that involve mitochondria affect mitochondrial permeability and the release of cytochrome c from mitochondria into the cytosol, which interacts with Apaf1 and procaspase 9, leading to the activation of caspase 9 and the downstream caspases (reviewed in reference 25). In contrast to the death receptor-mediated pathways, this process requires de novo mRNA and protein synthesis and involves the members of the bcl-2 gene family (25, 48). Thus, Bcl-2 and Bcl-xL inhibit the release of cytochrome c from the mitochondria and block apoptosis, while Bax and Bid, the proapoptotic members of the family, promote the release of cytochrome c from mitochondria (25, 48).
In the mammalian organism, hematopoietic cell growth is normally dictated by a group of growth factors known as cytokines. Recent studies have shown that cytokines not only mediate proliferation and differentiation of hematopoietic cells, but also enhance the survival of these cells by the suppression of apoptotic pathways (49, 65). Withdrawal of cytokines from the culture medium has been found to result in apoptosis of hematopoietic cells, which appears to require de novo RNA and protein synthesis and has been found to involve members of the bcl-2 gene family, suggesting the involvement of mitochondria (48). It has been known for some time that induction of proliferation of hematopoietic cells by cytokines leads to the induction of c-myc and c-myb expression, underlying the central role played by these two proto-oncogenes in hematopoietic cell growth (9, 12, 13). Intriguingly, however, it has been observed that under conditions of growth factor or cytokine deprivation, these two nuclear oncogenes promote apoptotic death of hematopoietic cells (2, 54). Thus, ectopic overexpression of c-myc in mammalian cells was found to result in the acceleration of programmed cell death following the withdrawal of growth factors or cytokines (2, 43). In a similar manner, ectopic overexpression of p75c-myb was found to accelerate transforming growth factor beta-mediated cell death of the M1 myeloid leukemia cell line (54). While the mechanisms associated with c-myc-mediated apoptosis have been studied in some detail (reviewed in reference 46), very few studies have been conducted to address the molecular mechanisms associated with c-myb-mediated apoptotic death of hematopoietic cells.
The c-myb proto-oncogene is the cellular homologue of v-myb, the transforming gene of avian myeloblastosis virus, which causes myeloblastic leukemia in chickens (reviewed in reference 42). The major translational product of the c-myb proto-oncogene is a 75-kDa nuclear protein which is expressed in most hematopoietic tissues (63). In addition to this 75-kDa protein product, another translational product of 89 kDa was found to be encoded by c-myb in several avian, murine, and human hematopoietic cells (11, 17). This 89-kDa protein is translated from an alternatively spliced mRNA encoded by the c-myb gene, which results in the addition of 363 bp between exons 9 and 10. This region has been designated exon 9A (50, 55). In addition, both proteins encode an N-terminal DNA-binding domain, a central transactivation domain, and a C-terminal negative regulatory domain. Both proteins are found in the nucleus (17) and function as transcriptional activators with sequence-specific DNA binding activities (5, 17, 52, 64, 66).
Although the effects of Myb proteins in hematopoiesis have been well studied, the molecular mechanisms by which Myb proteins regulate cellular events and the nature of the target genes through which these nuclear factors mediate their function are still unclear. It is also at present unclear whether the two isoforms of c-Myb perform identical functions or whether they mediate different biological effects.
To assess the role of the two isoforms of c-Myb in apoptotic death of hematopoietic cells, we expressed these two isoforms of c-myb under inducible conditions in the interleukin-3 (IL-3)-dependent myeloid precursor cell line 32Dcl3, which is derived from normal mouse bone marrow and is nontumorigenic (24, 37). This cell line was particularly useful for the studies outlined here becasue it is not predisposed to developing factor-independent clones (2). The results presented in this article show that the two isoforms of c-myb regulate apoptotic death of myeloid precursor cells in opposite ways and that they mediate these effects through the regulation of the glutathione S-transferase μ (GSTμ) gene, which in turn regulates intracellular levels of oxygen free radicals.
MATERIALS AND METHODS
Cell culture.
QT6 fibroblasts were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% HEPES buffer and 0.5% penicillin-streptomycin in a humidified incubator maintained at 37°C and circulated with 5% CO2. The murine myeloid progenitor cell line 32Dcl3 (2, 37, 51) was maintained in Iscove’s modified Dulbecco’s medium supplemented with 10% fetal bovine serum, 0.5% penicillin-streptomycin, and 10% WEHI-3B cell-conditioned medium as a source of IL-3 (68). FDCP1 cells were maintained in RPMI supplemented with 10% fetal bovine serum, 0.5% penicillin-streptomycin, and 10% WEHI-3B cell-conditioned medium as a source of IL-3.
Construction of plasmids.
The construction of reporter plasmids pTA3-luc and pT81-luc has been described previously (38). For expression in QT6, each effector myb construct, p75c-myb, p89c-myb, t-myb, and p75Δlz-c-myb, were inserted into expression vector pcDNA3.1 (Invitrogen), which placed the insert under the control of the cytomegalovirus immediate-early promoter. p75Δlz-c-myb was constructed by mutating Leu389 and Leu396 to alanines by site-directed mutagenesis and fusion PCR (26). The inducible vector pMT-neo has been described previously (41). Murine wild-type p75c-myb, p89c-myb, t-myb, and p75Δlz-c-myb were subcloned into the pMT-neo vector to express these genes in 32Dcl3 cells in a metal ion-inducible manner. Murine GSTμ (accession no. J04696) was cloned via reverse transcription-PCR from 32Dcl3 total RNA isolated 8 h after IL-3 withdrawal with oligonucleotides 5′-CTATGCCTATGACACTAGGTTAC-3′ and 5′-GGGCCAGCAGAGCACTCATGAG-3′, subcloned into pOPRSVI/MCS (Stratagene), and used with pCMVlacI repressor plasmid to express this gene in 32Dcl3 cells in an isopropylthiogalactopyranoside (IPTG)-inducible manner. The GSTμ cDNA was also cloned in pOPRSVI-Puro/MCS (31) to express GSTμ in 32D/c-myb p75 cells. The internal marker for transient transfection, pRL-CH110, was constructed by replacing the lacZ gene in pCH110 (Pharmacia) with the Renilla luciferase gene from pRL-null (Promega).
Transient transfections and luciferase assays.
For transfection into QT6 cells, cells were seeded into six-well plates at a density of 0.5 × 105 cells/well. The following day, DNA was transfected by the calcium phosphate method (Gibco-BRL) with the manufacturer's protocol. In each transfection, 5 μg of reporter and 5 μg of effector plasmid were transfected along with 0.5 μg of pRL-CH110 as an internal standard. Following incubation for 24 h, cells were washed and incubated in fresh medium for an additional 24 h. At 48 h posttransfection, cells were lysed in 500 μl of passive lysis buffer (Promega)/well. Luciferase activity was assayed with the dual-luciferase Reporter plasmid system (Promega). Luciferase activities were normalized against Renilla luciferase activity to determine relative luciferase activity, and the activation was calculated by setting the value of the empty vector control at 1.0.
For transfection of GSTμ antisense oligodeoxynucleotides in 32D cells, the folic acid-polylysine method was used as described previously (34). Briefly, folic acid-polylysine and GSTμ antisense oligodeoxynucleotide mixtures were prepared in 150 mM NaCl-20 mM HEPES (pH 7.3) and gently mixed and incubated at room temperature for 30 min. The transfection mixture was then added to 32D cells in six-well plates containing 1 M cells in 3 ml of medium. After 3 h, the cells were washed and viability was determined at the indicated time points.
Establishment of stable cell lines expressing transgenes.
For stable transfection of 32Dcl3 cells, exponentially growing 32Dcl3 cells were electroporated with various linearized plasmid DNAs with a Gene-Pulser (Bio-Rad) at a pulse of 230 V and 960 μF. The surviving cells were selected in 1 mg of G418 per ml (pMT-neo-based constructs), 2 μg of puromycin per ml (pOPRSVI-Puro/MCS-based constructs), or 1 mg of G418 and 1 mg of hygromycin per ml (pOPRSVI/lacI repressor-based constructs) for 2 to 3 weeks. To isolate single cell clones, mass cultures were serially diluted in 96-well plates in the presence of G418 and selected for clonal expansion. Since the expression of transgenes was leaky, unless specifically indicated, 100 μM ZnCl2 was added in all experiments with cell lines derived from pMT-neo-based constructs.
To generate stable FDCP1 transfectants, 10 μg of pMTp75c-myb, pMTp89c-myb or the empty vector (pMT-neo) was introduced into actively proliferating FDCP1 cells by electroporation (300 V, 960 μF) and selected in the presence of G418 (500 μg/ml). Single cell clones were obtained by limiting dilution.
Northern blot analysis.
Total RNA from each cell line in the presence of IL-3 or at the indicated time points following IL-3 withdrawal was isolated and purified with the Trizol reagent (Gibco-BRL). Then 20 μg of total RNA was used per sample and Northern blot analysis was performed. Full-length c-myb and GSTμ cDNAs were used to detect c-myb and GSTμ mRNA transcripts, respectively. As an internal control for RNA loading, blots were either stripped and reprobed with full-length glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA or stained with ethidium bromide after completion of RNA transfer onto the nitrocellulose filter to compare levels of 18S and 28S rRNAs across lanes.
Western blot analysis.
Anti-lamin B and anti-cytochrome c antibodies were purchased from Santa Cruz and PharMingen, respectively. To analyze protein products of the transfected c-myb genes in QT6 and 32Dcl3 cells, normalized amounts of proteins from each cell lysate were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and separated proteins were transferred to a nitrocellulose membrane in transfer buffer (10 mM CAPS, 10% methanol [pH 10.5]). The filter was blocked with 5% nonfat milk in TBS-T buffer (10 mM Tris [pH 7.5], 100 mM NaCl, 0.1% Tween 20) for 1 h at room temperature, incubated with primary anti c-Myb polyclonal antibody (41) overnight at 4°C, and washed three times in TBS-T solution. The secondary antibody reaction was performed by incubating the filters with horseradish peroxidase-conjugated anti-rabbit immunoglobulin (Amersham) and washed as described for the primary antibody reaction. The Amersham ECL detection system was used for visualization as specified by the manufacturer. To detect protein products of lamin B and cytochrome c, 3% nonfat milk was used in the protocol described above.
Fractionation of cell lysates.
For immunoblotting, cytosolic and mitochondrial fractions were prepared by selective plasma membrane permeabilization with digitonin (20). After treatment with 0.05% digitonin in isotonic sucrose buffer (composition in 250 nM sucrose, 10 nM HEPES, 10 nM KCl, 1.5 nM MgCl2, 1 nM EDTA and 1 nM EGTA, pH 7.1) for 1 min at room temperature, and soluble fractions of permeabilized cells containing cytosol were saved. Insoluble fractions were further extracted with ice-cold 0.5% Triton X-100 in isotonic sucrose buffer for 10 min to release membrane- and organelle-bound proteins, including mitochondrial cytochrome c. Protease inhibitors were included in all solubilization buffers. Cellular fractions sequentially solubilized by digitonin and Triton X-100 were centrifuged at 15,000 × g for 10 min at 4°C. Proteins in the resultant supernatants were resolved by SDS-PAGE and electroblotted onto polyvinylidene difluoride membranes for Western blot analysis with anti-cytochrome c monoclonal antibody (PharMingen).
Analysis of DNA fragmentation.
DNA fragments released from 10 × 106 cells were extracted and separated by electrophoresis in agarose gels (1). After experiments, cells were lysed in a buffer containing 0.5% Triton X-100, 50 mM Tris, 10 mM EDTA (pH 7.4), and 200 mg of proteinase K per ml. Cell lysates were then incubated at 42°C for 90 min, treated with 0.2 mg of RNase A per ml, incubated at 37°C for an additional 30 min, and centrifuged at 14,000 × g for 20 min. The resultant supernatant was then extracted twice with phenol-chloroform (1:1). DNA fragments were precipitated with 1/10 volume of 3 M sodium acetate and 2.5 volumes of ethanol at −20°C overnight and resuspended in 10 mM Tris-HCl-1 mM EDTA (pH 8.0). Then 2 mg of DNA was analyzed on a 2% agarose gel.
Flow cytometry.
Cell cycle determination by flow cytometry was performed on an Elite Coulter counter. For cell cycle analysis, 106 32Dcl3 cells were collected at the indicated time points following IL-3 withdrawal, washed three times with 1 ml of ice-cold 1% fetal calf serum in phosphate-buffered saline, resuspended in 0.5 ml of phosphate-buffered saline with 1% fetal calf serum, and fixed by addition of 4 ml of ice-cold absolute ethanol with slow vortexing. Following fixation, cells were washed once and resuspended in 0.8 ml of 1% fetal calf serum in phosphate-buffered saline, 0.1 ml of propidium iodide, and 0.1 ml of 10-mg/ml RNase A and incubated at 37°C for 30 min before being subjected to flow cytometric analysis.
Viability assay.
32Dcl3 and FDCP1 cells expressing exogenous c-Myb and GSTμ proteins were analyzed for cell viability in the absence of IL-3 at different time points. At the indicated time points, viable and nonviable cells were identified by trypan blue dye exclusion and counted in triplicate on a hemacytometer, and the percentage of viable cells was calculated.
Analysis of gene expression.
Atlas mouse cDNA expression array (Clontech catalog no. 7741-1) was used to analyze differential gene expression following IL-3 withdrawal in 32Dcl3 cells transfected with alternatively spliced c-myb gene products. Isolation of total RNA, cDNA synthesis, labeling of cDNA, and hybridization of cDNA probes to the Atlas array filters were performed according to the manufacturer's protocol (Clontech).
Caspase activity assay.
Following experimental incubation, cells were lysed in 1% Triton X-100 in 25 mM HEPES-115 mM NaCl-1 mM KH2PO4-4 mM KCl buffer (pH 7.4). To measure caspase activity, 50 μg of lysates was added to reaction mixtures containing 50 μM peptide substrate (DEVD-AFC [Asp-Glu-Val-Asp-7-amino-4-trifluoromethylcoumarin], LEHD-AFC, etc., purchased from Enzyme Systems Products), 100 mM HEPES, 10% sucrose, 0.1% CHAPS, 1 mM EDTA, and 10 mM dithiothreitol (pH 7.4) in a total volume of 200 μl and incubated at 37°C for 1 h. Production of AFC was monitored in a SpectroFluor plate reader (Tecan US Inc.) with an excitation wavelength of 360 nm and an emission wavelength of 530 nm.
RESULTS
Comparison of transactivation potential of p75 and p89 c-Myb proteins and their mutants.
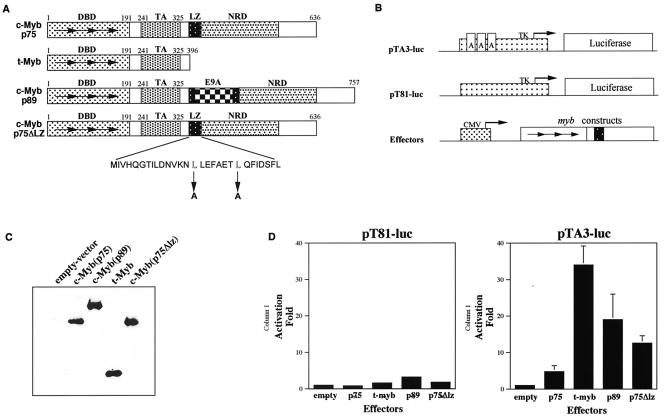
Figure 1A shows a structural comparison between the p75 and p89 c-Myb proteins. Both of these proteins contain an NH2-terminal DNA-binding domain, a central transactivation domain and a C-terminal negative regulatory domain (reviewed in references 42). One of the interesting structural motifs noted in the negative regulatory domain of p75 c-Myb is a putative leucine zipper motif with one isoleucine and three leucine residues. Interestingly, this leucine zipper is disrupted in p89 c-Myb because of the insertion of exon 9A sequences. This observation led to a suggestion that the leucine zipper domain in p75 c-Myb might mediate interactions with other leucine zipper proteins, which in turn might contribute to the negative regulation of the protein (19). Because p89 c-Myb did not contain the leucine zipper domain, it was proposed that this protein would not be able to participate in such protein interactions and thus not be subject to negative regulation.
FIG. 1.
Transcriptional transactivation by p75 c-Myb, p75Δlz c-Myb, p89 c-Myb, and t-Myb. (A) Schematic representations of wild-type p75 c-Myb and p89 c-Myb and mutants p75Δlz c-Myb and t-Myb are depicted. The numbers above each diagram are positions of amino acid residues in each corresponding region. Horizontal arrows represent the three 51- to 52-amino-acid repeats that constitute the DNA-binding domain. The stretch of amino acids below represents the putative leucine zipper motif present in wild-type p75 c-Myb. The downward arrows indicate the two leucine to alanine mutations made in order to construct p75Δlz c-Myb. DBD, DNA binding domain; TA, transactivation domain; NRD, negative regulatory domain; LZ, leucine zipper; E9A, exon 9A. (B) Schematics of the reporter and effector plasmids used in transient transactivation assays. The dotted box represents the promoters, and the arrows in the promoters represent the starting sites of transcription. The arrows in Myb represent the three 51- to 52-amino-acid repeats of the DNA binding domain, and the black box represents the transactivation domain. The boxes labeled A in pTA3-luc represent the three Myb-binding sites. TK, herpes simplex virus thymidine kinase promoter; CMV, immediate-early promoter for cytomegalovirus. (C) Transient expression of effector molecules. Cell lysates from QT6 cells transfected with different myb expression plasmids and expressing equal amounts of firefly luciferase activity were subjected to Western blot analysis and probed with an antibody raised against c-Myb. (D) Transcriptional activation by Myb proteins. Each myb expression plasmid was transfected into QT6 cells with reporter plasmid pTA3-luc or pT81-luc and the pRL-CH110 control plasmid as described in the text. After 2 days, the cells were harvested and assayed for firefly luciferase activity with the dual luciferase system (Promega). The luciferase activities were normalized to Renilla luciferase activity, and the activation was obtained by setting the value of empty vector at 1.0. Shown are the means of activation transcriptional potential from three independent experiments.
To verify this hypothesis and to investigate differences in the biological activities of the two proteins, we generated two additional mutants of p75 c-Myb, which were termed t-Myb and p75Δlz c-Myb. The t-Myb protein lacked the C-terminal negative regulatory domain and was previously shown to greatly enhance the transactivational potential of the protein (18). The p75Δlz c-Myb mutant was created following the introduction of two point mutations in the leucine zipper domain, where the leucines were replaced by alanines.
To gain an understanding of the relative transactivation potential of the four proteins in mammalian systems, we carried out transcriptional transactivation studies. Figure 1B shows the two reporter plasmids pTA3-luc and pT81-luc (18, 38). Of these, pT81-luc served as a control reporter and contained the thymidine kinase promoter linked to the luciferase gene (40). The reporter plasmid pTA3-luc contained three copies of Myb binding sites, cloned in tandem, upstream of the thymidine kinase promoter. The expression vectors for the Myb proteins were generated by cloning the four cDNAs into plasmid pRC/cytomegalovirus (Invitrogen), which places these genes under the control of the cytomegalovirus early promoter. Following transfection of the reporter and myb expression plasmids into the QT6 cell line, we analyzed the intracellular levels of the effector molecules and their relative transcriptional transactivation potentials.
The results presented in Fig. 1C show that the four Myb proteins studied here were expressed at equivalent levels in the transfected cells. The results presented in Fig. 1D show that while the p75 c-Myb transactivated the pTA3-luc promoter by approximately fivefold. p89 c-Myb showed considerably higher transactivation potential, which was approximately 20-fold. The leucine zipper mutant, p75Δlz c-Myb, showed a 12-fold transactivation potential, which was approximately half of that seen with the p89 c-Myb protein. In these experiments, t-Myb showed the highest levels of transcriptional transactivation, approximately 35-fold compared to the control vectors. These results suggest that the leucine zipper domain of p75 c-Myb exerts a negative regulatory effect and the insertion of exon 9A sequences results in elevation of the transactivation potential of this protein. These results also suggest that exon 9A sequences, in addition to disrupting the effects of the leucine zipper domain, may also exert an overall positive regulatory effect on the transactivation function of the c-Myb protein.
Ectopic overexpression of p75, p75Δlz, and p89 c-Myb proteins in 32Dcl3 myeloid cells.
Even though the p89 form of c-Myb was discovered over 15 years ago, no major differences in biological function have so far been described for the two proteins. To examine the role of the two isoforms of c-Myb in myeloid cell growth and apoptosis, we expressed these two proteins in the murine myeloid precursor cell line 32Dcl3. These cells, derived from normal mouse bone marrow, have been found to be strictly dependent on IL-3 for growth and undergo terminal differentiation when placed in an IL-3-free medium containing granulocyte colony-stimulating factor (G-CSF) (51, 61). However, when the cells were incubated in the absence of IL-3 or G-CSF, they were found to become arrested in the G1 phase of the cell cycle and rapidly entered apoptotic pathways, losing viability in 48 to 96 h (19). We had previously shown that overexpression of v-Myb or c-Myb in these cells results in a block to their ability to terminally differentiate into granulocytes in the presence of G-CSF (44, 45).
To determine the effects of p89 c-Myb expression on the differentiation and apoptotic programs of 32Dcl3 cells, we constructed inducible expression vectors where the two alternatively spliced forms of c-myb and the leucine zipper mutant of p75c-myb (p75Δlz) were placed under the control of the human metallothionein promoter (41). Following the construction of these vectors, the vector DNAs were transfected into 32Dcl3 cells by electroporation. As a negative control, empty vector DNA was similarly introduced into 32Dcl3 cells by electroporation. Following selection in G418, mass cultures as well as single cell clones were established from each stable transfection. The derived cell lines were tested for the expression of each transgene in the presence and absence of Zn2+, which acts as an inducer of transcription from the metallothionein promoter.
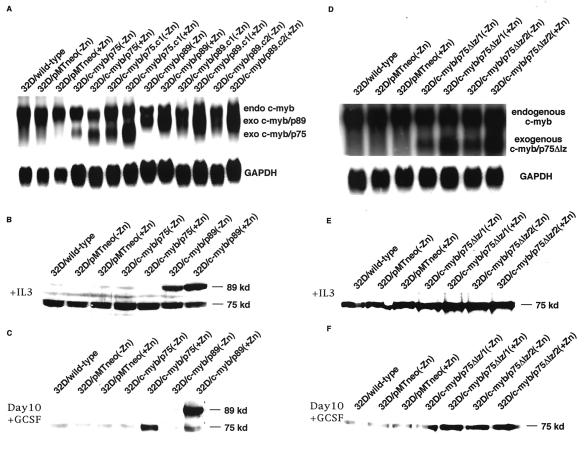
The profiles of RNA induction in these cell lines are shown in Fig. 2, panels A and D. Normal 32Dcl3 cells and cells transfected with the empty vector (32D/pMT/neo) showed the presence of a 3.4-kb endogenous c-myb band in the presence and absence of Zn2+. In mass cultures (32D/c-myb/p75 and 32D/c-myb/p89) as well as single cell clones (32D/c-myb/p75.c1, 32D/c-myb/p75.c2, 32D/c-myb/p89.c1, 32D/c-myb/p89.c2, 32D/c-myb/p75Δlz/1, and 32D/c-myb/p75Δlz/2) transfected with the three c-myb expression vectors, low levels of transgene expression were found to occur in the absence of Zn2+, which is seen as a shorter transcript. The smaller size of the transcript is due to the absence of 3′ untranslated sequence, which was deleted during the construction of the expression plasmids. In the presence of Zn2+, the levels of the p75 and p89 transgenic transcripts were elevated by approximately 3- to 10-fold. However, in the case of cell lines transfected with the leucine zipper mutant of p75, inducible expression of the transgene was not observed, and high-level expression of the transgenic transcript was seen in both the presence and absence of Zn2+.
FIG. 2.
Constitutive expression of p75 c-Myb, p89 c-Myb, and p75Δlz c-Myb in 32Dcl3 cells. p75c-myb, p89c-myb, and p75Δlz-c-myb cDNAs in the pMT-neo vector were transfected into 32Dcl3 cells. and mass cultures as well as single cell clones were established. (A and D) Northern blot analysis of total RNA extracted from different cell lines with a full-length c-myb cDNA probe. Endogenous c-myb transcript (upper band) and exogenous p75c-myb transcript (lower band in panel A), p89c-myb transcript (middle band in panel A), and p75Δlz-c-myb transcript (lower band in panel D) are shown. As an internal control for RNA loading, blots were stripped and reprobed with full-length GAPDH cDNA (shown below). (B and E) Expression of c-Myb protein in the presence of IL-3 in empty-vector-, p75c-myb-, and p89c-myb-transfected cells (B) and p75Δlz-c-myb-transfected cells (E). (C and F) Expression of c-Myb protein in the presence of G-CSF for 10 days in empty-vector-, p75c-myb- and p89c-myb-transfected cells (C) and p75Δlz-c-myb-transfected cells (F). −Zn and +Zn indicate the absence and presence, respectively, of 100 μM ZnCl2 used to induce the expression of different transgenes in the metallothionein promoter-based constructs.
When the same cultures were examined for the induction of c-Myb protein synthesis, we could readily demonstrate the expression of p89 c-Myb in cells transfected with the expression vector for p89c-myb (Fig. 2B). 32Dcl3 cells express very low levels of endogenous p89c-myb transcript, which allowed the detection of the transgenic protein relatively easily. However, when cells transfected with p75c-myb or p75Δlz-c-myb were examined for expression of the transgenic protein, it was difficult to distinguish the endogenous protein from that expressed from the transgenic expression vector because of their similar size (Fig. 2B and 2E).
In order to demonstrate that cells transfected with the p75c-myband p75Δlz-c-myb expression vectors did express the transgenic protein, we compared the c-Myb protein levels in cells that were cultured for 10 days in G-CSF. It had been shown previously that in 32Dcl3 cells grown in the presence of G-CSF, the endogenous levels of c-myb RNA and protein are downregulated and become undetectable by the 10th day of G-CSF treatment (44). Taking advantage of this observation, we analyzed the levels of c-Myb protein in control and p75c-myb and p75Δlz-c-myb expression vector-transfected cells following their incubation in G-CSF for 10 days. As shown in Fig. 2C and 2F, incubation of empty-vector-transfected cells in the presence of G-CSF for 10 days resulted in a complete downregulation of endogenous c-Myb protein levels, which became undetectable. In sharp contrast, in the p75c-myb and p89c-myb expression vector-transfected cells, a small amount of transgenic protein was detected in the absence of Zn2+, and these levels were markedly increased by Zn2+ treatment. In the case of p75Δlz-c-myb expression vector-transfected cells, transgenic protein expression was detected in the absence and presence of Zn2+ due to leaky mRNA expression of this transgene (Fig. 2F).
Effects of ectopic overexpression of p75, p75Δlz, and p89 c-Myb proteins on IL-3 withdrawal-induced apoptosis of 32Dcl3 cells.
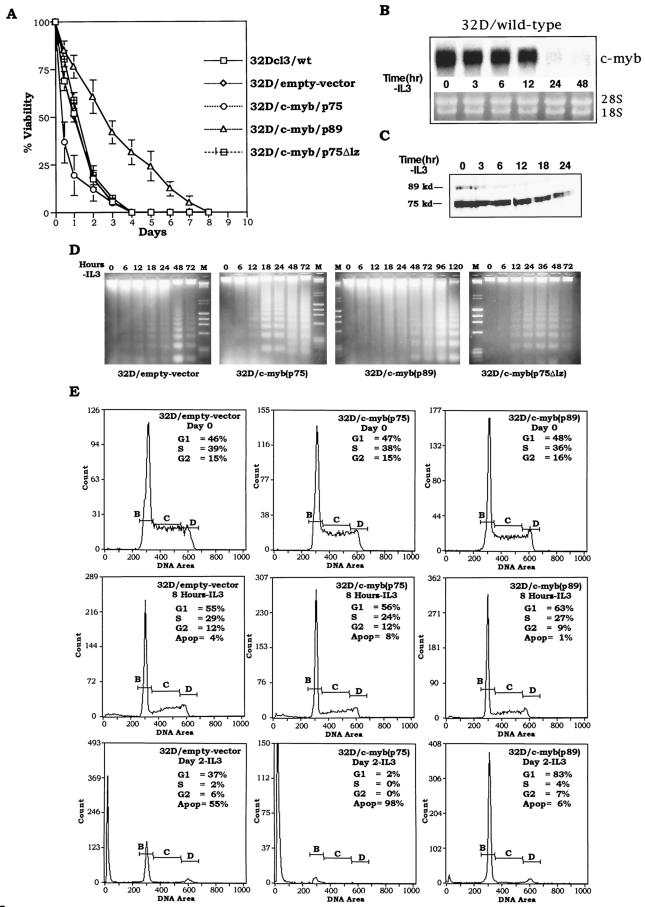
It has been shown previously that withdrawal of IL-3 from the incubation medium of 32Dcl3 cells results in the rapid downregulation of c-myc and c-myb mRNA expression, followed by accumulation of cells in the G1 phase of the cell cycle and eventual apoptotic cell death (2). It has also been demonstrated that ectopic overexpression of c-Myc in these cells results in accelerated apoptotic death of cells following IL-3 withdrawal (2). To study the effects of ectopic overexpression of c-Myb on apoptotic death initiated by IL-3 withdrawal, we examined the kinetics of cell death in wild-type 32Dcl3 cell line as well as the four clones derived from this cell line following transfection with pMT-neo or the p75c-myb, p89c-myb, and p75Δlz-c-myb expression vectors (Fig. 3A).
FIG.3.
Effect of p75, p75Δlz, and p89 c-Myb proteins on IL-3 withdrawal-induced apoptosis of 32Dcl3 cells. (A) Wild-type, mock-transfected (empty-vector pMTneo) and 32Dcl3 cells expressing exogenous p75, p75Δlz, and p89 c-Myb proteins were washed in IL-3-free medium and incubated up to 8 days. At each indicated time point, the cells were analyzed for viability by trypan blue exclusion. The curves represent a mean of three experiments. (B and C) Wild-type 32D cells were subjected to IL-3 withdrawal, and the quantity of c-myb mRNA and protein was measured at the indicated time points by Northern blot and Western blot analyses, respectively. As a control for RNA loading, filters were stained with ethidium bromide to compare the levels of 28S and 18S RNAs. (D) Analysis of DNA fragmentation. At the indicated times following IL-3 withdrawal, DNA fragments released from 10 × 106 cells from different 32D cell lines were extracted, separated by electrophoresis, and stained with ethidium bromide. (E) Cell cycle analysis of empty-vector, p75c-myb and p89c-myb cells at day 0, 8 h, and day 2 following IL-3 withdrawal. The cells were fixed and stained with propidium iodide, and DNA content was measured with a flow cytometer. The horizontal lines designated B, C, and D in the graphs represent the amount of DNA in the cells (1N, intermediate, and 2N, respectively) and therefore correspond to cells in G1, S, and G2 phases, respectively.
As has been described earlier, withdrawal of IL-3 from wild-type- or pMTneo vector-transfected cells resulted in a gradual loss of cell viability (2). At 24 h following IL-3 withdrawal, approximately 50% of the cells were found to be dead, and by 48 h, the cell viability was less than 20%. Cells expressing p75 c-Myb, on the other hand, exhibited an accelerated kinetics of cell death, with 75 to 80% loss of cell viability between 16 and 24 h. The kinetics of cell death in these cells was higher than that seen with 32Dcl3 cells that were transfected with c-myc expression vectors (data not shown). Interestingly, cells expressing the leucine zipper mutant, p75Δlz c-Myb, failed to show such an accelerated kinetics of death, suggesting that the leucine zipper domain plays a critical role in this process. Most interestingly, however, cells expressing p89 c-Myb showed considerably delayed kinetics of cell death, exhibiting minimal loss of cell viability in the first 24 h following IL-3 withdrawal. A 50% loss of cell viability was seen in these cells only after 72 h, and approximately 144 h of incubation in the absence of IL-3 was required to achieve 80% cell death (Fig. 3A). These results clearly suggest that the two isoforms of c-Myb perform opposite functions during the process of programmed cell death and the sequences encoded by exon 9A may play a positive role in maintaining cell viability under conditions of cytokine deprivation.
To verify that the above observations are not affected by the endogenous levels of c-myb in 32D cells, we subjected the wild-type 32D cells to IL-3 withdrawal and measured the mRNA and protein levels at different time points. The Northern and Western blots are shown in Fig. 3B and 3C, respectively. As expected, c-myb mRNA levels decreased with time following IL-3 withdrawal and were undetectable after 12 h. Similarly, the protein levels of c-myb p75 and p89 were also found to decrease following IL-3 withdrawal, consistent with the decrease in the mRNA level.
The phenomenon of apoptosis in the four cell lines was further verified by examining the degradation of DNA into oligonucleosomal fragments, which is shown in Fig. 3D. In empty-vector-transfected cells as well as cells transfected with the leucine zipper mutant of p75c-myb, the onset of DNA ladder formation did not occur for 24 h, and the most intense ladder formation was seen at the 48-h time point. In contrast, in 32Dcl3 cells transfected with p75c-myb, intense degradation of DNA was seen within 18 h following IL-3 withdrawal, which is consistent with the loss of viability seen in these cells. In cells transfected with p89c-myb, the DNA ladders appeared with delayed kinetics, with high-intensity ladders appearing only between 96 and 120 h following IL-3 withdrawal.
To determine the effects of ectopic overexpression of the p75 and p89 c-Myb isoforms on cell cycle progression following IL-3 withdrawal, we carried out flow cytometric determination of cell cycle distribution. Following ethanol fixation and propidium iodide staining, cells were fractionated with an Elite Coulter counter, and the percentage of cells distributed within the G0/G1, S, and G2/M phases of the cell cycle were determined. As illustrated in Fig. 3E, all three cell lines studied here showed similar profiles of cell cycle distribution at the zero time point, with approximately 50% of cells in the G0/G1 phase of the cell cycle and the rest in the S and G2/M phases of the cell cycle. Following the withdrawal of IL-3, the cells accumulated and arrested in the G0/G1 phase, as evident by the 8-h IL-3 time point. Subsequently, after 48 h, 60% of cells in empty-vector-transfected cells exhibited <2N DNA content, while the other 35% exhibited 2N DNA content, with very few showing >2N DNA content. On the other hand, >98% of cells expressing p75 c-Myb show <2N DNA content, further confirming the earlier observation that p75 c-Myb accelerates apoptotic death of 32Dcl3 cells. In sharp contrast, approximately 95% of cells expressing p89 c-Myb showed >2N DNA content, suggesting that ectopic expression of this protein facilitates the growth arrest of cells in the G0/G1 phase of the cell cycle and delays the onset of apoptosis induced by IL-3 deprivation.
Effects of ectopic overexpression of p75 and p89 c-Myb proteins on cytochrome c release following IL-3 withdrawal.
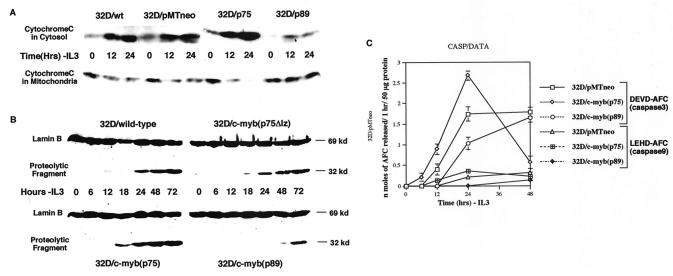
Previous studies have implicated mitochondrial damage and cytochrome c release in apoptotic cell death (29, 35, 67). We therefore assessed mitochondrial damage by examining the leakage of cytochrome c following IL-3 withdrawal in the four 32Dcl3 cell lines. To determine the subcellular distribution of cytochrome c, we prepared cytosolic and membrane (plus mitochondrial) fractions. Digitonin (0.05%) was used to selectively permeabilize the plasma membranes of cells to obtain the cytosol fractions (20). Digitonin-insoluble residues were further extracted with Triton X-100 to release membrane-associated and organelle-bound proteins. The two fractions were analyzed by SDS-PAGE and Western blotting. As shown in Fig. 4A, in the presence of IL-3 (0 h time point), most of the cytochrome c was detected in the membrane fraction (mitochondria) and very minimally in the cytosol. In wild-type- and empty-vector-transfected cells, at 12 and 24 h following IL-3 withdrawal, increasing amounts of cytochrome c were found in the cytosol. In comparison, the cytosol extracts from p75c-myb cells showed a much higher fraction of cytochrome c at 12 h following IL-3 withdrawal compared to that remaining in mitochondria, and by 24 h, most of the cytochrome c had leaked out of the mitochondria into the cytosol (Fig. 4A). Furthermore, cells transfected with p89c-myb showed little leakage of cytochrome c at 12 and 24 h following IL-3 withdrawal, and most of the cytochrome c was still detected in the mitochondria (Fig. 4A). These results further confirm our observations that p75 c-Myb and p89 c-Myb exhibit proapoptotic and antiapoptotic activities, respectively.
FIG. 4.
Analysis of caspase activity in p75-, p75Δlz-, and p89c-myb-transfected cells. (A) Cleavage of nuclear lamin B in wild-type, p75Δlz-c-myb, p75c-myb and p89c-myb cells subsequent to withdrawal of IL-3 from the medium. Cell lysates from different cell lines at the indicated time points were subjected to Western blotting with anti-lamin B antibody. The uncleaved lamin B is represented as a 69-kDa protein. A cleaved product of 32 kDa is detected secondary to lamin B breakdown by caspases. (B) Cytochrome c release during IL-3 withdrawal. Immunoblot analysis of cytosolic and organelle-bound fractions (as described in the text) after subjecting the various 32D cell lines to 0, 12, and 24 h of IL-3 withdrawal. The protein samples were analyzed by Western blotting with anti-cytochrome c monoclonal antibody. wt, wild-type; ev, empty vector. (C) Activation of DEVDase (caspase 3) and LEHDase (caspase 9) by IL-3 withdrawal. Empty-vector (pMTneo), p75c-myb and p89c-myb transfected cells were harvested at 0, 6, 12, 24, and 48 h following IL-3 withdrawal. After lysing the cells in Triton X-100-containing buffer as described in the text, the lysates were clarified by centrifugation, and the supernatants (50 μg of protein) were incubated with 50 μM substrate, including DEVD-AFC and LEHD-AFC, at 37°C for 1 h. Levels of released AFC were measured with a fluorescence microplate reader. Data are means of three experiments.
Effects of ectopic overexpression of p75, p75Δlz, and p89 c-Myb proteins on caspase activation following IL-3 withdrawal.
To demonstrate that the cell death seen in 32Dcl3 cells transfected with various plasmid constructs is due to apoptotic mechanisms, we examined the cleavage of nuclear lamin B by cell extracts, which has previously been shown to correlate with the activation of cellular caspases (33, 56). The results presented in Fig. 4B show that in wild-type cells, caspase activity appears around 24 h following IL-3 withdrawal, whereas extracts from p75c-myb-transfected cells show the appearance of this activity at an earlier time point. More strikingly, cells transfected with p89c-myb showed little or no caspase activity until about 72 h, further confirming our earlier observation that p89 c-Myb exhibits antiapoptotic activity. It is also interesting that cells transfected with the leucine zipper mutant of p75c-myb exhibit caspase activity identical to that seen with wild-type cells.
To determine the role of specific caspases in 32Dcl3 cells following IL-3 withdrawal, we measured the caspase activity in 32Dcl3 cells transfected with empty vector and with p75c-myb and p89c-myb. We examined caspase activities with the cleavage of tetrapeptides specific for various species. As shown in Fig. 4C, in mock-transfected cells, the DEVDase (caspase 3) activity progressively increased for 24 h following IL-3 withdrawal and then plateaued for the next 24 h. Strikingly, p75c-myb-transfected cells exhibited much higher kinetics of caspase 3 activity compared to empty-vector cells for the first 24 h following IL-3 withdrawal. This is consistent with the accelerated apoptotic cell death observed in these p75 c-Myb-expressing cells. The caspase 3 activity sharply declined from 24 to 48 h following IL-3 withdrawal, since much of the cell death had occurred in the first 24 h. Furthermore, p89c-myb-transfected cells exhibited a blunted activation of caspase-3 from 0 to 48 h following IL-3 withdrawal. Again, this is consistent with our earlier observation that p89 c-Myb expressing cells have a delayed apoptotic response. LETDase (caspase 9) showed a similar pattern of activation following IL-3 withdrawal but of much smaller magnitude (Fig. 4C). Other caspases (caspase 1, 4, 5, 6, 7, 8, and 10) did not show significant stimulation (data not shown). The activities of DEVDase and LEHDase were completely inhibited following IL-3 withdrawal by addition of VAD to the cell medium (data not shown).
Effects of ectopic overexpression of p75, p75Δlz, and p89 c-Myb proteins on IL-3 withdrawal-induced apoptosis of FDCP1 cells.
To confirm that the above observation about differential regulation of apoptosis by Myb isoforms upon cytokine withdrawal is not limited to 32Dcl3 cells, we used another cell line which was also derived from long-term murine bone marrow culture. We chose the FDCP1 cell line because of its phenotypic similarity to 32Dcl3 cells and because it represents early hematopoietic progenitor cells (14, 27). Like that of 32Dcl3 cells, the proliferation of FDCP1 cells is absolutely dependent on IL-3, and upon IL-3 withdrawal, these cells arrest in G0/G1 phase and subsequently undergo apoptosis. When cultured in the presence of G-CSF, these cells differentiate into monocytes (7).
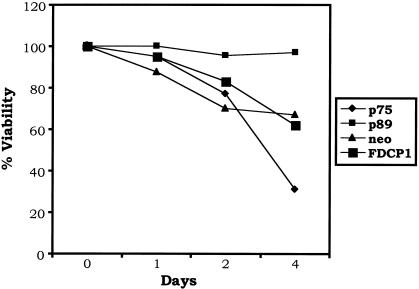
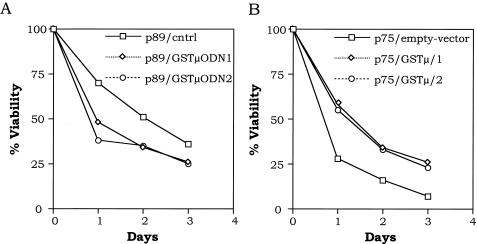
As described earlier with 32Dcl3 cells, we stably overexpressed p75 and p89 isoforms of c-Myb in FDCP1 cells and subjected these cells to IL-3 withdrawal. These results are presented in Fig. 5. By day 4 following cytokine withdrawal, approximately 40% of the wild-type- and the empty-vector-transfected cells underwent apoptotic cell death. However, the rate of programmed cell death in the p75 c-Myb-expressing cells was much more rapid; approximately 70% of the cells lost viability. On the other hand, less than 5% cell death was observed in p89 c-Myb-expressing cells by day 4. These results are consistent with those observed in 32Dcl3 cells and support our previous observations that p89c-myb confers a protective response to apoptotic stimuli, whereas p75c-myb promotes apoptosis upon factor withdrawal.
FIG. 5.
Effect of p75- and p89 c-Myb proteins on IL-3 withdrawal-induced apoptosis of FDCP1 cells. Wild-type, mock-transfected (empty-vector neo), and 32Dcl3 cells expressing exogenous p75 and p89 c-Myb proteins were washed in IL-3-free medium and incubated up to 4 days. At each indicated time point, the cells were analyzed for viability by trypan blue exclusion. The curves represent means of three experiments.
Analysis of nature of target genes induced/repressed by the two isoforms of c-Myb.
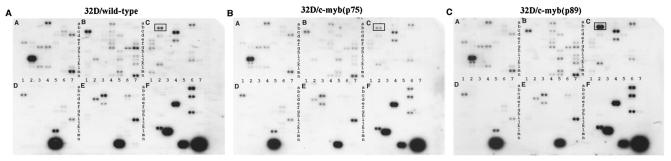
To gain an understanding of the molecular basis of the observed mode of action of the two isoforms of c-Myb, we first screened for gene expression changes with mouse cDNA expression arrays (Clontech). The Atlas mouse cDNA expression array used in this study contained 588 mouse cDNAs derived from members of oncogenes, stress response genes, apoptotic genes, transcription factors, and genes encoding growth factors, cytokine receptors, and signal transducing proteins. To identify genes that are regulated by p75c-myb and p89c-myb, mRNA was prepared from wild-type 32Dcl3 cells as well as cells ectopically expressing the p75 and p89 isoforms of c-Myb following IL-3 withdrawal for 9 h. 32P-labeled probes were made from each of the mRNA preparations, and the filters were hybridized with each of the labeled probes.
Analysis of multiple hybridization reactions revealed a consistent difference in the expression profile of a single gene encoding GSTμ (Fig. 6). This was an unexpected result as none of the other apoptotic response genes such as those belonging to the Bcl-2 or caspase gene families showed a significant alteration in their patterns of expression. RNase protection analysis carried out with probes for the Bcl-2 and caspase gene family members further confirmed this observation (data not shown). By quantitative analysis, a twofold reduction in the expression of GSTμ was observed in 32D/p75 cells compared to the wild-type cells. By contrast, a greater than fourfold increase was found in 32D/p89 cells compared to 32D/p75 cells and an approximately 2.5-fold increase compared to wild-type cells.
FIG. 6.
Gene expression profile of 32Dcl3 cells with constitutive expression of p75 c-Myb and p89 c-Myb in the absence of IL-3. Atlas mouse cDNA expression arrays (Clontech) were used to analyze the differential gene expression of 32Dcl3 cells transfected with alternatively spliced c-myb gene products 9 h following IL-3 withdrawal. The arrays include 588 mouse cDNAs, 9 housekeeping control cDNAs, and negative controls immobilized in duplicate on a nylon membrane. Each array is divided in six functional subclasses (A to F) of 98 genes each. The genes on each functional subclass are laid on a 7 by 14 grid (1 to 7 horizontally and a to n vertically). Following hybridization and washing, the array filters were scanned with a Fuji PhosphorImager. (A) Gene expression of empty-vector-transfected 32Dcl3 cells at 9 h afrter IL-3. (B) Gene expression of p75c-myb-transfected 32Dcl3 cells at 9 h after IL-3. (C) Gene expression of p89c-myb-transfected 32Dcl3 cells at 9 h after IL-3. The boxes in panels A, B, and C highlight the position and expression of GSTμ in the different cell lines.
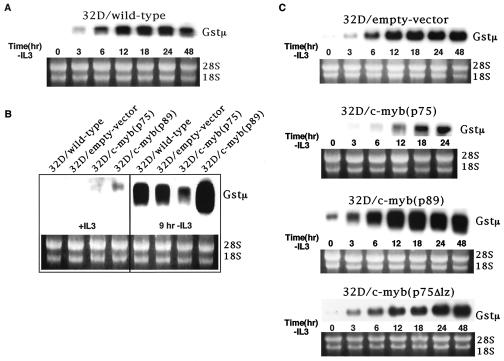
To verify whether the observation made with the cDNA array filters reflected the expression pattern of endogenous GSTμ mRNA levels, we examined the expression of GSTμ in 32Dcl3 cells during their proliferation in the presence of IL-3 as well as during the process of apoptosis seen following the withdrawal of IL-3. These results are presented in Fig. 7A, 7B and 7C and show that this gene is expressed at very low levels in 32Dcl3 cells proliferating in the presence of IL-3 and its expression is induced within 3 h following the withdrawal of IL-3 and the levels reached their peak between 12 and 18 h (Fig. 7A). Most interestingly, the levels of GSTμ mRNA were considerably diminished in cells expressing p75 c-Myb, while their levels were four- to fivefold higher in cells expressing p89 c-Myb compared to their wild-type controls or empty-vector transfected cells (Fig. 7C).
FIG. 7.
Activation of GSTμ in 32Dcl3 cells following IL-3 withdrawal. (A) Northern blot analysis of total RNA extracted at the indicated times following IL-3 withdrawal from wild-type 32Dcl3 cells with a full-length GSTμ cDNA probe. As an internal control for RNA loading, blots were stained with ethidium bromide after completion of RNA transfer onto the nitrocellulose filter to compare levels of 18S and 28S rRNAs across lanes (shown below). (B) Comparison of GSTμ expression by Northern blotting in wild-type-, empty-vector-, p75c-myb-, and p89c-myb-transfected cells in the presence of IL-3 and at 9 h following IL-3 withdrawal. (C) Expression pattern of GSTμ in empty-vector-, p75c-myb-, p89c-myb-, and p75Δlz-c-myb-transfected cells. Total RNA was isolated from each cell line at the indicated time points following IL-3 withdrawal and subjected to Northern blotting.
To directly compare the GSTμ mRNA levels among the different cell types, we performed Northern blot analysis at 9 h following IL-3 withdrawal (Fig. 7B). Quantitative analysis of the Northern blot revealed that there was a twofold reduction in GSTμ expression in 32D/p75 cells compared to wild-type cells. In addition, a greater than threefold increase in GSTμ expression was found in 32D/p89 cells compared to wild-type cells. These results are consistent with the microarray data. It is also interesting that the leucine zipper mutant had no effect on the expression of GSTμ, as the levels of this mRNA in these cells were equivalent to that seen in wild-type- and empty-vector-transfected cells.
Effect of ectopic overexpression of GSTμ on apoptotic program of 32Dcl3 cells.
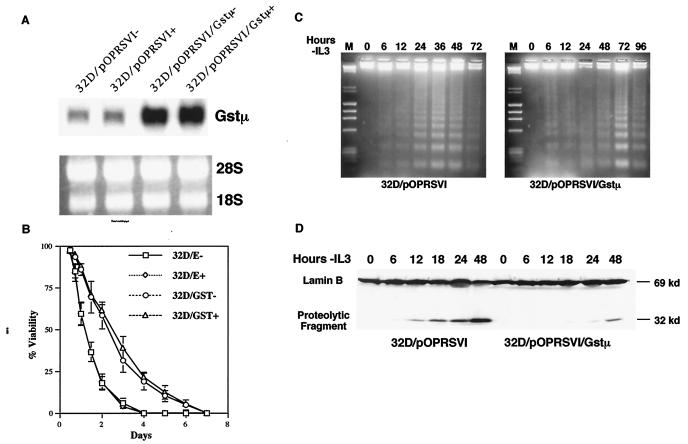
To determine whether the two isoforms of c-Myb exert their effects through the regulation of GSTμ, we created an inducible expression vector of GSTμ where the cDNA of murine GSTμ was placed under the control of the Rous sarcoma virus promoter. Following selection in hygromycin and G418, mass cultures as well as single cell clones were established from each stable transfection. The derived cell lines were tested for the expression of the transgene, which showed that the GSTμ mRNA is constitutively expressed in these cell lines even in the absence of the inducer (Fig. 8A). When cells ectopically overexpressing the GSTμ mRNA were subjected to IL-3 withdrawal, it was observed that they exhibited delayed kinetics of cell death (Fig. 8B), similar to that seen with p89 c-Myb-expressing 32Dcl3 cells.
FIG. 8.
Effect of GSTμ on IL-3 withdrawal mediated apoptosis of 32Dcl3 cells. (A) Constitutive expression of GSTμ cDNA in 32Dcl3 cells with the IPTG-inducible pOPRSVI/MCS vector. Total RNA was extracted from mock-transfected and GSTμ-transfected cells and probed with full-length GSTμ cDNA in the absence or presence of IPTG. As a control for RNA loading, filters were stained with ethidium bromide to compare the levels of 28S and 18S RNAs (shown below). (B) Analysis of the viability of GSTμ-transfected cells in the absence of IL-3. Mock-transfected (32D/E) and GSTμ-transfected (32D/GST) 32Dcl3 cells were washed in IL-3-free medium and incubated up to 8 days. At each indicated time point, the cells were analyzed for viability by trypan blue exclusion. The curves represent means of three experiments. (C) Analysis of DNA fragmentation. At the indicated times following IL-3 withdrawal, DNA fragments released from 107 cells from the two 32D cell lines were extracted and separated by electrophoresis and stained with ethidium bromide. (D) Analysis of caspase activity by determining the breakdown of nuclear lamin B. Mock-transfected and GSTμ-transfected cells were incubated in IL-3-free medium, and the cell lysates from the indicated time points were subjected to Western blotting with anti-lamin B antibody. The full-length lamin B is detected as a 69-kDa protein. The caspase activity is marked by lamin B cleavage, detected as a 32-kDa product.
This observation could be further verified by examining the degradation of DNA into oligonucleosomal fragments, which is shown in Fig. 8C. In empty-vector-transfected cells, the onset of DNA ladder formation did not occur for 24 h, and the most intense ladder formation was seen at 36 to 48 h. In contrast, in 32Dcl3 cells transfected with GSTμ, the appearance of DNA ladders was delayed, with high-intensity ladders appearing only after 72 h following IL-3 withdrawal. When these cell lysates were examined for caspase activity by determining the level of lamin B cleavage (Fig. 8D), it was determined that the caspase activity was delayed, a scenario which is very similar to that seen with 32Dcl3 cells transfected with expression vectors encoding p89c-myb. Taken together, these results suggest that p89 c-Myb exerts its antiapoptotic effects through the regulation of GSTμ.
Effect of inhibition of GSTμ in 32Dcl3/c-myb p89 cells.
To determine whether GSTμ mediates enhanced survival in 32Dcl3 cells expressing c-myb p89, we used GSTμ antisense oligodeoxynucleotides to suppress the expression of GSTμ in these cells. Selective and potent inhibitors of GSTμ have been described recently (57). We used GSTμODN1 (5′-CCACTGGCTTCGGTCATAGT-3′), shown to inhibit GSTμ expression up to 95%, and GSTμODN2 (5′-CAGGCCCTCAAAGCGACCCA-3′), shown to inhibit GSTμ expression up to 85% (57). As a negative control, we used a nonspecific oligonucleotide, 5′-TGAGAGCTGAAAGCAGGTCCAT-3′. The oligodeoxynucleotides were transfected in 32D/p89 cells with the folic acid-polylysine method as described earlier (34). Following transfection and incubation in growth medium for 3 h, the cells were washed in IL-3-free medium and subjected to cytokine withdrawal-mediated apoptosis. The results are shown in Fig. 9A. The p89 cells transfected with antisense GSTμODN1 and GSTμODN2 underwent cell death at a faster rate compared to that observed for the cells transfected with nonspecific control oligodeoxynucleotides. Thus, enhanced expression of GSTμ in p89 cells confers a protective effect against apoptotic cell death. Inhibition of GSTμ in these cells leads to increased susceptibility to apoptosis.
FIG. 9.
Effect of GSTμ on 32D/c-myb p75 and p89 cells. (A) 32D/c-myb p89 cells were transfected with GSTμ antisense oligodeoxynucleotides (ODN1 and ODN2) and a nonspecific oligonucleotide (cntrl). At 3 h following transfection, the cells were washed in IL-3-free medium and incubated for 3 days. At each indicated time point, the cells were analyzed for viability by trypan blue exclusion. The curves represent means of three experiments. (B) 32D/c-myb p75 cells were transfected either with an empty vector or with the GSTμ expression vector, and stable transfectants were selected (GSTμ/1 and GSTμ/2). Cells were washed in IL-3-free medium and incubated for 3 days. At each indicated time point, the cells were analyzed for viability by trypan blue exclusion. The curves represent means of three experiments.
Effect of ectopic expression of GSTμ in 32Dcl3/c-myb p75 cells.
As shown earlier, decreased levels of GSTμ expression were found in 32Dcl3 cells expressing the p75 isoform of c-Myb. To determine whether expression of GSTμ confers increased resistance to apoptosis in p75 cells, we transfected the GSTμ expression vector into these cells with a puromycin-based construct and selected stable clones. Following verification of GSTμ expression, we subjected the cells to IL-3 withdrawal and performed viability assays. The results are shown in Fig. 9B. As expected, mock-transfected 32D/p75 cells underwent cell death at an enhanced rate compared to the control cell line. However, p75 cells transfected with GSTμ (p75/GSTμ/1 and p75/GSTμ/2) exhibited decreased rates of cell death compared to empty-vector-transfected cells. Thus, downregulation of GSTμ confers enhanced susceptibility to 32D/p75 cells, and these cells can be rescued by enforced expression of GSTμ.
DISCUSSION
In this report, we describe a novel mechanism by which the two translational products of the c-myb proto-oncogene regulate programmed cell death of myeloid cells through differential transcriptional regulation of the GSTμ gene. The major translational product of the c-myb proto-oncogene is a 75-kDa nuclear protein with 636 amino acids which is expressed in most hematopoietic tissues (42, 63). In addition to this 75-kDa protein product, another translational product of 89 kDa was found to be encoded by c-myb in avian, murine, and human hematopoietic cells (11, 17). This 89-kDa protein is translated from an alternatively spliced mRNA encoded by the c-myb gene, which results in the addition of 363 bp between exons 9 and 10. This region has been designated exon 9A (50, 55).
These translated proteins were found to be localized in the nucleus (17) and to function as transcriptional activators with identical sequence-specific DNA binding activities (reviewed in reference 42). Both forms of c-Myb contain three functional domains: (i) the N-terminal DNA binding domain, (ii) the central transactivation domain, and (iii) the C-terminal negative regulatory domain.
One of the interesting characteristics noted in the negative regulatory domain of p75 c-Myb was a putative leucine zipper motif with one isoleucine and three leucine residues (5) resembling that of other leucine zipper domains seen in other transcription factors (32). While the precise role of this leucine zipper domain in c-Myb function is unclear, a role for this domain in the negative regulation of c-Myb has been proposed. This view is supported by the observation that mutation of one or more leucines to proline residues has been shown to result in increased transactivation and transformation activities of the mutant molecules, a phenomenon similar to that seen with C-terminal truncation (28). The precise mechanism by which the leucine zipper domain exerts its negative regulatory effect remains unresolved.
It has been suggested that this region might mediate the formation of Myb homodimers which are unable to bind DNA thereby contributing to a negative regulatory effect (39). A second model proposes a role for the leucine zipper region in mediating interaction with other cellular proteins, leading to the negative regulation of c-Myb (19). Recently, a gene that encodes a protein that can bind to the leucine zipper domain of c-Myb has been identified (58). In contrast to the above set of studies, results from another study show that deletion of the leucine zipper region has no effect on the transcriptional transactivation characteristics of the mutant protein. Instead, deletion of two other regions within the C-terminal domain abolished the negative regulatory effect of the C-terminal domain, leading to the suggestion that these domains might be more important for the negative regulation of c-Myb protein (16). It is interesting that the p89 form of the c-Myb protein lacks the putative leucine zipper domain because of the insertion of exon 9A-encoded sequences, resulting in its disruption. Results presented in this report as well as experiments carried out with chicken p89 c-Myb (66) show that p89 c-Myb exhibits higher transactivational activities compared to its p75 counterpart.
While the existence of the two translational products of c-myb has been known since 1986, the biological relevance of the existence of these two proteins has eluded an explanation. c-Myb is highly expressed in immature hematopoietic cells and is downregulated during cytokine- or chemically induced terminal differentiation of these cells (23, 30, 47, 63). Furthermore, constitutive expression of c-Myb in these cells was found to block the differentiation of these cell lines (6, 8, 36, 44, 45, 54, 60). These results suggest that downregulation of c-myb is required for terminal differentiation. With inducible expression of c-Myb, it was shown that the later phase of downregulation of c-myb is critical for the commitment of cells to terminal differentiation (6, 10). Past studies also demonstrate that the v-myb and c-myb gene products promote cell proliferation and suppress apoptosis under conditions that favor cell growth (21, 53). Thus, in the presence of cytokines such as IL-3 and IL-6, which promote growth, c-myb plays an indispensable role in the growth process (42, 54). Similarly, many transformed cell lines are dependent on c-myb for cell growth, as evidenced by the fact that antisense oligonucleotides directed against c-myb or dominant negative forms of myb block cell growth and induce apoptosis in these cells (53, 59). Under these conditions, the c-myb and v-myb genes have also been shown to regulate Bcl-2 expression (21, 53, 59). However, under conditions that favor apoptosis (such as treatment of cells with transforming growth factor beta or withdrawal of cytokines from the culture medium), expression of the p75 isoform of c-bmy has been shown to promote apoptosis (54). This is an analogous situation seen with c-myc, which is induced by growth factors and has been found to be necessary for cell proliferation mediated by these growth signals (2). Nevertheless, like c-myb, expression of c-myc under conditions that favor apoptosis (such as withdrawal of growth factors) has been found to result in the acceleration of apoptosis.
It is interesting that the results presented here show that the two isoforms of c-Myb exert opposite effects on myeloid cells undergoing programmed cell death following deprivation of cytokines from the growth medium. It has been shown earlier that withdrawal of IL-3 from the culture medium results in the arrest of 32Dcl3 cells in the G1 phase of the cell cycle, followed by their entry into the apoptotic phase. Ectopic overexpression of c-Myc in these cells was found to result in the acceleration of their entry into the death phase (2). Our results presented here show that, like c-Myc, ectopic overexpression of p75 c-Myb results in the acceleration of cell death. In fact, the rate of induction of cell death was higher in cells overexpressing p75 c-Myb than that observed in c-Myc-overexpressing cells (data not shown). In sharp contrast, ectopic overexpression of p89 c-Myb in these cells confers a protective effect on the onset of cell death.
Our studies presented here also show that the two isoforms of c-Myb exert these opposite effects on the apoptotic program of 32Dcl3 cells through transcriptional regulation of a stress response gene, GSTμ. Interestingly, one of the early responses that 32Dcl3 cells exhibit following IL-3 withdrawal is the upregulation of GSTμ expression, which is seen in the first 3 h following cytokine removal (Fig. 7). Expression of GSTμ reaches peak levels within 12 h, after which the RNA levels remain constant. Ectopic overexpression of p75 c-Myb appears to inhibit the expression of GSTμ by four- to sixfold, while a similar expression of p89 c-Myb seems to result in five- to sixfold overexpression of this gene. Thus, the two isoforms of c-Myb appear to exert opposite effects on the transcription of GSTμ. Most interestingly, the leucine zipper mutant of p75 c-Myb had no effect on the mRNA levels of GSTμ, implicating a role for the leucine zipper in the transcriptional regulation of this gene. It is interesting that 32Dcl3 cells growing in the presence of IL-3 did not express GSTμ. However, when the cells were transfected with expression vectors coding for p89 c-Myb, a low-level expression of GSTμ was seen in these cells (Fig. 7). These results suggest that IL-3 acts as an inhibitor of GSTμ expression and that p89 c-Myb can overcome some of the inhibitory effects of IL-3 on GSTμ expression.
We could also demonstrate that GSTμ directly regulates the apoptotic response of 32Dcl3 cells by transfection of expression vectors that code for this gene into 32Dcl3 cells. Cells ectopically expressing GSTμ were found to exhibit kinetics of cell death similar to those seen with cells expressing p89 c-Myb, further suggesting that c-Myb proteins modulate programmed cell death through the regulation of this gene. It is interesting that ectopic overexpression of p89 c-Myb as well as GSTμ was found to result in stabilization of the mitochondrial membrane, as seen by lack of cytochrome c release, which in turn appears to delay the activation of caspase activity in the cell. While the precise mechanism associated with stabilization of the mitochondrial membrane is at present unclear, it appears to be related to the accumulation of reactive oxygen species during programmed cell death resulting from the deprivation of cytokines. It is also possible that accumulation of reactive oxygen species is one of the events that activate loss of the mitochondrial membrane integrity and, consequently, leakage of cytochrome c, and this reaction is counterbalanced by the cell through the expression of free radical scavengers such as GSTμ. In 32Dcl3 cells overexpressing p75 c-Myb, there appears to be a destabilization of mitochondrial membrane resulting in a rapid release of cytochrome c and activation of caspase activity in the cell.
The results presented here demonstrate cross talk between three sets of genes, the c-myb proto-oncogene products, free-radical scavengers such as GSTμ, and proteins such as caspases that belong to the apoptotic machinery. It is also interesting that while c-myc and p75c-myb bring about similar effects on programmed cell death induced by cytokine withdrawal, the mechanisms of action of these two oncogenes appear to be completely different, since we did not see any effect on GSTμ transcription in 32Dcl3 cells ectopically overexpressing c-Myc (data not shown). Thus, different oncogenes appear to modulate cell growth and cell death through the modulation of different sets of genes, and a detailed understanding of these mechanisms is likely to lead to a better understanding of complex regulatory processes that bring about homeostasis.
Acknowledgments
We thank Pothana Saikumar for help with cell fractionation and caspase activity assays. We also thank Tecan US Inc. for the generous loan of the SpectroFluor plate reader. We are grateful to M. V. Ramana Reddy for preparation of the folic acid-polylysine conjugate.
This work was supported by grants from the National Cancer Institute (CA79086), and the Fels Foundation. Core facilities used in this study were supported by NIH R24 CA88261.
REFERENCES
- 1.Arends, M. J., R. G. Morris, and A. H. Wyllie. 1990. Apoptosis. The role of the endonuclease. Am. J. Pathol. 136:593-608. [PMC free article] [PubMed] [Google Scholar]
- 2.Askew, D. S., R. A. Ashmun, B. C. Simmons, and J. L. Cleveland. 1991. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene 6:1915-1922. [PubMed] [Google Scholar]
- 3.Baker, S. J., and E. P. Reddy. 1998. Modulation of life and death by the TNF receptor superfamily. Oncogene 17:3261-3270. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, A., J. Agapite, and H. Steller. 1998. Mechanisms and control of programmed cell death in invertebrates. Oncogene 17:3215-3223. [DOI] [PubMed] [Google Scholar]
- 5.Biedenkapp, H., U. Borgmeyer, A. E. Sippel, and K. H. Klempnauer. 1988. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature 335:835-837. [DOI] [PubMed] [Google Scholar]
- 6.Bies, J., R. Mukhopadhyaya, J. Pierce, and L. Wolff. 1995. Only late, nonmitotic stages of granulocyte differentiation in 32Dcl3 cells are blocked by ectopic expression of murine c-myb and its truncated forms. Cell Growth Differ. 6:59-68. [PubMed] [Google Scholar]
- 7.Bourette, R. P., G. M. Myles, K. Carlberg, A. R. Chen, and L. R. Rohrschneider. 1995. Uncoupling of the proliferation and differentiation signals mediated by the murine macrophage colony-stimulating factor receptor expressed in myeloid FDC-P1 cells. Cell Growth Differ. 6:631-645. [PubMed] [Google Scholar]
- 8.Clarke, M. F., J. F. Kukowska-Latallo, E. Westin, M. Smith, and E. V. Prochownik. 1988. Constitutive expression of a c-myb cDNA blocks Friend murine erythroleukemia cell differentiation. Mol Cell. Biol. 8:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleveland, J. L., M. Dean, N. Rosenberg, J. Y. Wang, and U. R. Rapp. 1989. Tyrosine kinase oncogenes abrogate interleukin-3 dependence of murine myeloid cells through signaling pathways involving c-myc: conditional regulation of c-myc transcription by temperature-sensitive v-abl. Mol Cell. Biol. 9:5685-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danish, R., O. el-Awar, B. L. Weber, J. Langmore, L. A. Turka, J. J. Ryan, and M. F. Clarke. 1992. c-myb effects on kinetic events during MEL cell differentiation. Oncogene 7:901-907. [PubMed] [Google Scholar]
- 11.Dasgupta, P., and E. P. Reddy. 1989. Identification of alternatively spliced transcripts for human c-myb: molecular cloning and sequence analysis of human c-myb exon 9A sequences. Oncogene 4:1419-1423. [PubMed] [Google Scholar]
- 12.Dean, M., J. L. Cleveland, U. R. Rapp, and J. N. Ihle. 1987. Role of myc in the abrogation of IL-3 dependence of myeloid FDC-P1 cells. Oncogene Res. 1:279-296. [PubMed] [Google Scholar]
- 13.Dean, M., R. A. Levine, W. Ran, M. S. Kindy, G. E. Sonenshein, and J. Campisi. 1986. Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J. Biol. Chem. 261:9161-9166. [PubMed] [Google Scholar]
- 14.Dexter, T. M., J. Garland, D. Scott, E. Scolnick, and D. Metcalf. 1980. Growth of factor-dependent hemopoietic precursor cell lines. J. Exp. Med. 152:1036-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dragovich, T., C. M. Rudin, and C. B. Thompson. 1998. Signal transduction pathways that regulate cell survival and cell death. Oncogene 17:3207-3213. [DOI] [PubMed] [Google Scholar]
- 16.Dubendorff, J. W., L. J. Whittaker, J. T. Eltman, and J. S. Lipsick. 1992. Carboxy-terminal elements of c-Myb negatively regulate transcriptional activation in cis and in trans. Genes Dev. 6:2524-2535. [DOI] [PubMed] [Google Scholar]
- 17.Dudek, H., and E. P. Reddy. 1989. Identification of two translational products for c-myb. Oncogene 4:1061-1066. [PubMed] [Google Scholar]
- 18.Dudek, H., R. V. Tantravahi, V. N. Rao, E. S. Reddy, and E. P. Reddy. 1992. Myb and Ets proteins cooperate in transcriptional activation of the mim-1 promoter. Proc. Natl. Acad. Sci. USA 89:1291-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Favier, D., and T. J. Gonda. 1994. Detection of proteins that bind to the leucine zipper motif of c-Myb. Oncogene 9:305-311. [PubMed] [Google Scholar]
- 20.Fiskum, G., S. W. Craig, G. L. Decker, and A. L. Lehninger. 1980. The cytoskeleton of digitonin-treated rat hepatocytes. Proc. Natl. Acad. Sci. USA 77:3430-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frampton, J., T. Ramqvist, and T. Graf. 1996. v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes Dev. 10:2720-2731. [DOI] [PubMed] [Google Scholar]
- 22.Gaozza, E., S. J. Baker, R. K. Vora, and E. P. Reddy. 1997. AATYK: a novel tyrosine kinase induced during growth arrest and apoptosis of myeloid cells. Oncogene 15:3127-3135. [DOI] [PubMed] [Google Scholar]
- 23.Gonda, T. J., and D. Metcalf. 1984. Expression of myb, myc and fos proto-oncogenes during the differentiation of a murine myeloid leukaemia. Nature 310:249-251. [DOI] [PubMed] [Google Scholar]
- 24.Greenberger, J. S., R. J. Eckner, M. Sakakeeny, P. Marks, D. Reid, G. Nabel, A. Hapel, J. N. Ihle, and K. C. Humphries. 1983. Interleukin 3-dependent hematopoietic progenitor cell lines. Fed. Proc. 42:2762-2771. [PubMed] [Google Scholar]
- 25.Gross, A., J. M. McDonnell, and S. J. Korsmeyer. 1999. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 13:1899-1911. [DOI] [PubMed] [Google Scholar]
- 26.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension with the polymerase chain reaction. Gene. 77:51-59. [DOI] [PubMed] [Google Scholar]
- 27.Ihle, J. N., and D. Askew. 1989. Origins and properties of hematopoietic growth factor-dependent cell lines. Int. J. Cell Cloning 7:68-91. [DOI] [PubMed] [Google Scholar]
- 28.Kanei-Ishii, C., E. M. MacMillan, T. Nomura, A. Sarai, R. G. Ramsay, S. Aimoto, S. Ishii, and T. J. Gonda. 1992. Transactivation and transformation by Myb are negatively regulated by a leucine-zipper structure. Proc. Natl. Acad. Sci. USA 89:3088-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kluck, R. M., E. Bossy-Wetzel, D. R. Green, and D. D. Newmeyer. 1997. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 275:1132-1136. [DOI] [PubMed] [Google Scholar]
- 30.Kuehl, W. M., T. P. Bender, J. Stafford, D. McClinton, S. Segal, and E. Dmitrovsky. 1988. Expression and function of the c-myb oncogene during hematopoietic differentiation. Curr. Top. Microbiol. Immunol. 141:318-323. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, A., C. M. Lee, and E. P. Reddy. 2003. c-Myc is essential but not sufficient for c-Myb-mediated block of granulocytic differentiation. J. Biol. Chem. 278:11480-11488. [DOI] [PubMed] [Google Scholar]
- 32.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 33.Lazebnik, Y. A., A. Takahashi, R. D. Moir, R. D. Goldman, G. G. Poirier, S. H. Kaufmann, and W. C. Earnshaw. 1995. Studies of the lamin proteinase reveal multiple parallel biochemical pathways during apoptotic execution. Proc. Natl. Acad. Sci. USA 92:9042-9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leopold, L. H., S. K. Shore, T. A. Newkirk, R. M. Reddy, and E. P. Reddy. 1995. Multi-unit ribozyme-mediated cleavage of bcr-abl mRNA in myeloid leukemias. Blood 85:2162-2170. [PubMed] [Google Scholar]
- 35.Liu, X., C. N. Kim, J. Yang, R. Jemmerson, and X. Wang. 1996. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86:147-157. [DOI] [PubMed] [Google Scholar]
- 36.McClinton, D., J. Stafford, L. Brents, T. P. Bender, and W. M. Kuehl. 1990. Differentiation of mouse erythroleukemia cells is blocked by late up-regulation of a c-myb transgene. Mol Cell. Biol. 10:705-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Migliaccio, G., A. R. Migliaccio, V. Broudy, B. Kreider, G. Rovera, and J. W. Adamson. 1989. Selection of lineage-restricted cell lines immortalized at different stages of hematopoietic differentiation from the murine cell line 32 D. Prog. Clin. Biol. Res. 183-196. [PubMed]
- 38.Ness, S. A., A. Marknell, and T. Graf. 1989. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 59:1115-1125. [DOI] [PubMed] [Google Scholar]
- 39.Nomura, T., N. Sakai, A. Sarai, T. Sudo, C. Kanei-Ishii, R. G. Ramsay, D. Favier, T. J. Gonda, and S. Ishii. 1993. Negative autoregulation of c-Myb activity by homodimer formation through the leucine zipper. J. Biol. Chem. 268:21914-21923. [PubMed] [Google Scholar]
- 40.Nordeen, S. K. 1988. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques 6:454-458. [PubMed] [Google Scholar]
- 41.Oh, I. H., and E. P. Reddy. 1997. Murine A-myb gene encodes a transcription factor, which cooperates with Ets-2 and exhibits distinctive biochemical and biological activities from c-myb. J. Biol. Chem. 272:21432-21443. [DOI] [PubMed] [Google Scholar]
- 42.Oh, I. H., and E. P. Reddy. 1999. The myb gene family in cell growth, differentiation and apoptosis. Oncogene 18:3017-3033. [DOI] [PubMed] [Google Scholar]
- 43.Packham, G., and J. L. Cleveland. 1994. Ornithine decarboxylase is a mediator of c-Myc-induced apoptosis. Mol Cell. Biol. 14:5741-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel, G., B. Kreider, G. Rovera, and E. P. Reddy. 1993. v-myb blocks granulocyte colony-stimulating factor-induced myeloid cell differentiation but not proliferation. Mol Cell. Biol. 13:2269-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel, G., R. Tantravahi, I. H. Oh, and E. P. Reddy. 1996. Transcriptional activation potential of normal and tumor-associated myb isoforms does not correlate with their ability to block GCSF-induced terminal differentiation of murine myeloid precursor cells. Oncogene 13:1197-1208. [PubMed] [Google Scholar]
- 46.Prendergast, G. C. 1999. Mechanisms of apoptosis by c-Myc. Oncogene 18:2967-2987. [DOI] [PubMed] [Google Scholar]
- 47.Ramsay, R. G., K. Ikeda, R. A. Rifkind, and P. A. Marks. 1986. Changes in gene expression associated with induced differentiation of erythroleukemia: protooncogenes, globin genes, and cell division. Proc. Natl. Acad. Sci. USA 83:6849-6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rathmell, J. C., and C. B. Thompson. 1999. The central effectors of cell death in the immune system. Annu. Rev. Immunol. 17:781-828. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez-Tarduchy, G., M. Collins, and A. Lopez-Rivas. 1990. Regulation of apoptosis in interleukin-3-dependent hemopoietic cells by interleukin-3 and calcium ionophores. EMBO J. 9:2997-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosson, D., D. Dugan, and E. P. Reddy. 1987. Aberrant splicing events that are induced by proviral integration: implications for myb oncogene activation. Proc. Natl. Acad. Sci. USA 84:3171-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rovera, G., B. Kreider, N. Shirsat, D. Venturelli, G. Naso, and F. Mavilio. 1989. Alteration of the program of terminal differentiation caused by oncogenes in the hemopoietic progenitor cell line 32D C13 (G). Ann. N. Y. Acad. Sci. 567:154-164. [DOI] [PubMed] [Google Scholar]
- 52.Sakura, H., C. Kanei-Ishii, T. Nagase, H. Nakagoshi, T. J. Gonda, and S. Ishii. 1989. Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc. Natl. Acad. Sci. USA 86:5758-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salomoni, P., D. Perrotti, R. Martinez, C. Franceschi, and B. Calabretta. 1997. Resistance to apoptosis in CTLL-2 cells constitutively expressing c-Myb is associated with induction of BCL-2 expression and Myb-dependent regulation of bcl-2 promoter activity. Proc. Natl. Acad. Sci. USA 94:3296-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selvakumaran, M., H. K. Lin, R. T. Sjin, J. C. Reed, D. A. Liebermann, B. Hoffman. 1994. The novel primary response gene MyD118 and the proto-oncogenes myb, myc, and bcl-2 modulate transforming growth factor beta 1-induced apoptosis of myeloid leukemia cells. Mol Cell. Biol. 14:2352-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen-Ong, G. L. 1987. Alternative internal splicing in c-myb RNAs occurs commonly in normal and tumor cells. EMBO J. 6:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimizu, T., C. X. Cao, R. G. Shao, and Y. Pommier. 1998. Lamin B phosphorylation by protein kinase calpha and proteolysis during apoptosis in human leukemia HL60 cells. J. Biol. Chem. 273:8669-8674. [DOI] [PubMed] [Google Scholar]
- 57.t Hoen, P. A., B. S. Rosema, J. N. Commandeur, N. P. Vermeulen, M. Manoharan, T. J. van Berkel, E. A. Biessen, and M. K. Bijsterbosch. 2002. Selection of effective antisense oligodeoxynucleotides with a green fluorescent protein-based assay. Discovery of selective and potent inhibitors of glutathione S-transferase Mu expression. Eur. J. Biochem. 269:2574-2583. [DOI] [PubMed] [Google Scholar]
- 58.Tavner, F. J., R. Simpson, S. Tashiro, D. Favier, N. A. Jenkins, D. J. Gilbert, N. G. Copeland, E. M. Macmillan, J. Lutwyche, R. A. Keough, S. Ishii, and T. J. Gonda. 1998. Molecular cloning reveals that the p160 Myb-binding protein is a novel, predominantly nucleolar protein which may play a role in transactivation by Myb. Mol Cell. Biol. 18:989-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor, D., P. Badiani, and K. Weston. 1996. A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev. 10:2732-2744. [DOI] [PubMed] [Google Scholar]
- 60.Todokoro, K., R. J. Watson, H. Higo, H. Amanuma, S. Kuramochi, H. Yanagisawa, and Y. Ikawa. 1988. Down-regulation of c-myb gene expression is a prerequisite for erythropoietin-induced erythroid differentiation. Proc. Natl. Acad. Sci. USA 85:8900-8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valtieri, M., D. J. Tweardy, D. Caracciolo, K. Johnson, F. Mavilio, S. Altmann, D. Santoli, and G. Rovera. 1987. Cytokine-dependent granulocytic differentiation. Regulation of proliferative and differentiative responses in a murine progenitor cell line. J. Immunol. 138:3829-3835. [PubMed] [Google Scholar]
- 62.Varadhachary, A. S., and P. Salgame. 1998. CD95 mediated T cell apoptosis and its relevance to immune deviation. Oncogene 17:3271-3276. [DOI] [PubMed] [Google Scholar]
- 63.Westin, E. H., R. C. Gallo, S. K. Arya, A. Eva, L. M. Souza, M. A. Baluda, S. A. Aaronson, and F. Wong-Staal. 1982. Differential expression of the amv gene in human hematopoietic cells. Proc. Natl. Acad. Sci. USA 79:2194-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weston, K., and J. M. Bishop. 1989. Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-myb. Cell 58:85-93. [DOI] [PubMed] [Google Scholar]
- 65.Williams, G. T., C. A. Smith, E. Spooncer, T. M. Dexter, and D. R. Taylor. 1990. Haemopoietic colony stimulating factors promote cell survival by suppressing apoptosis. Nature 343:76-79. [DOI] [PubMed] [Google Scholar]
- 66.Woo, C. H., L. Sopchak, and J. S. Lipsick. 1998. Overexpression of an alternatively spliced form of c-Myb results in increases in transactivation and transforms avian myelomonoblasts. J. Virol. 72:6813-6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, J., X. Liu, K. Bhalla, C. N. Kim, A. M. Ibrado, J. Cai, T. I. Peng, D. P. Jones, and X. Wang. 1997. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 275:1129-1132. [DOI] [PubMed] [Google Scholar]
- 68.Ymer, S., W. Q. Tucker, C. J. Sanderson, A. J. Hapel, H. D. Campbell, and I. G. Young. 1985. Constitutive synthesis of interleukin-3 by leukaemia cell line WEHI-3B is due to retroviral insertion near the gene. Nature 317:255-258. [DOI] [PubMed] [Google Scholar]