Abstract
p38 mitogen activated protein kinase (MAPK) is essential for T-cell activation. Here we demonstrated that nuclear factor of activated T cells (NFAT) is a direct target of p38 MAPK. Inhibition of p38 MAPK led to selective inactivation of NFAT in T cells. We further linked a strict requirement of p38 MAPK to activation of NFATc. A stimulatory effect of p38 MAPK on at least four other stages of NFATc activation was found. First, the p38 MAPK cascade activated the NFATc promoter and induced the transcription of NFATc mRNA. Second, p38 MAPK mildly increased the mRNA stability of NFATc. Third, p38 MAPK enhanced the translation of NFATc mRNA. Fourth, p38 MAPK promoted the interaction of NFATc with the coactivator CREB-binding protein. In contrast, p38 MAPK moderately enhanced the expulsion of NFATc from the nucleus in T cells. Therefore, p38 MAPK has opposite effects on different stages of NFATc activation. All together, the overall effect of p38 MAPK on NFATc in T cells is clear activation.
Mitogen-activated protein kinase (MAPK) mediates signal transduction from extracellular stimulation to the nucleus. p38 MAPK, initially identified in response to inflammation and cellular stresses, is known to be involved in development, cell growth, cell differentiation, and cell death (for reviews, see references 27, 35, and 38). p38 MAPK is specifically activated by MAPK kinase 3 (MKK3), MKK4, or MKK6. In T lymphocytes, T-cell receptor (TCR) engagement activates p38 MAPK, and inhibition of p38α prevents the expression of interleukin 2 (IL-2) (17, 19, 20, 33, 54). Of the two major isoforms of p38 MAPK in T cells, p38α and p38δ (16, 47), TCR stimulates mainly p38α (19). p38 MAPK activates downstream effectors through different mechanisms. The activation of transcription factors, including CREB, ATF-1, ATF-2, p53, Sap-1a, C/EBPβ, and CHOP, by p38 MAPK is mediated by direct phosphorylation (38). Alternatively, p38 MAPK induces the production of inflammation-related cytokines through increased mRNA stabilization or enhanced mRNA translation (25-28, 35, 38, 49).
The nuclear factor of activated T cells (NFAT) is one of the major transcription factors binding to IL-2 gene promoters. At least five members of NFAT have been identified: NFATc (NFAT2), NFATp (NFAT1), NFAT3, NFAT4, and NFAT5 (24, 42). In T cells, NFATc and NFATp are the major NFAT isoforms involved during T-cell activation (5, 24, 39, 42). In the early phase of T-cell activation, NFATp is dephosphorylated and translocated into the nucleus immediately after TCR ligation (30). These steps are followed by NFATc synthesis and nuclear entry during the later phase of T-cell activation. NFATc is critical for the expression of IL-2 and other cytokines (5, 10, 39, 41, 46, 51). Microarray analysis has further identified many new NFAT downstream targets (15, 32).
The activation of NFAT can be divided into different stages. For an inducible NFAT isoform, such as NFATc, mRNA expression and translation are initiated upon T-cell activation. A critical stage for all NFATs, either inducible or preexisting, is the translocation of NFAT from the cytosol to the nucleus (9, 36). Once in the nucleus, NFAT binds to specific DNA motifs in the promoter regions of the target gene, often with the coordinated presence of other transcription factors, such as AP-1 (31). NFAT then interacts with transcription coactivators, such as CREB-binding protein (CBP) and p300 (1, 12, 14). Calcineurin, which promotes the nuclear entry of NFAT through the dephosphorylation of NFAT, is the most well-characterized signaling molecule in NFAT activation (8, 9, 34, 36), but many other T-cell signaling molecules have also been linked to NFAT activation. Ras and protein kinase C stimulate the synthesis and activation of Jun/Fos (31) for the full activation of the NFAT-AP-1 complex. c-Raf and Rac have been shown to promote an NFAT-CBP interaction (1). In contrast, the phosphorylation of NFATc by glycogen synthase kinase 3 leads to the nuclear export of NFATc (3). By blocking NFAT activation, glycogen synthase kinase 3 has been shown to be a negative regulator of T-cell activation (37). Among different MAPKs, c-Jun N-terminal kinase inhibits the targeting of calcineurin to NFATc in T cells (6), and extracellular signal-regulated kinase increases the nuclear export of NFATc (27). More recently, p38 MAPK was demonstrated to phosphorylate NFATp and NFAT3 and to promote their nuclear export (13, 40, 50).
In the present study, we found that NFATc is one of the major targets of p38 MAPK in T cells. Our results suggest that p38 MAPK promotes the nuclear expulsion of NFATc in T cells. However, during the other steps of NFATc activation, p38 MAPK activates the NFATc promoter, stabilizes NFATc mRNA, increases NFATc translation, and promotes NFATc-CBP binding. The overall effect of p38 MAPK therefore is the activation of NFATc. Our results also illustrate a scenario in which the same kinase may regulate the different activation steps of a transcription factor in opposite directions yet still have a clear stimulatory effect.
MATERIALS AND METHODS
Reagents and plasmids.
A23187, tetradecanoyl phorbol acetate (TPA), and concanavalin A (ConA) were purchased from Sigma Chemical Co. (St. Louis, Mo.). DEAE-dextran (molecular weight, 5 × 105) was purchased from Pharmacia (Uppsala, Sweden). SB 203580 was purchased from Calbiochem (La Jolla, Calif.).
Human NFATc and catalytic calcineurin subunit A cDNA were gifts from Gerald Crabtree (Stanford University, Stanford, Calif.) and were obtained through Hsiou-Chi Liou (Cornell University Medical College, New York, N.Y.). Human regulatory calcineurin subunit B was obtained by reverse transcription (RT)-PCR with 5′ primer CAG CCG CGG CGC AAC ACT TCT CCG AGC CAG and 3′ primer AGA GAG AAT TCC TGG ACG TCT TGA and was subcloned into pcDNA3 (Invitrogen, Carlsbad, Calif.). NFATp was a gift from Anjana Rao (Harvard Medical School, Boston, Mass.). NFAT-CAT, containing five tandem repeats of the murine IL-2 promoter-distal NFAT-binding site (−280 bp), was previously described (18). The murine NFATc promoter (−752 to −21 bp) was isolated by PCR with 5′ primer TGC AAT CTG TTA GTA ATT TAG CGG GA and 3′ primer GTT CGG AAC CTC TCG GTC TCA and was subcloned into luciferase reporter vector pGL2-Basic (Promega, Madison, Wis.). pGAL-CBP, encoding a fusion protein containing Gal residues 1 to 147 (Gal1-147) and full-length CBP, and RSV-CBP were gifts from Richard H. Goodman (Oregon Health Sciences University, Portland). pG5B-CAT was obtained from Mark Ptashne (Harvard University, Cambridge, Mass.). pNFAT(TAD)-VP16 was constructed by ligation of the N-terminal transactivation domain (TAD; amino acids 113 to 205) of NFATc [(NFAT(TAD)] to VP16. The dominant-negative mutant of p38α [p38α(Ala180, Phe182); p38α(AF)] (21) and the active mutant of MKK3b [MKK3b(Glu189, Glu193); MKK3b(E)] (23) were gifts from Jiahuai Han (Scripps Research Institute, La Jolla, Calif.).
Transgenic mice.
p1017, containing the lck 3.2-kb proximal promoter and the 3′ untranslated region of human growth hormone (minigene exons 1 to 5), was a gift from Roger Perlmutter (University of Washington, Seattle). p38α(AF) was subcloned into the BamHI site of vector p1017, generating p1017-p38α(AF). Transgenic mice were generated at the Transgene/Knockout Core Facility of the Institute of Molecular Biology, Academia Sinica (Taipei, Taiwan). p1017-p38α(AF) was injected into the pronuclei of C57BL/6 zygotes. Four independent founders were obtained. All transgenic mice were maintained at the specific-pathogen-free mouse facility of the Institute of Molecular Biology, Academia Sinica. All animal experiments were conducted with the approval of the Experimental Animal Committee, Academia Sinica. The phenotypes of p38α(AF)-transgenic mice were similar to those previously reported (11, 43). In brief, the number of p38α(AF)-transgenic thymocytes was reduced by 25%. The fractions of thymic CD4− CD8−, CD4+ CD8+, CD4+ CD8−, and CD4− CD8+ populations were 3, 85, 9, and 3% for p38α(AF)-transgenic mice and 3, 82, 11, and 4% for normal littermate control (NLC) mice, respectively. The fractions of splenic CD4+ and CD8+ populations were 21 and 11.4% for transgenic mice and 23 and 12.3% for NLC mice. Splenic T cells were purified by panning twice against anti-mouse immunoglobulin. Because the T-cell subpopulations were similar for p38α(AF)-transgenic and NLC mice, equal numbers of thymocytes and splenic T cells were used for each experiment without adjustments.
Cell cultures and transfection.
T cells were cultured in RPMI medium with 10% fetal calf serum (both from GIBCO, Grand Island, N.Y.), 10 mM glutamine, 100 U of penicillin/ml, 100 U of streptomycin/ml, and 2 × 10−5 M 2-mercaptoethanol. Dulbecco modified Eagle medium was used instead of RPMI medium for culturing 293T cells. For DEAE-dextran transfection, 1.6 × 107 T cells were washed once with STBS (25 mM Tris-HCl [pH 7.4], 137 mM NaCl, 5 mM KCl, 0.6 mM Na2HPO4, 0.7 mM CaCl2, 0.5 mM MgCl2) and incubated with a total of 10 μg of DNA in 1.2 ml of STBS containing 0.5 mg of DEAE-dextran/ml for 20 min at room temperature. Enhanced green fluorescent protein (EGFP; 1 μg; Clontech, Palo Alto, Calif.) was included in all transfections. T cells were then treated with 15% dimethyl sulfoxide for 3 min and washed once with STBS. 293T cells were transfected by the calcium phosphate method. The transfection efficiency was determined 24 h after transfection by counting the fraction of cells showing green fluorescence. The cells were activated and harvested 6 to 24 h after activation. Chloramphenical acetyltransferase (CAT) activities were measured as described previously (18) and normalized with the transfection efficiency. For luciferase activity, the production of light through the oxidation of luciferin in the presence of ATP was measured with a Luminometer.
Nuclear extracts and total cell extracts.
T cells were lysed in 10 mM Tris-HCl (pH 7.4)-10 mM NaCl-3 mM MgCl2-0.5% Nonidet P-40. The lysates were incubated on ice for 5 min and centrifuged at 200 × g for 5 min at 4°C. The pellet (nuclei) was suspended in hypertonic buffer, containing 20 mM HEPES (pH 7.9), 1.5 mM MgCl2, 0.3 M KCl, 0.5 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 20% glycerol, and 0.4 mM EDTA, and the suspension was rocked at 4°C for more than 30 min. The mixture was centrifuged at 12,000 × g for 10 min. The supernatant was mixed with 2 volumes of the above-described buffer (without KCl) and immediately frozen. Total cell extracts were prepared by resuspending cells in hypotonic buffer, containing 10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.5 mM PMSF, 100 μg of aprotinin/ml, and 1.25 μg of leupeptin/ml, and lysing cells by three cycles of freezing and thawing. A 1/10 volume of 3 M KCl was added, and the lysates were rocked at 4°C for 30 min. The same isolation procedures as those used for the nuclear extracts were followed. The protein concentration was determined by the Bradford assay (Bio-Rad, Richmond, Calif.).
Immunoblotting.
Cell extracts (20 to 50 μg) were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore) for 4 h at 20 V. Membranes were washed in rinse buffer (phosphate-buffered saline with 2% Tween 20) at room temperature for 15 min and incubated in blocking buffer (5% nonfat milk in rinse buffer) for 1.5 h. Membranes were then incubated with 1:1,000 anti-NFATc or anti-NFATp antibodies (BD-PharMingen, San Diego, Calif.). After being washed, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit immunoglobulin antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) in blocking buffer for 1.5 h. After three washes, the membranes were developed with enhanced chemiluminescence Western blot detection reagents (Amersham, Little Chalfont, Buckingshamshire, United Kingdom). The developed membranes were detected with X-ray film and quantitated with a densitometer (Molecular Dynamics).
Polysomal RNA preparation.
293T cells were collected 24 h after transfection and lysed in lysis buffer (50 mM Tris [pH 7.5], 250 mM NaCl, 25 mM MgCl2, 0.5% Triton X-100, 200 U of RNasin/ml, 20 μg of cycloheximide/ml, 2 mM DTT). Cell debris and nuclei were removed by microcentrifugation. The supernatant was layered onto 12-ml 15 to 50% sucrose gradients. Centrifugation was conducted at 217,400 × g for 2.5 h with an SW41 rotor. The gradients were then fractionated with a Brandel density gradient fractionator at a rate of 0.375 ml/min. During fractionation, the absorbance of the flowthrough was recorded at 254 nm. RNA from each collected fraction was prepared by ethanol precipitation after proteinase K digestion and phenol-chloroform extraction. The amount of RNA was determined by RT-PCR followed by analysis with real-time PCR.
Real-time PCR.
RNA was reverse transcribed in a total volume of 20 μl with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and oligo(dT). Aliquots (0.5 μl) of the RT products were added to a 20-μl reaction mixture containing 2 μl of LightCycler DNA Master SYBR Green I (Roche, Mannheim, Germany), 0.5 μM primer, and 3 mM MgCl2. Quantitative PCR was performed with a LightCycler (Roche), with initial incubation at 95°C for 60 s, followed by 40 cycles of denaturation at 95°C for 0.5 s, annealing at 62°C for 5 s, and elongation at 72°C for 22 s. The amount of DNA was determined by the incorporation of SYBR Green I at each cycle.
RESULTS
NFAT is a major target of p38 MAPK on the IL-2 gene promoter.
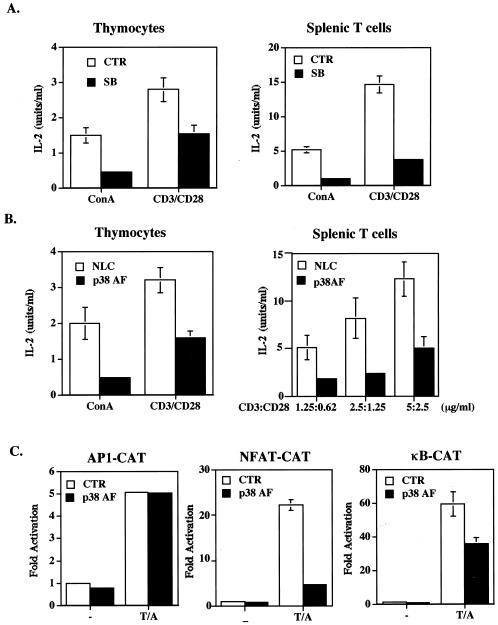
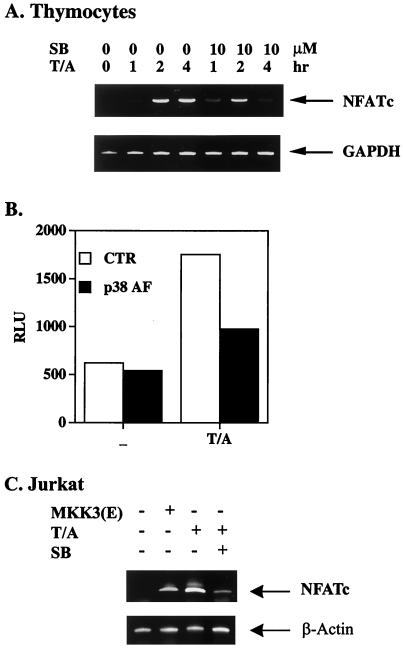
p38 MAPK is involved in T-cell activation. Using the p38 MAPK inhibitor SB 203580, various investigators have shown that p38 MAPK is required for IL-2 expression (14, 18, 20, 31, 54). This finding was illustrated by the blockage of activation-induced IL-2 production by SB 203580 in both immature (thymocytes) and mature (splenic) T cells (Fig. 1A). We also used transgenic expression of p38α(AF) (20, 21), the dominant-negative form of p38α, to assess the participation of p38 MAPK in T-cell activation. C57BL/6 transgenic mice with T-cell-specific expression of p38α(AF) were generated. The expression of p38α(AF) inhibited TCR-stimulated p38 activity by 50% (data not shown). Similar to the inhibitory effect of SB 203580, IL-2 production stimulated by ConA, by CD3 and CD28, or by TPA and A23187 was largely suppressed in p38α(AF)-transgenic thymocytes and splenic T cells (Fig. 1B and data not shown for TPA and A23187). The expression of IL-2 in naive T cells is determined primarily by transcription activation of the IL-2 gene promoter, containing essential elements, including AP-1, NFAT, and NF-κB (22, 44). EL4 T cells were transfected with CAT reporters containing AP-1, NFAT, or NF-κB elements in the absence or presence of 1 μg of p38α(AF). A profound inhibition of NFAT-CAT was observed when T cells were cotransfected with p38α(AF), while AP-1-CAT was not affected (Fig. 1C). The activation of NF-κB-CAT was moderately (40%) inhibited by p38α(AF). Therefore, among the transcription elements dictating IL-2 expression, NFAT was most sensitive to the inhibition of p38, suggesting that NFAT is a major target of p38 MAPK during T-cell activation.
FIG. 1.
NFAT is a target of p38 MAPK on the IL-2 gene promoter. (A) SB 203580 inhibited IL-2 production. Thymocytes and splenic T cells (5 × 105/well) from C57BL/6 mice were stimulated with ConA (2.5 μg/ml) plus anti-CD3 (10 μg/ml) or anti-CD28 (2.5 μg/ml) in the presence (SB) or absence (CTR [control]) of SB 203580 (10 μM). IL-2 produced was quantitated 24 h later with indicator cell line HT-2. (B) Transgenic p38α(AF) suppressed IL-2 production in T cells. Thymocytes and splenic T cells (5 × 105/well) from p38α(AF)-transgenic mice and NLC mice were stimulated with ConA (2.5 μg/ml) and CD3 plus CD28 at the concentrations indicated. IL-2 levels were determined 24 h after activation. (C) p38α(AF) inhibited the activation of NFAT-CAT in T cells. EL4 T cells were transfected with CAT reporters containing AP-1, NFAT, or NF-κB elements (18) and with or without 1 μg of p38α(AF) by the DEAE-dextran method. T cells were stimulated with TPA (10 ng/ml) plus A23187 (80 ng/ml) (T/A) 24 h later (or not stimulated [−]), and CAT activities were determined after another 8 h. Data are reported as means and standard errors of the means.
Inhibition of p38 interferes with NFATc activation.
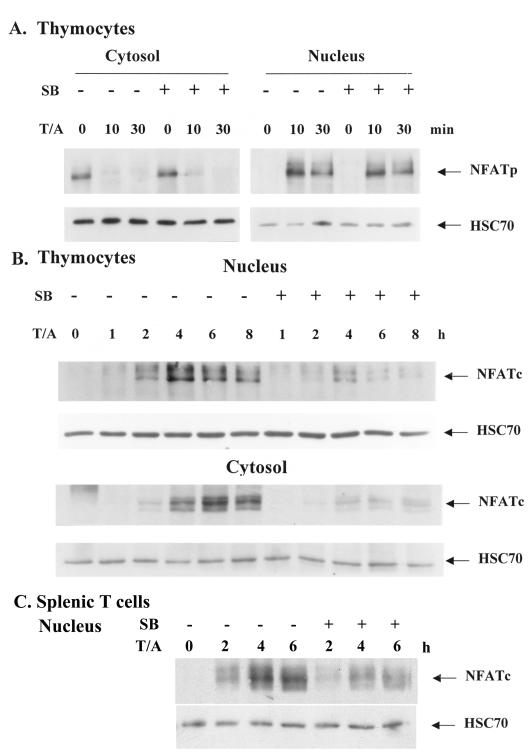
NFAT activation in T cells is determined by an early phase of NFATp nuclear translocation followed by a late phase of NFATc synthesis (9, 24, 30). We examined the effect of SB 203580 on NFAT activation in thymocytes stimulated with TPA and A23187 by monitoring the levels of NFATp and NFATc in both the cytoplasm and the nucleus. NFATp was constitutively present in the cytosol before T-cell activation and was translocated from the cytosol to the nucleus after stimulation (Fig. 2A). The translocation was nearly complete 10 min after T-cell activation, as NFATp was mostly nucleus located. Nuclear entry of NFATp was not affected when p38 MAPK was inhibited by SB 203580 (Fig. 2A). In contrast to NFATp, NFATc was not detectable in the cytosol of resting T cells (Fig. 2B). The appearance of NFATc in the cytosol and nucleus was observed 2 h after T-cell activation (Fig. 2B). The addition of SB 203580 suppressed the induction of NFATc in both the nucleus and the cytosol of thymocytes. An inhibition of the appearance of NFATc in the nuclear and cytoplasmic fractions of splenic T cells was also evident (Fig. 2C and data not shown for cytoplasm). Similar results were found when EL4 T-lymphoma and Jurkat T-lymphoma cells were used (data not shown). These findings indicate that TCR-induced NFATc expression is one of the stages of NFATc activation that requires full p38 MAPK activity.
FIG. 2.
p38 MAPK is required for the activation of NFATc but not NFATp. Thymocytes (A and B) and splenic T cells (C) were stimulated with TPA (10 ng/ml) and A23187 (80 ng/ml) (T/A) in the absence or presence of SB 203580 (10 μM) (SB), and cytosol extracts and nuclear extracts were prepared. (A) Nuclear translocation of NFATp was not affected by p38 MAPK suppression. Cytoplasmic and nuclear NFATp contents in thymocytes 10 and 30 min after activation were determined by immunoblotting with anti-NFATp (4G6-G5; BD-PharMingen). (B) Inhibition of p38 MAPK reduced nuclear and cytoplasmic presence of NFATc in thymocytes. Nuclear and cytosolic NFATc contents of thymocytes isolated 1, 2, 4, 6, and 8 h after TPA and A23187 activation were determined with anti-NFATc (7A6; BD-PharMingen). (C) SB 203580 inhibited the nuclear appearance of NFATc in splenic T cells. The amounts of NFATc in nuclear extracts of splenic T cells activated for 2, 4, and 6 h were determined as described for panel B.
p38 MAPK mediates the nuclear export of NFATc but less exclusively in T cells than in 293T cells.
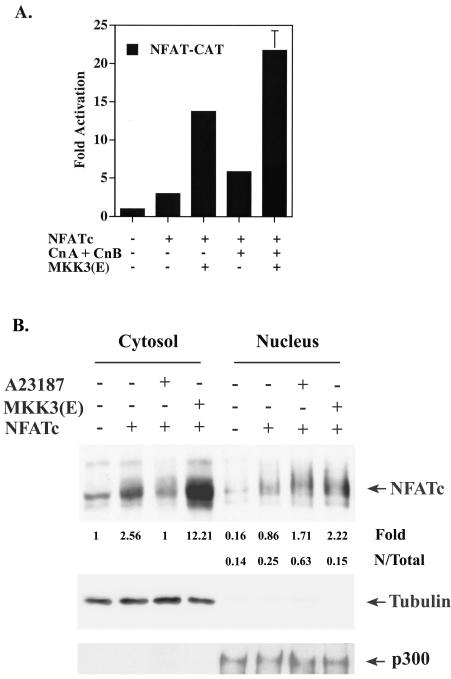
One of the critical stages in the activation of NFATc is the nuclear translocation of NFATc. p38 MAPK was reported to promote the nuclear export of NFATc in COS cells but to display a minimum effect on the translocation of NFATc in HeLa cells (13, 50). We therefore examined the direct effect of p38 MAPK on the distribution of NFATc in T cells. Due to the involvement of p38 MAPK in NFATc synthesis (Fig. 2), whether p38 MAPK participates in NFATc nuclear entry could not clearly addressed. We used CMV-NFATc to circumvent the participation of p38 MAPK in NFATc synthesis. CMV-NFATc transfection led to moderate activation of NFAT-CAT (Fig. 3A). Cotransfection with MKK3b(E), an activator of p38 MAPK (23), resulted in significant enhancement of NFAT-CAT activation. NFATc-mediated transcription activation was also augmented, to a lesser extent than that of MKK3b(E), by the overexpression of both calcineurin catalytic A and calcineurin regulatory B subunits (8, 34) (Fig. 3A). Optimum NFAT-CAT activation was found with the coexpression of both MMK3b(E) and calcineurin.
FIG. 3.
p38 MAPK promotes NFATc activation but increases nuclear export of NFATc in T cells. (A) NFAT transcription activation mediated by NFATc was enhanced by MKK3b(E). EL4 T cells were transfected with NFAT-CAT in the presence of CMV-NFATc, calcineurin catalytic A subunit plus regulatory B subunit (CnA + CnB), and MKK3b(E) as indicated. CAT activities were determined 24 later. Data are reported as means and standard errors of the means. (B) EL4 T cells were similarly transfected with CMV-NFATc and/or MKK3b(E) or treated with A23187. Cytosolic and nuclear extracts were prepared 24 h after transfection, and the amounts of NFATc in the nucleus and cytoplasm were assessed by immunoblotting. α-Tubulin was used as a marker for the cytosol, and p300 was used as a marker for the nucleus. The quantity of NFATc was determined by densitometry and normalized by densitometric reading of the respective internal control (α-tubulin or p300). For ease of comparison, the quantity of cytoplasmic NFATc in untreated T cells was set at 1. N/Total, ratio of nuclear NFATc content to total (nucleus plus cytosol) NFATc content.
Western analysis was then used to examine the cytosolic and nuclear distributions of NFATc in the presence of MKK3b(E) cotransfection. α-Tubulin and p300 were used as markers for the cytoplasm and the nucleus, respectively (Fig. 3B). Transfection of EL4 T cells with CMV-NFATc alone led to some nuclear entry of NFATc despite the presence of a larger fraction of NFATc residing in the cytosol (Fig. 3B). Treatment of T cells with A23187 promoted the nuclear translocation of NFATc. MKK3b(E) expression dramatically increased the quantity of cytosolic NFATc (see below) but did not significantly enhance the nuclear localization of NFATc (Fig. 3B). The ratio of nuclear NFATc to total NFATc (cytoplasmic plus nuclear) was calculated based on the protein level detected by immunoblotting. The fraction of nuclear NFATc after transfection with CMV-NFATc alone was 0.24; this fraction was increased to 0.63 by A23187 treatment (Fig. 3B). Coexpression with MKK3b(E), however, reduced the fraction of nucleus-situated NFATc to 0.15, suggesting that constitutive p38 MAPK activation stimulated the expulsion of NFATc from the nucleus.
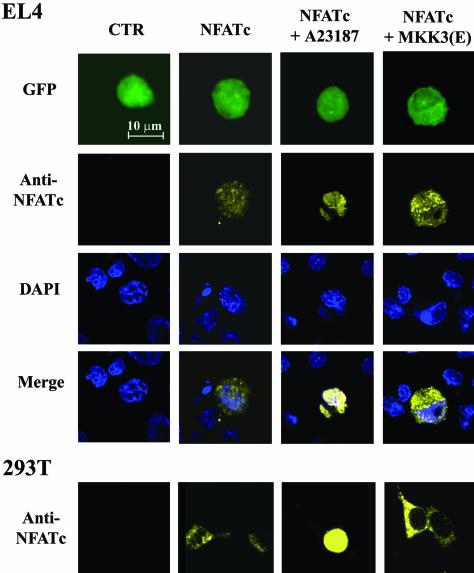
Confocal microscopy was also used to monitor the intracellular localization of transfected NFATc. Transfected cells were marked with EGFP. The nuclei were labeled with 4′,6′-diamidino-2-phenylindole (DAPI). The nuclear localization of NFATc was assessed by superimposing the NFATc image over the DAPI image (Fig. 4, merge). There were small differences between the immunoblot and the confocal analyses in the amounts of nucleus-situated NFATc (Fig. 3B and 4), likely due to differences in detection sensitivity. NFATc was not detected in nontransfected EL4 T cells (Fig. 4, CTR [control]). NFATc transfection led to the appearance of NFATc protein in both the cytosol and the nucleus in EL4 T cells (Fig. 4, NFATc). Treatment with A23187 led to the nuclear localization of NFATc (Fig. 4, NFATc + A23187). Cotransfection with MKK3b(E) resulted in a large increase in the cytosolic NFATc level and a very small increase in the nuclear NFATc level [Fig. 4, NFATc + MKK3(E)], suggesting an increase in the nuclear export of NFATc. An identical effect of MKK3b(E) on the distribution of NFATc was found when Jurkat cells were used (data not shown). Therefore, the microscopy results also support the notion that active p38 MAPK increased the nuclear exclusion of NFATc. The effect of p38 MAPK on the nuclear export of NFATc in T cells, however, was not as profound as that in 293T cells. Coexpression of CMV-NFATc with MKK3b(E) in 293T cells led to the exclusive cytosolic localization of NFATc, as determined by microscopy (Fig. 4, 293T), while a small amount of NFATc remained nucleus situated in T cells (Fig. 4, EL4). Therefore, p38 MAPK activation did not lead to a complete exclusion of NFATc from the nucleus, allowing the operation of NFATc-mediated transcription activation in T cells.
FIG. 4.
Confocal image of intracellular distribution of NFATc stimulated with MKK3(E) or A23187. EL4 T cells and 293T cells were transfected with EGFP only (CTR [control]); EGFP and CMV-NFATc (NFATc); EGFP, CMV-NFATc, and A23187 (NFATc + A23187); and EGFP, CMV-NFATc, and MKK3b(E) [NFATc + MKK3(E)]. At 24 h after transfection, cells were fixed with 3.7% paraformaldehyde, followed by methanol permeabilization. The cells were stained with DAPI (DAPI) and with anti-NFATc followed by phycoerythrin-conjugated anti-mouse immunoglobulin (anti-NFATc). The NFATc expression of cells was analyzed by use of Zeiss confocal laser scanning microscope LSM 510 with a ×63 objective lens. Green cells (GFP) indicate transfected cells, while DAPI indicates the nucleus. The nuclear localization of NFATc was examined by overlapping the anti-NFATc-stained image with the DAPI-stained image (merge).
Induction of NFATc during T-cell activation requires p38 MAPK.
We next examined the individual stage of NFATc activation that is likely to be positively regulated by p38 MAPK. The possible involvement of p38 MAPK in TCR-stimulated NFATc expression (Fig. 2) was examined by analysis of NFATc mRNA expression. T cells (thymocytes, splenic T cells, EL4 cells, and Jurkat cells) were activated with TPA and A23187 in the absence or presence of SB 203580 (10 μM). The amount of NFATc mRNA was determined by RT-PCR. NFATc mRNA expression, prominent at 2 h after T-cell activation, further increased at 4 h after activation (Fig. 5A and data not shown for splenic T cells, EL4 cells, and Jurkat cells). SB 203580 significantly decreased NFATc mRNA induction at 4 h poststimulation. To examine the direct dependence of NFATc transcription on p38 MAPK, we further isolated the NFATc promoter (−752 to −21 bp) by PCR (7, 55). T-cell activation by TPA and A23187 led to activation of the NFATc promoter, as indicated by the induction of luciferase activity (Fig. 5B). Activation of the NFATc promoter was suppressed when p38 MAPK activity was inhibited by the coexpression of p38α(AF). Transfection of MKK3b(E), a constitutive activator of p38, also led to a small but significant induction of NFATc in Jurkat cells and 9C12.7 T-cell hybridomas (Fig. 5C and data not shown for 9C12.7 cells). We failed to detect NFATc expression in MKK3b(E)-transfected EL4 cells (data not shown). The induction of NFATc mRNA by MKK3b(E) was less prominent than that by TPA and A23187 and was not observed in all T cells, yet it demonstrated the capacity of p38 MAPK to direct NFATc expression.
FIG. 5.
NFATc mRNA induction requires p38 MAPK. (A) NFATc mRNA induction in thymocytes was inhibited by SB 203580. Thymocytes were activated with TPA and A23187 (T/A) in the absence or presence of SB 203580 (10 μM) (SB). Thymocytes were harvested for RNA isolation 1, 2, and 4 h after activation. The amount of NFATc mRNA was determined by RT-PCR. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B) Activation of the NFATc promoter was p38 MAPK dependent. A pGL2 reporter containing the NFATc promoter (−752 to −21 bp) was transfected into EL4 T cells without (CTR [control]) or with p38α(AF). T cells were activated with TPA and A23187 24 h later, and luciferase activity was determined 24 h after activation. −, no activation. RLU, relative light units. (C) MKK3b(E) alone induced NFATc expression in Jurkat cells. Jurkat T cells were transfected with active MKK3b(E) or pcDNA3 as a control. Cells transfected with pcDNA3 were then left untreated or were treated with TPA amd A23187 24 h later. The amount of NFATc mRNA was determined by RT-PCR.
NFATc mRNA stability is moderately increased by p38 MAPK.
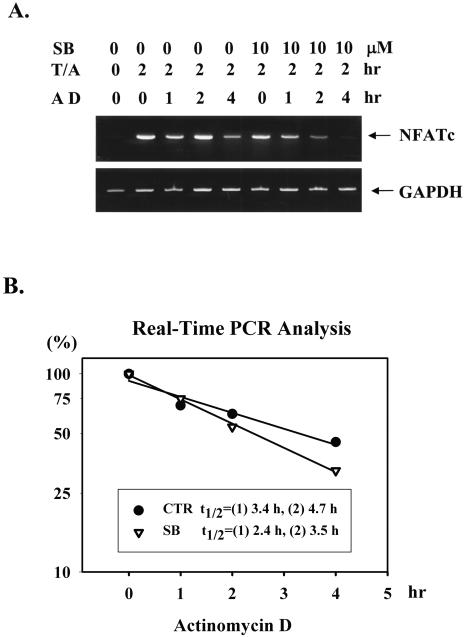
p38 MAPK is well known for its ability to stimulate the expression of IL-6, IL-8, and cyclooxygenase through enhancement of the stability of their mRNAs (28, 49). In addition to direct transcriptional activation of the NFATc promoter by p38 MAPK, elevated steady-state levels of NFATc mRNA (Fig. 5A) may also result from posttranscriptional mechanisms. Therefore, the ability of p38 MAPK to increase the stability of NFATc mRNA was examined. RNA from T cells treated with TPA and A23817 for 2 h was used as the starting material for the stability determination, because at this time the difference in NFATc mRNA quantities between SB 203580-treated and untreated thymocytes was small (Fig. 5A). NFATc mRNA stability was assessed by determining the rate of mRNA degradation after new mRNA synthesis was blocked with actinomycin D (Fig. 6A). The amount of NFATc mRNA was determined by RT-PCR (Fig. 6A) and real-time PCR (Fig. 6B) at different times after actinomycin D addition. From the kinetics of mRNA degradation, the half-lives of NFATc mRNA for activated thymocytes and SB 203580-treated thymocytes were 3.4 and 2.4 h for the first measurement and 4.7 and 3.5 h for the second measurement (Fig. 6B). Inhibition of p38 MAPK increased NFATc mRNA decay by about 30%. Therefore, p38 MAPK does contribute to the mRNA stability for NFATc, but not as profoundly as it contributes to the mRNA stabilities for IL-6, IL-8, and cyclooxygenase.
FIG. 6.
NFATc mRNA stability was moderately enhanced by p38 MAPK. (A) RNA stability was determined by adding actinomycin D (10 μg/ml) (AD) to thymocyte cultures preactivated with TPA and A23187 (T/A) for 2 h and with or without SB 203580 (10 μM) (SB). The RNA was isolated 1, 2, and 4 h after actinomycin D addition. (B) The kinetics of NFATc mRNA degradation were plotted, and the half-life (t1/2) was calculated with a second-order polynomial curve fit on CA-Cricket Graph III (Computer Associates, Islandia, N.Y.). CTR, control. Half-lives of NFATc mRNA from two independent experiments are indicated. The amounts of NFATc mRNA were determined by RT-PCR (A) and by real-time PCR (B).
p38 MAPK promotes NFATc translation.
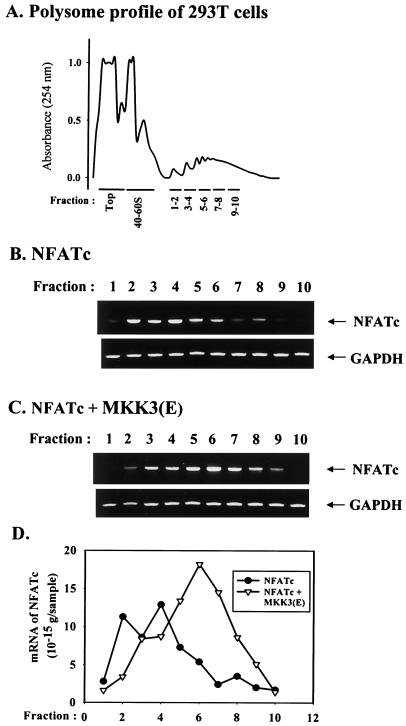
We detected a large increase in NFATc protein levels when CMV-NFATc was cotransfected with MKK3b(E) (Fig. 3B). p38 MAPK has been shown to stimulate tumor necrosis factor alpha (TNF-α) production by augmented translation of TNF-α mRNA (25, 26). The regulatory sites are on the AU-rich elements located in the 3′ untranslated region of TNF-α mRNA. The construct CMV-NFATc, containing the complete 3′ untranslated region of NFATc mRNA and AU-rich elements, could also be a target of p38 MAPK for enhanced translation. Transfection of CMV-NFATc was performed with 293T cells because of their high transfection efficiency (>75%), which allowed subsequent biochemical analysis. Extracts from 293T cells were fractionated on sucrose gradients to separate the polysomal fractions from 40S to 60S ribosomes (Fig. 7A). The heavy polysomal fraction, representing the translating mRNA with ribosomes assembled on it, corresponded to fractions of higher numbers (2). RNA was isolated from each polysomal fraction, and the content of NFATc mRNA was determined by RT-PCR (Fig. 7B and C) and real-time PCR (Fig. 7D). The total amount of NFATc mRNA in polysomal fractions, measured as the area under the curve in Fig. 7D, was significantly larger for cells cotransfected with MKK3b(E). There was also a clear shift in NFATc mRNA to the heavier polysomal fractions in 293T cells cotransfected with MKK3b(E), indicating that p38 MAPK enhanced the translation initiation of NFATc mRNA (Fig. 7B to D). As a control, the contents of glyceraldehyde-3-phosphate dehydrogenase transcripts associated with polysomes were nearly identical in 293T cells transfected with or without MKK3(E) (Fig. 7B and C).
FIG. 7.
Increased NFATc mRNA translation by p38 MAPK. 293T cells were collected 24 h after transfection with CMV-NFATc or CMV-NFATc plus MKK3b(E). Cell lysates were analyzed by sucrose gradient centrifugation. (A) Typical profile of the sucrose gradient monitored by measuring the absorbance at 254 nm. The top of the gradient, the 40S and 60S fractions, and polysome-containing fractions (1 to 10) are indicated. (B and C) Amounts of NFATc mRNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA in a polysome-containing fraction from 293T cells transfected with NFATc (B) or MKK3(E) plus NFATc (C) were determined by RT-PCR. (D) The distribution of the NFATc mRNA polysome-containing fraction was enhanced by p38 MAPK. NFATc mRNA in each polysome fraction was quantitated by real-time PCR. The quantity of NFATc mRNA was then plotted against the number of the polysome fraction.
Interaction of NFATc and CBP is p38 dependent.
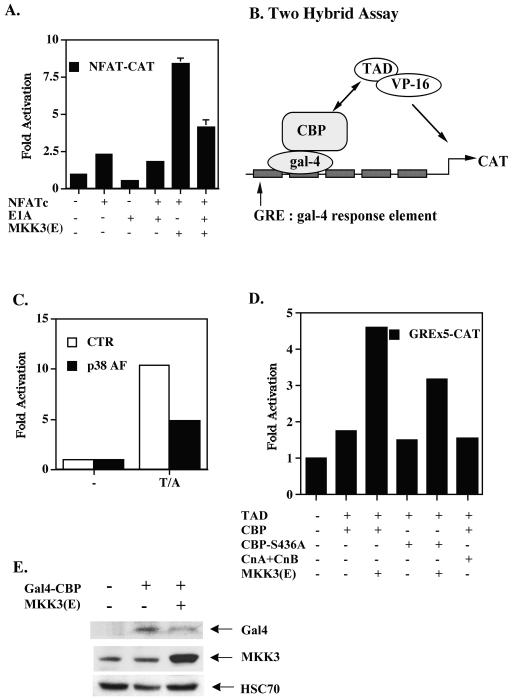
NFAT is known to interact with CBP/p300, and this interaction requires signals from both Raf and Rac (1, 12). We further examined whether p38 MAPK, downstream of Rac activation, participated in any interaction of CBP with NFATc. The activation of NFAT-CAT by CMV-NFATc and MKK3(E) was inhibited by E1A (Fig. 8A), a competitor for CBP, indicating that p38 MAPK-induced NFAT activation is CBP dependent. We next used the binding of pGAL-CBD to pVP16-NFAT(TAD) to measure the interaction between NFATc and CBP. pGAL-CBP carries a fusion of Gal1-147 and full-length CBP, and pVP16-NFAT(TAD) carries a fusion of VP16 and the TAD of NFATc (Fig. 8B). The activation of pG5B-CAT was used as an indicator for the binding of CBP to NFAT(TAD). Activation of 293T cells resulted in the interaction of NFAT(TAD) with CBP (Fig. 8C, CTR). The role of p38 MAPK in promoting the NFAT-CBP association was demonstrated by a prominent reduction in NFAT-CBP binding when p38α(AF) was cotransfected (Fig. 8C, p38 AF). The NFATc-CBP interaction was also promoted by direct stimulation with p38 MAPK in Jurkat cells. Cotransfection of Jurkat cells with MKK3b(E) but not with calcineurin A chain and B chain stimulated NFAT-CBP binding (Fig. 8D). The enhanced NFATc-CBP interaction was not due to MKK3b(E)-promoted expression of the two-hybrid component proteins. The level of Gal4-CBP was not increased by MKK3b(E) cotransfection (Fig. 8E).
FIG. 8.
p38 MAPK promotes the interaction between NFATc and CBP. (A) E1A inhibited p38 MAPK-stimulated NFAT activation. EL4 cells were transfected with NFAT-CAT in the presence of NFATc, E1A, or MKK3(E) as indicated. Cells were harvested 24 h after transfection, and CAT activities were determined. Data are reported as means and standard errors of the means. (B) Interaction of CBP and NFATc, as determined by the illustrated binding of pGAL-CBP to pVP16-NFAT(TAD) and the activation of pG5B-CAT (containing five tandem repeats of the GRE). (C) The NFAT-CBP interaction was p38 MAPK dependent. 293T cells were transfected with pGAL-CBP, pVP16-NFAT(TAD), or pG5B-CAT, with or without (CTR [control]) p38α(AF). After 24 h, 293T cells were stimulated with TPA and A23187 (T/A), and CAT activities were determined after another 24 h. −, no stimulation. (D) MKK3(E) stimulated NFATc-CBP binding. Jurkat T cells were transfected with pGAL-CBP or pGAL-CBP(S436A), pVP16-NFAT(TAD), pG5B-CAT, 1 μg of EGFP with or without calcineurin (both A and B subunits) (CnA+CnB), or MKK3(E) by the DEAE-dextran method. CAT activation was quantitated 24 h later. (E) Enhanced NFATc-CBP interaction was not due to MKK3(E)-stimulated Gal4-CBP expression. Levels of Gal4-CBP and MKK3(E) in panel D were quantitated with anti-Gal4 and anti-MKK3.
We also attempted to identify the phosphorylation sites on NFATc and CBP that are targeted by p38 MAPK. A previous study (1) illustrated that mutation of the five potential MAPK phosphorylation sites (serine-proline motif) in NFAT(TAD) does not affect NFAT(TAD)-CBP binding. Mutation of serine 317 and serine 436 in CBP, located in the NFAT-binding domain of CBP, however, affects growth factor-dependent CBP recruitment to the transcription complex (53). We constructed pGAL-CBP(S317A) and pGAL-CBP(S436A) and tested their association with NFAT(TAD). The CBP-NFAT interaction was similar to that of CBP(S436A)-NFAT in unstimulated Jurkat T cells (Fig. 8D). In contrast, p38 MAPK-mediated binding of CBP to NFAT(TAD) was partially inhibited by mutation of serine 436 in CBP (Fig. 8D), suggesting the involvement of serine 436 phosphorylation in the p38 MAPK-induced interaction with NFAT(TAD). Mutation of serine 317 in CBP did not interfere with the binding of CBP to NFAT (data not shown).
DISCUSSION
In the present study, we demonstrated that p38 MAPK plays a critical role in the activation of NFATc in T cells. We identified at least four distinct stages of NFATc activation that are highly p38 MAPK-dependent: the activation of the NFATc promoter (Fig. 5), the stabilization of NFATc mRNA (Fig. 6), the translation of NFATc mRNA (Fig. 7), and the binding of NFATc to CBP (Fig. 8). A combination of these stimulatory effects results in an absolute requirement of p38 MAPK for NFATc activation.
Among the several new observations made in this study, we first identified a direct link between p38 MAPK and NFAT activation. Previous studies had established the notion that p38 MAPK is indispensable for IL-2 expression (17, 19, 20, 33, 54), as was also observed here for thymocytes and splenic T cells with attenuated p38 MAPK activity (Fig. 1A and B). Transcriptional activation is known to be the primary regulatory mechanism for IL-2 expression in naive T cells (22). By using reporters containing AP-1, NFAT, and NF-κB, dominant elements on the IL-2 promoter, we mapped NFAT as the major element that is targeted by p38 MAPK.
We next identified the NFATc promoter as one of the targets of p38 MAPK. The NFATc promoter contains binding elements for CREB, NFATp, and NF-κB (7, 55). It was previously shown that p38 MAPK is one of the kinase pathways that are essential for the full activation of CREB in T cells (52). The requirement of p38 MAPK for the activation of NFATc promoter (Fig. 5B) could be explained in part by the direct activation of CREB by p38 MAPK. Full activation of p38 MAPK requires the engagement of both CD3 and CD28 (52, 54), representing primary signal integration of TCR and costimulatory molecules. NFAT activation has also been used as an indicator of the successful integration of T-cell activation signals from both TCR and costimulatory molecules, such as CD28 (9). Therefore, full T-cell activation is already required at the stage of transcriptional activation of the NFATc promoter, further extending the integrative role of NFATc in T-cell activation.
The half-life of NFATc mRNA decreased by 30% in the presence of SB 203580 (Fig. 6). In contrast, the half-lives of mRNAs for IL-6, IL-8, and cyclooxygenase differ by a minimum of twofold in the presence versus the absence of p38 MAPK (28, 49). The mRNA stabilization effect of p38 MAPK on NFATc, compared to that on IL-6, IL-8, and cyclooxygenase 2, is relatively weak. AU-rich elements located in the 3′ untranslated region of mRNA are critical for regulating specific RNA stability (4). The mRNA stabilization effect of p38 MAPK is linked to the AUUUA sequence (28, 49). Human NFATc mRNA contains a single AUUUA element, while murine NFATc mRNA contains two AUUUA elements. In contrast, there are 4 and 22 copies of the AUUUA motif in the 3′ untranslated regions of IL-8 and cyclooxygenase 2 mRNAs, respectively (28, 49). Whether differences in the numbers of AU-rich elements result in the differential stabilization effects of p38 MAPK on NFATc mRNA and IL-8 or cyclooxygenase mRNA remains to be determined. It must be noted that the molecular basis underlying the p38 MAPK-mediated mRNA stabilization is complicated by other factors, in addition to the presence of AUUUA elements.
We also observed a large increase in NFATc mRNA translation when p38 MAPK was activated (Fig. 3A and 7). At least two kinases downstream of p38 MAPK, Mnk1 and MAPK-activated protein (MAPKAP) kinase 2, have been demonstrated to enhance the translation of specific mRNA (29, 48). The activation of Mnk1 by p38 MAPK leads to the phosphorylation of eukaryotic initiation factor 4E, linking p38 MAPK to the initiation of protein translation. p38 MAPK also promotes TNF-α translation through the AU-rich element on the 3′ untranslated of TNF-α mRNA (25, 26), a process which is mediated by MAPKAP kinase 2 (29). The exact mechanism by which p38 MAPK stimulates NFATc mRNA translation is currently being investigated.
We found another direct stimulatory effect of p38 MAPK—on the binding of NFATc to CBP/p300 (Fig. 8). The activation of NFAT requires the recruitment of coactivators CBP/p300 (1, 12). A previous study by Avots et al. (1) showed that an interaction between NFATc and CBP/p300 depends on the integration of Raf and Rac signals. The activation of p38 MAPK through the costimulation of CD3 and CD28 is known to occur downstream of Rac and Ras activation (27, 38, 45). The observation that p38 MAPK promoted the binding of NFATc(TAD) to CBP (Fig. 8) is in full agreement with the signal requirement for such an interaction (1). Therefore, the coordinated activation of Ras and Rac, resulting from the engagement of both TCR and CD28, leads to the full activation of p38 MAPK and NFATc-CBP binding. We also conducted a preliminary search for the phosphorylation sites on NFAT and CBP that are the targets of p38 MAPK. The study by Avots et al. (1) demonstrated that mutation of the five serine-proline motifs, potential phosphorylation sites for p38 MAPK, in NFATc(TAD) did not affect binding to CBP. We therefore examined the potential phosphorylation sites of CBP. A few phosphorylation events associated with CBP/p300 have been documented (14). A prominent role for serine 436 in growth factor-induced CBP binding to a transcriptional complex has been established, concomitant with a much weaker contribution from serine 317 (53). Consistent with the observations of Zanger et al. (53), mutation of serine 436 but not serine 317 reduced the p38 MAPK-mediated binding of CBP to NFAT(TAD) (Fig. 8 and data not shown for S317A). Whether or not serine 436 in CBP is directly phosphorylated by p38 MAPK needs further biochemical investigation. The partial effect of the S436A mutation on CBP-NFAT binding (Fig. 8D), in contrast to its profound inhibition of the CBP-AP-1 interaction (53), suggests that additional serine residues in the NFAT-binding domain of CBP may contribute to p38 MAPK inducibility. We are currently examining such a possibility.
In contrast to the stimulatory effect of p38 MAPK on the expression of NFATc and on the NFATc-CBP interaction, p38 MAPK appeared to oppose the nuclear localization of NFATc in T cells (Fig. 3 and 4). In analogy to the report that p38 MAPK increases NFATc nuclear export in COS cells (40), MKK3b(E)-p38 MAPK stimulated nearly complete cytosolic localization of NFATc in 293T cells (Fig. 4). The activation of p38 MAPK by MKK3b(E) in T cells did not lead to a complete exclusion of NFATc from the nucleus, as was the case in 293T cells (Fig. 3 and 4). Therefore, our results support p38 MAPK-mediated nuclear export of NFATc, similar to that of NFATp and NFAT3 (13, 40, 50). The observed differences in the efficiencies of p38-dependent NFATc expulsion between T cells and 293T cells may help reconcile the reported discrepancies between COS cells and HeLa cells in p38-mediated NFATc localization (13, 50).
Our results also illustrate that stimulatory effects could override inhibitory effects on a transcription factor from the same kinase. p38 MAPK increased the nuclear exclusion of NFATc (Fig. 3 and 4), one of the critical steps during the activation of NFATc, leading to the blockage of NFATc-mediated transcription. The reduction in the nuclear entry of NFATc, however, was compensated for by p38 MAPK-mediated enhancement of other NFATc activation steps. NFATc expression was induced by p38 MAPK-containing activation signals in T cells (Fig. 5). NFATc mRNA was stabilized by 30% in the presence of p38 MAPK (Fig. 6). NFATc mRNA translation was also augmented by p38 MAPK activity (Fig. 7). The combination of these effects resulted in a large excess of cytosolic NFATc protein and allowed a significant level of NFATc molecules to remain in the nucleus (Fig. 3 and 4). Once NFATc was resident in the nucleus, NFATc-mediated transcription was further enhanced by p38 MAPK-mediated CBP binding (Fig. 8). Therefore, for the overall activation of NFATc, the requirement of p38 MAPK is absolute necessary. Despite the p38 MAPK-mediated nuclear expulsion of NFATc (Fig. 3), there was no activation of NFATc in the absence of p38 MAPK (Fig. 1 and 2). The complicated level of regulation of NFATc by p38 MAPK could be extended to other signaling molecules. The activation of NFATc is dependent on multiple signaling cascades, with some known to participate in different stages of NFATc activation. For example, calcineurin is required for the dephosphorylation of NFATc for nuclear entry and is also required for maintaining NFATc in the nucleus (9). Thus, multiple signals modulate NFATc at different steps, constituting the complicated network involved in the activation of a single transcription factor.
Acknowledgments
This project was supported by grants NSC89-2320-B001-050 and NSC 90-2320-B001-070 from the National Science Council and by a grant from Academia Sinica.
We thank Jiahuai Han, Gerald Crabtree, Anjara Rao, Laurie H. Glimcher, Richard Goodman, Roger Perlmutter, and Hsiou-Chi Liou for plasmids; Chi-Kuang Leo Wang for transgenic mouse construction; and Ken Deen for editorial correction of the manuscript.
REFERENCES
- 1.Avots, A., M. Buttmann, S. Chuvpilo, C. Escher, U. Smola, A. J. Bannister, U. R. Rapp, T. Kouzarides, and E. Serfling. 1999. CBP/p300 integrates Raf/Rac-signaling pathways in the transcriptional induction of NF-ATc during T cell activation. Immunity 10:515-524. [DOI] [PubMed] [Google Scholar]
- 2.Ballinger, D. G., and M. L. Pardue. 1983. The control of protein synthesis during heat shock in Drosophila cells involved altered polypeptide elongation rate. Cell 33:103-144. [DOI] [PubMed] [Google Scholar]
- 3.Beals, C. R., C. M. Sheridan, C. W. Turck, P. Gardner, and G. R. Crabtree. 1997. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science 275:1930-1934. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 5.Chow, C.-W., M. Rincón, and R. J. Davis. 1999. Requirement for transcription factor NFAT in interleukin-2 expression. Mol. Cell. Biol. 19:2300-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow, C. W., C. Dong, R. A. Flavell, and R. J. Davis. 2000. c-Jun NH2-terminal kinase inhibits targeting of the protein phosphatase calcineurin to NFATc1. Mol. Cell. Biol. 20:5227-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuvpilo, S., E. Jankevics, D. Tyrsin, A. Akimzhanov, D. Moroz, M. K. Jha, J. Schulze-Luehrmann, B. Santner-Nanan, E. Feoktistova, T. Konig, A. Avots, E. Schmitt, F. Berberich-Siebelt, A. Schimpl, and E. Serfling. 2002. Autoregulation of NFATc1/A expression facilitates effector T cells to escape from rapid apoptosis. Immunity 16:881-895. [DOI] [PubMed] [Google Scholar]
- 8.Clipstone, N. A., and G. R. Crabtree. 1992. Identification of calcineurin as a key signaling enzyme in T-lymphocyte activation. Nature 357:695-697. [DOI] [PubMed] [Google Scholar]
- 9.Crabtree, G. R., and E. N. Olson. 2002. NF-AT signaling: choreographing the social lives of cells. Cell 109:S67-S79. [DOI] [PubMed]
- 10.Cron, R. Q., S. J. Bort, Y. Wang, M. W. Brunvand, and D. B. Lewis. 1999. T cell priming enhances IL-4 gene expression by increasing nuclear factor of activated T cells. J. Immunol. 162:860-870. [PubMed] [Google Scholar]
- 11.Diehl, N. L., H. Enslen, K. A. Fortner, C. Merritt, N. Stetson, C. Charland, R. A. Flavell, R. J. Davis, and M. Rincón. 2000. Activation of the p38 mitogen-activated protein kinase pathway arrests cell cycle progression and differentiation of immature thymocytes in vivo. J. Exp. Med. 191:321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Rodriguez, C., and A. Rao. 1998. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein. J. Exp. Med. 187:2031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez del Arco, P., S. Martinez-Martinez, J. L. Maldonado, I. Ortega-Perez, and J. M. Redondo. 2000. A role for the p38 MAP kinase pathway in the nuclear shuttling of NFATp. J. Biol. Chem. 275:13872-13878. [DOI] [PubMed] [Google Scholar]
- 14.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 15.Graef, I. A., F. Chen, L. Chen, A. Kuo, and G. R. Crabtree. 2001. Signals transduced by Ca(2+)/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell 105:863-875. [DOI] [PubMed] [Google Scholar]
- 16.Hale, K. K., D. Trollinger, M. Rihanek, and C. L. Manthey. 1999. Differential expression and activation of p38 mitogen-activated protein kinase α, β, γ, and δ in inflammatory cell lineages. J. Immunol. 162:4246-4252. [PubMed] [Google Scholar]
- 17.Hehner, S. P., T. G. Hofmann, O. Dienz, W. Drοge, and M. L. Schmitz. 2000. Tyrosine-phosphorylated Vav1 as a point of integration for T-cell receptor- and CD28-mediated activation of JNK, p38, and interleukin-2 transcription. J. Biol. Chem. 275:18160-18171. [DOI] [PubMed] [Google Scholar]
- 18.Ho, H.-Y., H.-H. Lee, and M.-Z. Lai. 1997. Overexpression of mitogen-activated protein kinase kinase kinase reversed cAMP inhibition of NF-kappaB in T cells. Eur. J. Immunol. 27:222-226. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, S.-C., M. Gavrilin, M.-H. Tsai, J. Han, and M.-Z. Lai. 1999. p38 mitogen activated protein kinase is involved in Fas ligand expression. J. Biol. Chem. 274:25769-25776. [DOI] [PubMed] [Google Scholar]
- 20.Hsu, S.-C., C.-C. Wu, J. Han, and M.-Z. Lai. 2003. Involvement of p38 mitogen activated protein kinase in different stages of thymocyte development. Blood 101:970-976. [DOI] [PubMed] [Google Scholar]
- 21.Huang, S., Y. Jiang, Z. Li, E. Nishida, P. Mathias, S. Lin, R. J. Ulevitch, G. R. Nemerow, and J. Han. 1997. Apoptosis signaling pathway in T cells is composed of ICE/CED-3 family proteases and MAP kinase kinase 6b. Immunity 6:739-749. [DOI] [PubMed] [Google Scholar]
- 22.Jain, J., C. Loh, and A. Rao. 1995. Transcriptional regulation of the IL-2 gene. Curr. Opin. Immunol. 7:333-342. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, Y., C. Chen, Z. Li, W. Guo, J. A. Gegner, S. Lin, and J. Han. 1996. Characterization of the structure and function of a new mitogen-activated protein kinase (p38β). J. Biol. Chem. 271:17920-17926. [DOI] [PubMed] [Google Scholar]
- 24.Kiani, A., A. Rao, and J. Aramburu. 2000. Manipulating immune responses with immunosuppressive agents that target NFAT. Immunity 12:359-372. [DOI] [PubMed] [Google Scholar]
- 25.Kontoyiannis, D., M. Pasparakis, T. T. Pizarro, F. Cominelli, and G. Kollias. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10:387-398. [DOI] [PubMed] [Google Scholar]
- 26.Kontoyiannis, D., A. Kotlyarov, E. Carballo, L. Alexopoulou, P. J. Blackshear, M. Gaestel, R. Davis, R. Flavell, and G. Kollias. 2001. Interleukin-10 targets p38 MAPK to modulate ARE-dependent TNF mRNA translation and limit intestinal pathology. EMBO J. 20:3760-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kyriakis, J. M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807-869. [DOI] [PubMed] [Google Scholar]
- 28.Lasa, M., K. R. Mahtani, A. Finch, G. Brewer, J. Saklatvala, and A. R. Clark. 2000. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol. Cell. Biol. 20:4265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehner, M. D., F. Schwoebel, A. Kotlyarov, M. Leist, M. Gaestel, and T. Hartung. 2002. Mitogen-activated protein kinase-activated protein kinase 2-deficient mice show increased susceptibility to Listeria monocytogenes infection. J. Immunol. 168:4667-4673. [DOI] [PubMed] [Google Scholar]
- 30.Loh, C., J. A. Carew, J. Kim, P. G. Hogan, and A. Rao. 1996. T-cell receptor stimulation elicits an early phase of activation and a later phase of deactivation of the transcription factor NFAT1. Mol. Cell. Biol. 16:3945-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macian, F., C. Lopez-Rodriguez, and A. Rao. 2001. Partners in transcription: NFAT and AP-1. Oncogene 20:2476-2489. [DOI] [PubMed] [Google Scholar]
- 32.Macian, F., F. Garcia-Cozar, S. H. Im, H. F. Horton, M. C. Byrne, and A. Rao. 2002. Transcriptional mechanisms underlying lymphocyte tolerance. Cell 109:719-731. [DOI] [PubMed] [Google Scholar]
- 33.Matsuda, S., T. Moriguchi, S. Koyasu, and E. Nishida. 1998. T lymphocyte activation signal for interleukin-2 production involve activation of MKK6-p38 and MKK7-SAPK-JNK signaling pathways sensitive to cyclosporin A. J. Biol. Chem. 273:12378-12382. [DOI] [PubMed] [Google Scholar]
- 34.Milan, D., J. Griffith, M. Su, E. R. Price, and F. McKeon. 1994. The latch region of calcineurin B in both immnosuppressant-immunophilin complex docking and phosphatase activation. Cell 79:437-447. [DOI] [PubMed]
- 35.Nebreda, A. R., and A. Porras. 2000. p38 MAP kinase: beyond the stress response. Trends Biochem. Sci. 25:257-260. [DOI] [PubMed] [Google Scholar]
- 36.Neilson, J., K. Stankunas, and G. R. Crabtree. 2001. Monitoring the duration of antigen-receptor occupancy by calcineurin/glycogen-synthase-kinase-3 control of NF-AT nuclear shuttling. Curr. Opin. Immunol. 13:346-350. [DOI] [PubMed] [Google Scholar]
- 37.Ohteki, T., M. Parsons, A. Zakarian, R. G. Jones, L. T. Nguyen, J. R. Woodgett, and P. S. Ohashi. 2000. Negative regulation of T cell proliferation and interleukin 2 production by the serine threonine kinase GSK-3. J. Exp. Med. 192:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono, K., and J. Han. 2000. The p38 signal transduction pathway: activation and function. Cell. Signal. 12:1-13. [DOI] [PubMed] [Google Scholar]
- 39.Peng, S. L., A. J. Gerth, A. M. Ranger, and L. H. Glimcher. 2001. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity 14:13-20. [DOI] [PubMed] [Google Scholar]
- 40.Porter, C. M., M. A. Havens, and N. A. Clipstone. 2000. Identification of amino acid residues and protein kinases involved in the regulation of NFATc subcellular localization. J. Biol. Chem. 275:3543-3551. [DOI] [PubMed] [Google Scholar]
- 41.Ranger, A. M., M. R. Hodge, E. M. Gravallese, M. Oukka, L. Davidson, F. W. Alt, F. C. de la Brousse, T. Hoey, M. Grusby, and L. H. Glimcher. 1998. Delayed lymphoid repopulation with defects in IL-4-driven responses produced by inactivation of NF-ATc. Immunity 8:125-134. [DOI] [PubMed] [Google Scholar]
- 42.Rao, A., C. Luo, and P. G. Hogan. 1997. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15:707-747. [DOI] [PubMed] [Google Scholar]
- 43.Rincón, M., H. Enslen, J. Raingeaud, M. Recht, T. Zapton, M. S.-S. Su, L. A. Penix, R. J. Davis, and R. A. Flavell. 1998. Interferon-γ expression by Th1 effector T cells mediated by the p38 MAP kinase signaling pathway. EMBO J. 17:2817-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rooney, J. W., M. R. Hodge, P. G. McCaffrey, A. Rao, and L. H. Glimcher. 1994. A common factor regulates both Th1- and Th2-specific cytokine gene expression. EMBO J. 13:625-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salojin, K. V., J. Zhang, and T. L. Delovitch. 1999. TCR and CD28 are coupled via ZAP-70 to the activation of the Vav/Rac-1-/PAK-1/p38 MAPK signaling pathway. J. Immunol. 163:844-853. [PubMed] [Google Scholar]
- 46.Todd, M. D., M. J. Grusby, J. A. Lederer, E. Lacy, A. H. Lichtman, and L. H. Glimcher. 1993. Transcription of the interleukin 4 gene is regulated by multiple promoter elements. J. Exp. Med. 177:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, X. S., K. Diener, C. L. Manthey, S. Wang, B. Rosenzweiz, J. Bray, J. Delaney, C. N. Cole, P.-Y. Chan-Hui, N. Mantlo, H. S. Lichenstein, M. Zukowski, and Z. Yao. 1997. Molecular cloning and characterization of a novel p38 mitogen-activated protein kinase. J. Biol. Chem. 272:23668-23674. [DOI] [PubMed] [Google Scholar]
- 48.Waskiewicz, A. J., A. Flynn, C. G. Proud, and J. A. Cooper. 1997. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16:1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C. Y. Chen, A. B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18:4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, T. T., Q. Xiong, H. Enslen, R. J. Davis, and C. W. Chow. 2002. Phosphorylation of NFATc4 by p38 mitogen-activated protein kinases. Mol. Cell. Biol. 11:3892-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida, H., H. Nishina, H. Takimoto, L. E. Marengere, A. C. Wakeham, D. Bouchard, Y. Y. Kong, T. Ohteki, A. Shahinian, M. Bachmann, P. S. Ohashi, J. M. Penninger, G. R. Crabtree, and T. W. Mak. 1998. The transcription factor NF-ATc1 regulates lymphocyte proliferation and Th2 cytokine production. Immunity 8:115-124. [DOI] [PubMed] [Google Scholar]
- 52.Yu, C.-T., H.-M. Shih, and M.-Z. Lai. 2001. Multiple signals required for cyclic AMP-responsive element binding protein (CREB) binding protein interaction induced by CD3/CD28 costimulation. J. Immunol. 166:284-292. [DOI] [PubMed] [Google Scholar]
- 53.Zanger, K., S. Radovick, and E. F. Wondisford. 2001. CREB binding protein recruitment to the transcription complex requires growth factor-dependent phosphorylation of its GF box. Mol. Cell 7:551-558. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, J., K. V. Salojin, J. X. Gao, M. J. Cameron, I. Bergerot, and T. Delovitch. 1999. p38 Mitogen-activated protein kinase mediates signal integration of TCR/CD28 costimulation in primary murine T cells. J. Immunol. 162:3819-3829. [PubMed] [Google Scholar]
- 55.Zhou, B., R. Q. Cron, B. Wu, A. Genin, Z. Wang, S. Liu, P. Robson, and H. S. Baldwin. 2002. Regulation of the murine Nfatc1 gene by NFATc2. J. Biol. Chem. 277:10704-10711. [DOI] [PubMed] [Google Scholar]