Abstract
As a shuttling protein, p53 is constantly transported through the nuclear pore complex. p53 nucleocytoplasmic transport is carried out by a bipartite nuclear localization signal (NLS) located at its C-terminal domain and two nuclear export signals (NES) located in its N- and C-terminal regions, respectively. The role of nucleocytoplasmic shuttling in p53 ubiquitination and degradation has been a subject of debate. Here we show that the two basic amino acid groups in the p53 bipartite NLS function collaboratively to import p53. Mutations disrupting individual amino acids in the NLS, although causing accumulation of p53 in the cytoplasm to various degrees, reduce but do not eliminate the NLS activity, and these mutants remain sensitive to MDM2 degradation. However, disrupting both parts of the bipartite NLS completely blocks p53 from entering the nucleus and causes p53 to become resistant to MDM2-mediated degradation. Similarly, mutations disrupting four conserved hydrophobic amino acids in the p53 C-terminal NES block p53 export and prohibit it from MDM2 degradation. We also show that colocalization of a nonshuttling p53 with MDM2 either in the nucleus or in the cytoplasm is sufficient for MDM2-induced p53 polyubiquitination but not degradation. Our data provide new insight into the mechanism and regulation of p53 nucleocytoplasmic shuttling and degradation.
The tumor suppressor protein p53 plays a pivotal role in preventing damaged or abnormal cells from becoming malignant, and its loss of function is associated with a majority of human cancers (23). p53 activity is not required during normal cell growth, and the protein must be kept at low levels and inactive. This is accomplished by the proto-oncoprotein MDM2, either through ubiquitin-dependent p53 degradation in the cytoplasm (11, 13, 15) or repression of p53 transcriptional activity in the nucleus (19, 30). The MDM2 gene is in turn transcriptionally activated by p53, constituting a feedback regulatory loop (2, 33).
Growing evidence has shown that p53 is regulated primarily by its protein stability (1, 31). A major mechanism of p53 stabilization and activation is triggered by DNA damage, which induces protein phosphorylation. Through a cascade of activity of protein kinases, DNA damage, such as that caused by ionizing radiation, induces multiple-site phosphorylation of p53 at its N and C termini. Phosphorylation of the N terminus of p53 affects its affinity for MDM2 and subsequent degradation (10, 18, 25). A recent study demonstrates that phosphorylation at the p53 N terminus inhibits its nuclear export, underscoring the importance of export in controlling p53 function (38). Another mechanism of p53 stabilization is triggered by aberrant growth signals, such as oncogenic Ras or Myc, and is mediated by a small protein called p14ARF (p19ARF in the mouse) encoded by DNA at the p16INK4a locus (24). Upon induction, ARF binds to MDM2 (21, 39), inhibits its E3 ligase activity (14), and leads to p53 stabilization. This is achieved, at least in part, by blocking the nuclear export of both p53 and MDM2 (29, 37), again indicating the importance of controlling nuclear export in p53 regulation.
Both MDM2 and p53 are nuclear proteins that shuttle constantly through the nuclear pore complex. MDM2 and p53 are translocated between the cytoplasm and the nucleus by their intrinsic nuclear localization signal (NLS) and nuclear export signals (NES) sequences (6, 17, 22, 27). Blocking their nuclear export by mutations in the NES or by leptomycin B leads to their stabilization, indicating that both MDM2 and p53 are degraded in the cytoplasm (7, 22). The nuclear membrane provides a barrier effectively separating the location of the function (in the nucleus) and the destruction (in the cytoplasm) of both MDM2 and p53. However, the role of MDM2 in the regulation of p53 nuclear export and degradation remains a subject of debate (36).
One model, based on the observation that MDM2 shuttles between the nucleus and the cytoplasm via its NES, whose mutation abolishes both MDM2 nuclear export and p53 degradation, suggests that MDM2 binds p53 in the nucleus and shuttles it into the cytoplasm (22, 28). A second model, based on evidence that p53-green fluorescent protein (GFP) fusion protein undergoes active nuclear export in mouse embryo fibroblast cells deficient in both MDM2 and p53 and that mutations in the p53 NES prevent the exit of p53, proposes that p53 can leave the nucleus via its own NES located in the C-terminal domain, independent of MDM2 (27, 38). A third model suggests that MDM2 assists p53 nuclear export by promoting its ubiquitination in the nucleus. This is based on evidence that mutations in the C-terminal RING finger domain of MDM2 eliminate its E3 ligase activity and trap p53 in the nucleus (5, 9). To further complicate the issue, a series of recent studies suggest that MDM2-mediated p53 ubiquitination and degradation take place in the nucleus, in the cytoplasm, or in both; accordingly, various mechanisms have been proposed (5, 9, 26, 34, 35).
In an attempt to clarify the role of nuclear import and export in regulating p53 degradation, we relied on p53 mutations disrupting its NLS and NES instead of using leptomycin B, a pharmacological drug that could potentially affect p53 transport and degradation by multiple mechanisms. We showed that the activities of p53 nuclear import and export were dynamically counterbalanced and were both required for p53 degradation but not for p53 ubiquitination. We also found that MDM2-mediated p53 ubiquitination takes place efficiently in the cytoplasm and that p53 ubiquitination can be uncoupled from its degradation. Our data provide new insight into the regulation of p53 nucleocytoplasmic transport, ubiquitination, and degradation.
MATERIALS AND METHODS
Plasmids.
Full-length human p53 and HDM2 cDNAs were described previously (39). HDM2 and p53 mutations were introduced by site-directed mutagenesis with the Quick-Change mutagenesis kit (Stratagene) and verified by DNA sequencing.
Cell lines, culture conditions and cell transfection.
Human U2OS and H1299 cells were obtained from the American Type Culture Collection, and mouse cells lacking MDM2 and p53 (2KO) were a gift from G. Lozano (M. D. Anderson Cancer Center). All cells were cultured in a 37°C incubator with 5% CO2 in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Transfections were done as described elsewhere (37, 39).
Protein analysis and indirect immunofluorescence.
p53 DO1 (Invitrogen), HDM2 Ab5 (Invitrogen), SMP14 (Santa Cruz), and GFP Ab2 (NeoMarkers) antibodies were purchased. To assess HDM2 and p53 levels, proteins from whole-cell extracts were separated by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting as described previously in detail (38). Procedures for immunoprecipitation and immunoblotting have been described previously (37). Indirect immunofluorescence and construction of the GFP fusion protein were described previously in detail (38). Antibodies to human p53 and HDM2 were previously described (37). Texas Red- and fluorescein isothiocyanate-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) were purchased commercially.
In vivo ubiquitin ligase activity assays.
The procedure for in vivo assay of the HDM2-associated ubiquitin ligase activity was essentially the same as described previously (8). For the in vivo p53 ubiquitination assay, U2OS cells on a 100-mm dish were transfected with plasmids expressing HDM2, p53, and heamagglutinin (HA)-tagged ubiquitin. The total amount of plasmid DNA in each transfection was adjusted with pcDNA3 empty vector when needed. Twenty hours after transfection, cells were treated with the proteasome inhibitor MG132 at 25 μM for 6 h. Cells were then collected, pelleted by centrifugation, lysed in 200 μl of boiled SDS lysis buffer (50 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 1% SDS, 1 mM dithiothreitol), and boiled for an additional 10 min. Lysates were clarified by centrifugation at 14,000 rpm on a microcentrifuge for 10 min. The supernatant was diluted with 0.5% NP-40 buffer and resolved on SDS-12.5% PAGE, followed by immunoblotting with anti-HDM2 antibody (1 μg/ml) or anti-p53 antibody (1 μg/ml).
RESULTS
Disruption of both parts but not individual basic amino acids in the bipartite NLS abolishes p53 nuclear import.
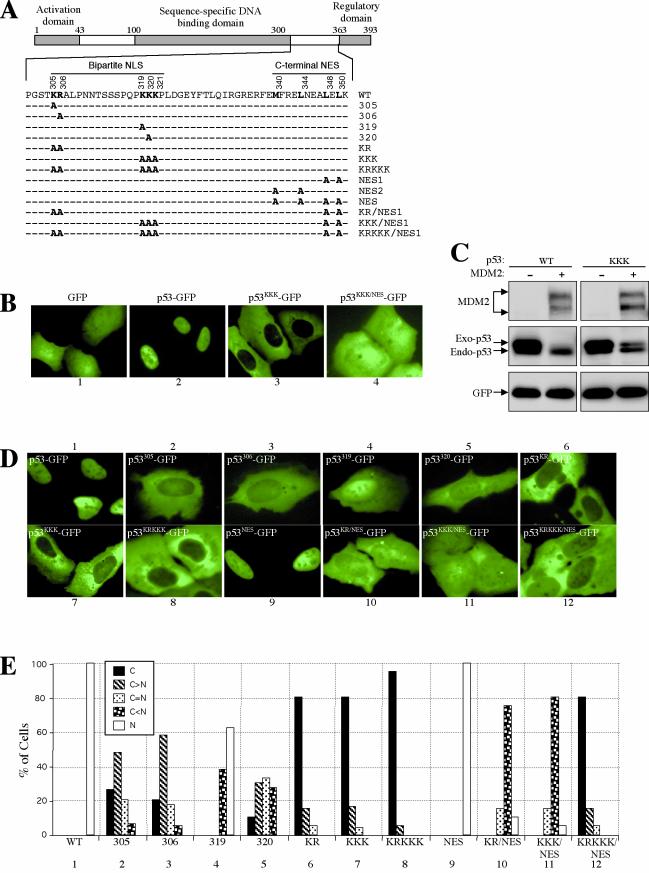
To determine the mechanism underlying the nuclear import of p53, a p53 mutant was constructed in which three consecutive lysine residues in the NLS were replaced with alanine (p53KKK), and the mutant p53 was fused with the jellyfish green fluorescent protein (GFP) to determine its localization in living U2OS cells. Lacking both NLS and NES, naked GFP diffused throughout the nucleus and the cytoplasm (Fig. 1B, panel 1). Driven by a p53 NLS, the p53-GFP fusion protein localized exclusively to the nucleus (Fig. 1B, panel 2). Confirming a previous report (17), the p53KKK-GFP mutant was localized exclusively to the cytoplasm, indicating that the mutation disrupted the NLS function (Fig. 1B, panel 3). Immunofluorescent staining of the mutant p53KKK without a GFP fusion showed identical cytoplasmic localization (data not shown). However, although p53KKK showed nuclear exclusion, it was nearly as susceptible to MDM2 degradation as the wild-type p53 (Fig. 1C). The cytoplasmic localization and susceptibility to MDM2 degradation of p53KKK suggest either that nuclear import is not required for p53 degradation or that p53KKK is able to enter the nucleus but with reduced efficiency, such that a changed balance of nuclear import and export favors cytoplasmic accumulation.
FIG.1.
Disruption of both basic amino acid parts but not individual amino acids in the bipartite NLS abolishes p53 nuclear import. (A) Diagram of p53 with positions and amino acid sequences of the bipartite NLS and C-terminal NES indicated. Amino acid substitutions used in the study are indicated in bold face. (B) Combining an NES mutation revealed a nuclear import activity in the p53KKK mutant. U2-OS cells were transfected with the indicated plasmids, and pictures were taken 24 h after transfection with living cells. (C) Degradation of p53KKK mutant by MDM2. U2-OS cells were transiently transfected with indicated p53 and MDM2 plasmid DNAs along with a GFP plasmid. Twenty-four hours after transfection, total cell lysate was prepared from each transfected cell population, electrophoretically separated by SDS-PAGE, and immunoblotted with antibodies specific to MDM2 (SMP14), p53 (DO1), and GFP (Ab2). (D) Various contributions of the basic amino acids in the bipartite NLS to the nuclear import of p53. The conserved basic amino acids in the bipartite NLS of p53 were replaced with alanine residues individually or in combination as indicated in panel A. The plasmid DNA was transfected into U2-OS cells, and the pictures were taken of living cells 24 h after transfection. (E) Quantification of GFP-positive cells. The GFP fluorescence pattern of each transfected cell population expressing the indicated p53 constructs was scored for at least 200 cells. The graph shows the percentage of cells with the indicated GFP patterns.
If there is a weak nuclear import activity in p53KKK-GFP, it may be revealed by disrupting the p53 NES so that the protein will remain in the nucleus after entering it. To test this possibility, the p53 C-terminal NES was mutated in p53KKK-GFP, and the localization of the mutant (p53KKK/NES-GFP) was examined. Interestingly, p53KKK/NES-GFP displayed diffused localization in both the nucleus and the cytoplasm, indicating the mutant protein entered the nucleus (Fig. 1B, panel 4). Because the size of the p53-GFP fusion protein (≈80 kDa) is well above the size (≈40 kDa) of a protein which can enter the nucleus by passive diffusion (20), the entry of p53KKK/NES-GFP into the nucleus must therefore be driven by a nuclear import activity. The nuclear exclusion of p53KKK-GFP is thus due to reduced rather than eliminated nuclear import activity and a shifted balance of protein nucleocytoplasmic shuttling in favor of nuclear export.
p53 contains a bipartite NLS consisting of two basic amino acid groups, Lys-Arg (KR) and Lys-Lys-Lys (KKK), separated by a spacer of 12 amino acid residues (17) (Fig. 1A). The fact that p53KKK-GFP retains a nuclear import activity indicates that one part of the bipartite NLS (e.g., KR) may function individually as a weaker NLS. Alternatively, another NLS may exist that contributes to p53 nuclear import. To evaluate the contributions of each basic amino acid residue in the bipartite NLS to p53's import function, the arginine and lysine residues were replaced with alanine individually or in combination (Fig. 1A), and the cellular localization of these mutants was examined (Fig. 1D). Single Ala substitutions of Lys305 or Arg306 greatly reduced the NLS function, and the fusion protein exhibited predominant cytoplasmic localization, while a small fraction of the protein entered the nucleus (Fig. 1D, panels 2 and 3; also see statistics in Fig. 1E, panels 2 and 3). Changing Lys319 to Ala, on the other hand, had only a subtle effect on NLS function; the fusion protein remained mostly in the nucleus (Fig. 1D, panel 4; Fig. 1E, panel 4). Mutation of Lys320 showed a stronger effect than that of Lys319, presumably because Lys320 is in the middle of a three-lysine residue cluster, mutation of which may have a more detrimental effect on the integrity of the NLS (Fig. 1D, panel 5).
Double mutation of Lys305 and Arg306 (p53KR-GFP) had a effect similar to that of the triple mutation of Lys319, -320, and -321 (p53KKK-GFP), in that both mutants exhibited complete nuclear exclusion (Fig. 1D, panels 6 and 7). Similar to the p53KKK/NES-GFP mutant, combining the NES mutation with p53KR-GFP (p53KR/NES-GFP) enabled it to show nuclear localization, indicating that p53KR-GFP also retained certain nuclear import activity (Fig. 1D, panels 10 and 11). Interestingly, a p53 mutated in both parts of the NLS (p53KRKKK-GFP) not only showed a nuclear exclusion, but also, unlike p53KR/NES-GFP and p53KKK/NES-GFP, the nuclear exclusion could not be altered by combining an NES mutation (p53KRKKK/NES-GFP) (Fig. 1D, panels 8 and 12), indicating that the nuclear import activity was reduced to an undetectable level. Together, our data showed that the two basic amino acid clusters in the NLS contribute cooperatively to p53 nuclear import; disrupting one part of it reduced but did not eliminate p53 nuclear import, whereas disrupting both parts blocked p53 import. These data suggest that the bipartite NLS is likely the only NLS in p53. In addition, previous studies of p53 transport used mutants in which only one part of the NLS was disrupted; interpretation of data obtained with such mutants hence requires caution.
Nuclear import of p53 is essential for MDM2-mediated degradation.
The above finding prompted us to examine the requirement of nuclear import of p53 for MDM2-induced degradation. We found that GFP tagging did not appreciably affect localization of wild-type and mutant p53, nor did it affect p53 ubiquitination and degradation by MDM2. Nevertheless, all experiments examining p53 ubiquitination and degradation were performed with untagged p53. Plasmids expressing each of the p53 NLS mutants were coexpressed with MDM2, and the steady-state levels of p53 were determined by Western blotting. We noticed that fusion of GFP at the C terminus of p53 did not affect its degradation by MDM2 (data not shown). Nevertheless, p53 mutants without the GFP moiety were used in most degradation analyses. Consistent with previous reports (34, 35), altering one lysine or one arginine residue in the NLS, although showing a diverse degree of nuclear exclusion, did not block MDM2-induced p53 degradation (data not shown).
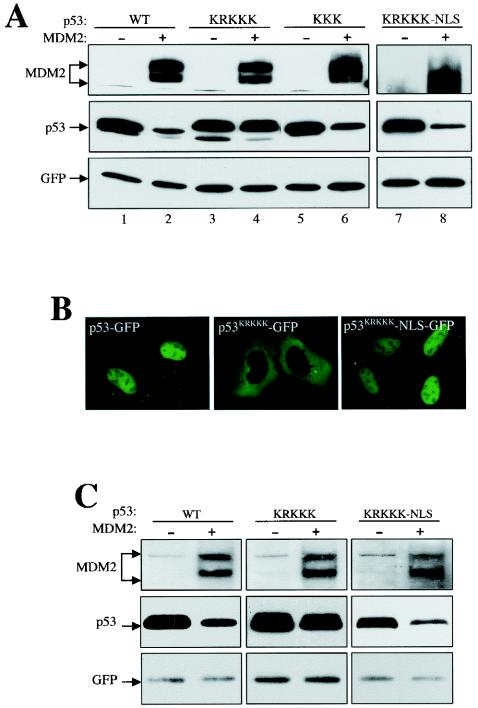
In line with their ability to enter the nucleus, p53KKK and p53KR were susceptible to MDM2 degradation (Fig. 2A, lane 6, and data not shown), whereas p53KRKKK was resistant to MDM2 degradation (Fig. 2A, lane 4), indicating that nuclear import is required for p53 degradation. The p53KRKKK protein migrated slightly slower than the wild-type and other p53 mutants, presumably because a change of multiple basic amino acids affected the overall charge of the protein in a denaturing SDS gel. To rule out the possibility that functions other than nuclear import that could contribute to its resistance to MDM2 might have been disrupted in the p53KRKKK mutant, a simian virus 40 NLS (3) was attached to the C terminus of the p53KRKKK (p53KRKKK-NLS), and its susceptibility to MDM2 was tested. The attachment of the simian virus 40 NLS not only resumed the nuclear localization of the mutant p53 (Fig. 2B), but also restored its sensitivity to MDM2 degradation (Fig. 2A, lane 8). Identical results were also obtained in mouse embryo fibroblast cells lacking both MDM2 and p53 (2KO), ruling out a cell type-specific effect (Fig. 2C). Together, our data are consistent with the notion that nuclear import of p53 is required for its degradation by MDM2.
FIG. 2.
Nuclear import of p53 is required for MDM2-mediated degradation. (A) U2-OS cells were transiently transfected with plasmid DNA expressing various p53 and MDM2 mutants, as indicated. Cell lysates were collected 24 h after transfection, separated by SDS-PAGE, and transferred onto a nitrocellulose membrane. Different portions of the membrane were blotted with the indicated antibodies. (B) Attachment of the simian virus 40 NLS to the C terminus of the nuclear import-deficient p53KRKKK restored its nuclear localization and its sensitivity to MDM2 degradation (A). (C) Mouse embryo fibroblast cells lacking both MDM2 and p53 (2KO) were transiently transfected, and the cell lysate was blotted as for panel A.
Nuclear export of p53 is essential for MDM2-mediated degradation.
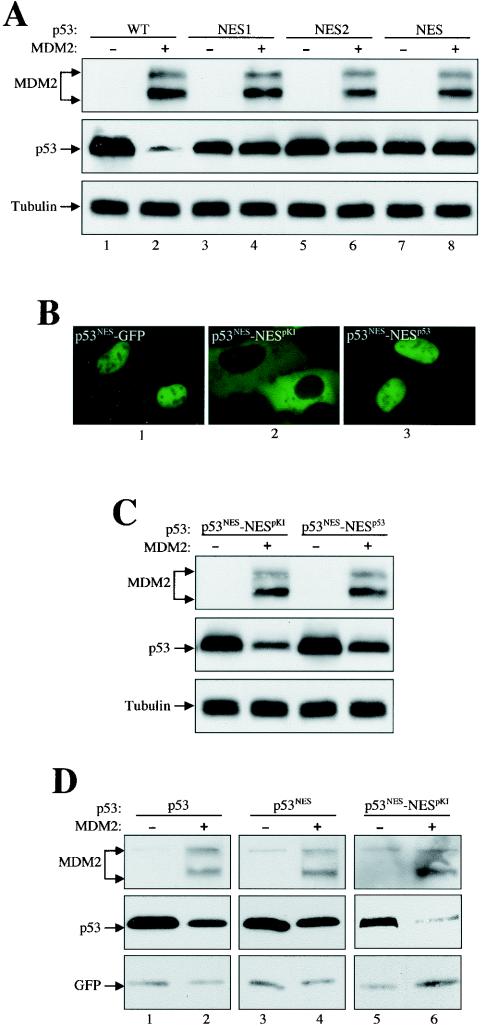
We next determined the requirement of nuclear export of p53 for its sensitivity to MDM2 degradation. Point mutations were introduced into the functionally conserved hydrophobic amino acids in the p53 C-terminal NES (see Fig. 1A). The p53 N-terminal NES was not examined because mutations in the N-terminal NES simultaneously disrupt MDM2 binding, making results difficult to interpret. Nevertheless, it has been shown that disruption of either the N-terminal or the C-terminal NES alone is sufficient to block p53 nuclear export (27, 38). Consistent with previous reports (5, 9), double substitutions of Leu 348 and Leu 350 (p53NES1) in the NES inhibited p53 degradation (Fig. 3A, lane 4). Double substitutions of Met 340 and Leu 344 (p53NES2), on the other hand, exhibited substantial but incomplete inhibition of p53 degradation, indicating that these two hydrophobic amino acids are also important for NES function (Fig. 3A, lane 6). When all four hydrophobic amino acids in the NES were replaced with alanines (p53NES), the mutant was completely resistant to MDM2 degradation (Fig. 3A, lane 8).
FIG. 3.
Nuclear export of p53 is also required for MDM2-mediated degradation. (A) The NES mutant p53 was resistant to MDM2 degradation. Plasmid DNAs encoding wild-type and various NES mutant p53s were either singly transfected or cotransfected with MDM2 into U2-OS cells. Cell lysates were prepared 24 h after transfection, separated by SDS-PAGE, and transferred onto a nitrocellulose membrane. Different portions of the membrane were blotted with antibodies, as indicated. (B) Localization of p53-GFP fusion proteins. The pKI NES or p53 NES was attached to the C terminus of the p53NES mutant to create p53NES-NESpKI and p53NES-NESp53, respectively. The constructs were then fused C-terminally with GFP and expressed in U2-OS cells. Pictures were taken of living cells 24 h after transfection. (C) Attachment of the pKI NES or p53 NES to the to the C terminus of the p53NES mutant restored its sensitivity to MDM2 degradation. (D) 2KO cells were transfected and the cell lysate was blotted as for panel A.
To rule out the possibility that the resistance of the p53 NES mutants to MDM2 degradation was due to disruption of functions other than a loss of nuclear export, the NES from the cyclic AMP-dependent protein kinase inhibitor (pKI) (32) or from p53 itself (12, 38) was attached to the C terminus of the p53NES mutant (p53NES-NESpKI and p53NES-NESp53, respectively), and their sensitivity to MDM2 degradation was examined. As expected, fusion of the p53 NES to the C terminus of p53NES did not alter its nuclear localization (Fig. 3B, panel 3). Fusion of the pKI NES, on the other hand, caused p53 to be excluded from the nucleus, indicating that a robust pKI NES overpowers the p53 NLS activity (Fig. 3B, panel 2). Corresponding to the restored nuclear export function, both p53NES-NESpKI and p53NES-NESp53 exhibited restored sensitivity to MDM2 degradation (Fig. 3C). Once again, identical results were obtained in 2KO cells, ruling out a cell type-specific effect (Fig. 3D). Thus, these results demonstrate that the nuclear export of p53 is also required for its degradation by MDM2. Together with Fig. 2, the data show that nucleocytoplasmic shuttling of p53 is essential for MDM2-induced degradation.
Colocalization of a nonshuttling p53 with MDM2 either in the cytoplasm or in the nucleus is not sufficient for p53 degradation.
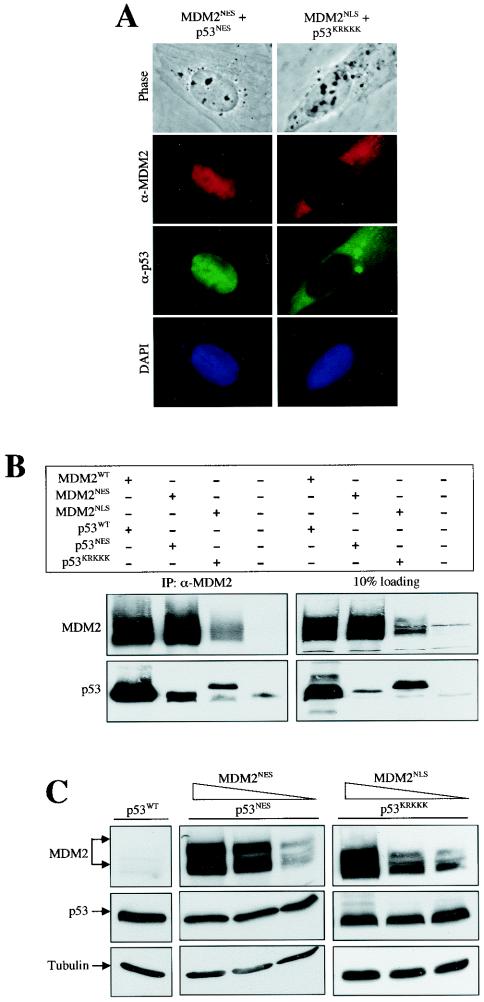
A caveat of the experiment showing that p53KRKKK is resistant to wild-type MDM2 degradation is that the p53 mutant is localized to the cytoplasm, whereas the MDM2, though it shuttles across the nuclear membrane, accumulates in the nucleus. The resistance of p53KRKKK to MDM2 degradation can also be interpreted as a lack of stable interaction of p53 with MDM2 in the same subcellular compartment. To determine whether colocalization of MDM2 with p53, either in the cytoplasm or in the nucleus, can induce p53 degradation, an MDM2NES mutant was coexpressed with p53NES and an MDM2NLS mutant was coexpressed with p53KRKKK. Although colocalized in the same subcellular compartment (Fig. 4A) and existing in an immunocomplex (Fig. 4B), the shuttling-inert mutant p53 remained resistant to MDM2 degradation even though MDM2 expression was relatively abundant (Fig. 4C). Hence, colocalization of MDM2 with a nonshuttling p53, either in the nucleus or in the cytoplasm, is not sufficient for inducing p53 degradation.
FIG. 4.
Colocalization with MDM2 either in the nucleus or in the cytoplasm is not sufficient to mediate nonshuttling p53 degradation. (A) Colocalization of MDM2 mutants with p53 mutants. U2-OS cells were transiently cotransfected with the indicated MDM2 and p53 DNAs. Twenty-four hours after transfection, cells were fixed and coimmunostained with a mouse anti-MDM2 antibody (SMP14) and a rabbit anti-p53 antibody (FL393) followed by staining with Texas red-conjugated donkey anti-mouse and fluorescein isothiocyanate-conjugated donkey anti-rabbit immunoglobulin antibodies. Phase contrast and 4′,6′-diamidino-2-phenylindole (DAPI) pictures are also shown. (B) Interaction of mutant p53 and MDM2 in vivo. U2-OS cells were transiently transfected with the indicated plasmid, and cells were lysed and immunoprecipitated with antibodies recognizing MDM2. Different portions of the membrane were blotted with antibodies, as indicated. (C) Resistance of nonshuttling p53 to MDM2 degradation. Plasmid DNA encoding nuclear export-deficient p53NES or nuclear import-deficient p53KRKKK were coexpressed with decreasing amountsof the nuclear export-deficient MDM2NES or nuclear import-deficient MDM2NLS mutant in U2-OS cells, respectively. Cell lysates were prepared 24 h after transfection, separated by SDS-PAGE, and transferred onto a nitrocellulose membrane. Different portions of the membrane were blotted with antibodies, as indicated.
Nucleocytoplasmic shuttling of MDM2 is not essential for inducing p53 degradation.
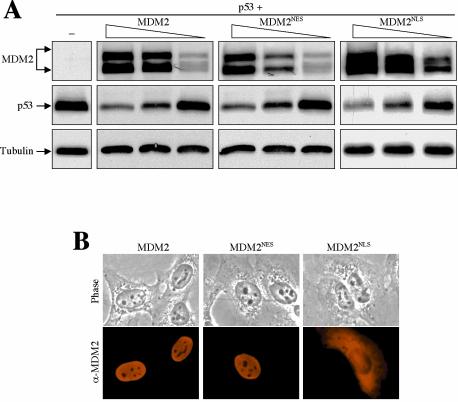
MDM2 shuttles through the nuclear membrane via its intrinsic NLS and NES (22). Whether the nucleocytoplasmic shuttling of MDM2 is required for its function to induce p53 degradation has been controversial (36). To gain insight into the role of MDM2 shuttling in controlling p53 degradation, MDM2 mutations that disrupted either the NES (L205A and I208A; MDM2NES) or the NLS (RRR161, 162,163AAA; MDM2NLS) were generated, and their ability to degrade p53 was determined. Interestingly, both the MDM2NLS and MDM2NES mutants exhibited p53 degradation activity comparable to that of wild-type MDM2 (Fig. 5A). However, we found that the MDM2NES mutant retained a weak nuclear export activity when examined in a sensitive heterokaryon assay (data not shown), and the MDM2NLS mutant showed visible nuclear accumulation (Fig. 5B, panel 3), suggesting an incomplete disruption of the NES and NLS functions by the mutations. We are currently seeking other NES and NLS sequences in MDM2. Nevertheless, as shown in Fig. 5, the efficiency of these MDM2 mutants in degrading p53 was apparently similar to that of wild-type MDM2, suggesting that nucleocytoplasmic shuttling of MDM2 is not essential for its function in mediating wild-type p53 degradation.
FIG. 5.
Nucleocytoplasmic shuttling of MDM2 is not essential for promoting p53 degradation. (A) Plasmid DNA (1 μg) encoding wild-type p53 was coexpressed with decreasing amounts (5, 3, and 1 μg) of a wild-type, an NES mutant, or an NLS mutant MDM2 in U2-OS cells. Cell lysates were prepared 24 h after transfection, separated by SDS-PAGE, and transferred onto a nitrocellulose membrane. Different portions of the membrane were blotted with antibodies, as indicated. (B) Localization of MDM2. U2-OS cells were transiently transfected with the indicated MDM2 DNAs. Twenty-four hours after transfection, cells were fixed and immunostained with an anti-MDM2 antibody (SMP14) and a Texas red-conjugated donkey anti-mouse immunoglobulin antibody. Phase contrast pictures are also shown.
p53 is efficiently ubiquitinated by MDM2 in the cytoplasm.
To determine whether the resistance of nonshuttling p53 to MDM2 degradation is due to a disruption of p53 polyubiquitination, we performed an in vivo ubiquitination assay by cotransfecting p53 and MDM2 into U2OS cells and lysing the cells after 6 h of MG132 treatment. Surprisingly, both the p53KRKKK and p53NES mutants, although resistant to MDM2 degradation, were ubiquitinated by MDM2 (Fig. 6). This was especially unexpected for the p53KRKKK mutant, which was localized exclusively in the cytoplasm.
FIG. 6.
Nonshuttling, nondegradable p53 mutants can be ubiquitinated by MDM2. U2-OS cells were transiently transfected with plasmid DNA encoding wild-type MDM2, wild-type p53, or various p53 mutants, as indicated. Twenty-four hours after transfection, cells were treated with 10 μM MG132 for 6 h and then harvested in SDS lysis buffer. The cell lysate was separated by SDS-PAGE and transferred onto a nitrocellulose membrane. Different portions of the membrane were blotted with antibodies to MDM2 (SMP14), p53 (DO1), and β-tubulin, as indicated.
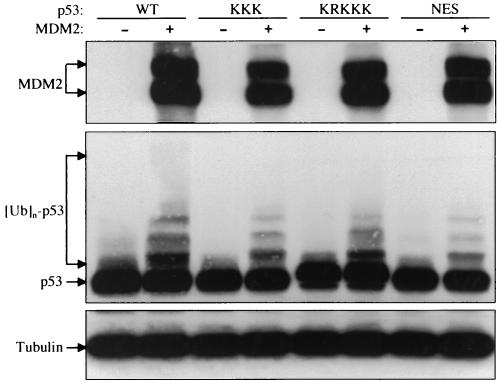
To test whether p53 can be ubiquitinated by MDM2 in the cytoplasm, we used the MDM2NLS and MDM2NES mutants in a p53 ubiquitination assay. Consistent with the results that the MDM2 shuttling mutants are able to mediate wild-type p53 degradation (Fig. 5A), both MDM2NLS and MDM2NES were able to ubiquitinate a wild-type p53, whereas a MDM2 RING finger mutant (MDM2C464A) was completely inactive (Fig. 7A). Interestingly, the MDM2NLS mutant exhibited a higher p53 ubiquitination activity than either the wild-type MDM2 or the MDM2NES mutant (Fig. 7A, lane 3). The ubiquitination ladder was apparently higher than a few ubiquitin molecules, suggesting that this cytoplasmic MDM2NLS-induced p53 ubiquitination was a polyubiquitination. These data indicated that the MDM2-mediated p53 ubiquitination occurs most efficiently in the cytoplasm.
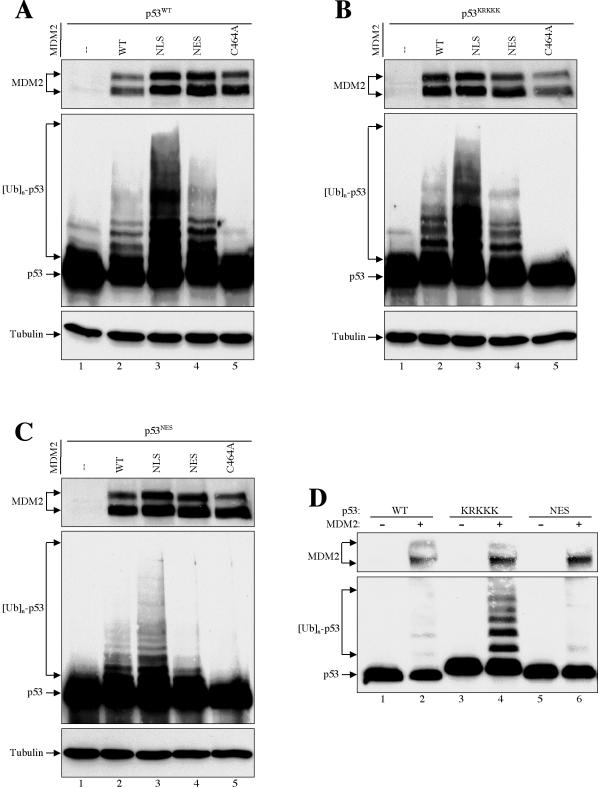
FIG. 7.
MDM2-mediated p53 ubiquitination occurs preferentially in the cytoplasm. (A to C) Wild-type and mutant MDM2s were cotransfected with plasmid DNA encoding wild-type p53 (A), nuclear import mutant p53KRKKK (B), and nuclear export mutant p53NES (C) into U2-OS cells. Twenty-four hours after transfection, cells were treated with 10 μM MG132 for 6 h and then harvested in SDS lysis buffer. The cell lysate was separated by SDS-PAGE and transferred onto a nitrocellulose membrane. Different portions of the membrane were blotted with antibodies to MDM2 (SMP14), p53 (DO1), and β-actin, as indicated. (D) H1299 cells were transiently transfected with plasmid DNA encoding wild-type MDM2, wild-type p53, and various p53 mutants, as indicated. Twenty-four hours after transfection, cells were lysed with 0.5% NP-40 lysis buffer. The cell lysate was separated by SDS-PAGE and transferred onto a nitrocellulose membrane. Different portions of the membrane were blotted with antibodies to MDM2 (SMP14) and p53 (DO1), as indicated.
To further confirm this cytoplasmic p53 ubiquitination, the cytoplasm-localized p53KRKKK was coexpressed with MDM2 (Fig. 7B). Consistently, the p53KRKKK was most efficiently ubiquitinated by a cytoplasmic MDM2NLS (Fig. 7B, lane 3), even though both proteins were localized in the cytoplasm (Fig. 4A). Corroborating this MDM2-mediated cytoplasmic p53 ubiquitination, the nucleus-confined p53NES was poorly ubiquitinated by either the wild-type or mutant MDM2 proteins (Fig. 7C). Finally, we reasoned that if p53KRKKK could be efficiently ubiquitinated by MDM2 but not degraded, we might be able to detect a polyubiquitinated form of p53 without with the proteasome inhibitor MG132 or lysing the cells in SDS. As shown in Fig. 7D, a polyubiquitinated p53KRKKK could be detected in transfected H1299 cells lysed with 0.5% NP-40 without MG132 treatment (Fig. 7D, lane 4), demonstrating that MDM2-mediated p53 ubiquitination occurs in the cytoplasm and that p53 ubiquitination can be uncoupled from its degradation. In contrast, polyubiquitinated species were not detected from the nucleus-confined p53NES under the same condition (Fig. 7D, lane 6), indicating either that p53 polyubiquitination does not occur in the nucleus (16) or that there exists a high deubiquitination activity in the nucleus. Polyubiquitinated species were also not detected from wild-type p53 (Fig. 7D, lane 2), presumably because they were quickly degraded. Together, these data strongly argue that MDM2-mediated p53 polyubiquitination occurs more efficiently, if not exclusively, in the cytoplasm and that p53 polyubiquitination can be uncoupled from its degradation.
DISCUSSION
p53 is controlled primarily by its protein stability through MDM2-mediated ubiquitination and degradation. Both MDM2 and p53 shuttle back and forth between the nucleus and the cytoplasm. A conceivable physiological role of the shuttling is that the nuclear membrane provides a barrier that effectively separates the location of the function (in the nucleus) and the destruction (in the cytoplasm) of both MDM2 and p53. Ample evidence has suggested that p53 nucleocytoplasmic translocation is linked to its ubiquitination and destruction. However, the exact mechanism involved remains unclear, and multiple models have been proposed attempting to justify data obtained from individual studies.
In an attempt to clarify the role of nucleocytoplasmic shuttling in p53 degradation, we focused on p53 point mutants defective in the function of the NLS and NES instead of using leptomycin B, a pharmacological drug that could potentially affect p53 transport and degradation by multiple mechanisms. There are several new findings from our study. (i) We found that mutations disrupting only one part of the p53 bipartite NLS can severely reduce but not eliminate its nuclear import activity. (ii) With a set of novel p53 NLS and NES mutants, we demonstrated that both nuclear import and export activities are required for p53 degradation. (iii) MDM2 nucleocytoplasmic shuttling appears to be unimportant for inducing p53 degradation. MDM2 mutants defective in nuclear import or export functions are similarly active in degrading p53. (iv) We provide evidence arguing that MDM2-mediated p53 polyubiquitination occurs most efficiently in the cytoplasm and that p53 polyubiquitination can be uncoupled from its degradation.
p53 has a C-terminal bipartite NLS containing two clusters of basic amino acids (Fig. 1A) (17). Previous studies involving p53 nuclear import often used mutations disrupting one of the two parts of the NLS, which is sufficient to illustrate nuclear exclusion of the mutated protein by immunofluorescence staining or by GFP tag. However, both techniques are limited by only providing evidence of the location of a protein at a steady-state level, and neither can reveal dynamic traffic. By combining mutations in the NES and the NLS, we have shown a previously unrecognized weak nuclear import activity of mutant p53 in which only one part of the NLS is disrupted (Fig. 1). We propose that the nuclear exclusion of such a p53 mutant (e.g., p53KR and p53KKK) is the result of a much reduced but not eliminated nuclear import function of the NLS, so that the activity of the NES becomes relatively stronger and the balance of the mutant p53 shuttling favors cytoplasmic accumulation. In combination with mutations of the NES, we show that disrupting both parts of the NLS (p53KRKKK) blocks the nuclear import of p53 as well as the degradation induced by MDM2.
It has been reported that p53 degradation can occur in either the nucleus or the cytoplasm, provided it is colocalized with MDM2 (34, 35). Our data show that blocking p53 nuclear import or export by multiple mutations in the NLS or NES abolishes its degradation. A possible explanation for the discrepancy is the use of different p53 mutants. We found that the individual point mutations in the p53 NLS, though variably affecting p53 nuclear import, did not block it, and the p53 mutants remained sensitive to MDM2 degradation. Only mutations that completely blocked p53 nuclear import (e.g., p53KRKKK) rendered p53 resistant to MDM2 degradation. On the other hand, p53 confined in the nucleus is also nondegradable by MDM2.
Our data are consistent with a model in which p53 is transported into the nucleus to function as a transcription activator. The nuclear p53 then needs to be exported to the cytoplasm for degradation. Although p53 degradation appears to occur exclusively in the cytoplasm, without involving the nucleus, a constantly cytoplasm-bound p53 cannot be degraded. This mechanism effectively separates the locations of function and destruction of p53 to avoid premature degradation by MDM2 before p53 reaches its target in the nucleus. Based on our data, we propose that p53 needs to be “primed” in the nucleus prior to its degradation. The nature of the potential nuclear factor(s) responsible for the priming remains unclear, but it apparently excludes MDM2-mediated ubiquitination.
A surprising finding of our study is that MDM2-induced p53 ubiquitination appears to occur more efficiently in the cytoplasm. It has been proposed that MDM2 promotes p53 mono- or short ubiquitination in the nucleus, which facilitates p53 nuclear export (5, 35). Once in the cytoplasm, more ubiquitin can be continuously added on to this mono- or short-ubiquitinated p53 required for proteasomal degradation. Although our data do not formally exclude the possibility of a conformational change-induced effect from the p53KRKKK or MDM2NLS mutants and do not exclude the possibility of p53 nuclear ubiquitination, they do indicate that p53 ubiquitination can occur efficiently in the cytoplasm.
It is postulated that a polyubiquitinated p53 is guided to the 26S proteasome and degraded immediately. However, our data indicate that p53 can be polyubiquitinated in the cytoplasm (the polyubiquitin ladder shown in Fig. 7 is apparently higher than mono- or multisite monoubiquitin), and the polyubiquitination of p53 can be uncoupled from degradation. A shuttling-deficient p53 (e.g., p53KRKKK and p53NES) remaining either in the nucleus or in the cytoplasm can be polyubiquitinated but not degraded by MDM2. This uncoupling of polyubiquitination and degradation of p53 has been observed previously with an MDM2 mutant with two mutated phosphorylatable serine residues (S251 and 254A) (4). Thus, it appears that polyubiquitination is required but not sufficient for p53 degradation. We speculate that modifications, other than ubiquitination of p53, which occur only in the nucleus are required for p53's degradation. Without this modification, even a polyubiquitinated p53 cannot be degraded.
Acknowledgments
We thank Koji Itahana and Krishna Bhat for helpful discussions and critical reading of the manuscript and Aiwen Jin for technical support.
Y.Z. is the recipient of a Career Award in Biomedical Science from the Burroughs Wellcome Fund and a Howard Temin Award from the National Cancer Institute. This study was supported by the M. D. Anderson Research Trust Fund and an NIH grant (to Y.Z.).
The first two authors contributed equally to this work.
REFERENCES
- 1.Ashcroft, M., and K. H. Vousden. 1999. Regulation of p53 stability. Oncogene 18:7637-7643. [DOI] [PubMed] [Google Scholar]
- 2.Barak, Y., T. Juven, R. Haffner, and M. Oren. 1993. Mdm-2 expression is induced by wild-type p53 activity. EMBO J. 12:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benditt, J. O., C. Meyer, H. Fasold, F. C. Barnard, and N. Riedel. 1989. Interaction of a nuclear location signal with isolated nuclear envelopes and identification of signal-binding proteins by photoaffinity labeling. Proc. Natl. Acad. Sci. USA 86:9327-9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, C., T. Hay, D. W. Meek, and D. P. Lane. 2002. Hypophosphorylation of Mdm2 augments p53 stability. Mol. Cell. Biol. 22:6170-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, S. D., K. Y. Tsai, and T. Jacks. 2000. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2:563-568. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., J. Lin, and A. J. Levine. 1995. Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol. Med. 1:142-152. [PMC free article] [PubMed] [Google Scholar]
- 7.Freedman, D. A., and A. J. Levine. 1998. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 18:7288-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furukawa, M., Y. Zhang, J. McCarville, T. Ohta, and Y. Xiong. 2000. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20:8185-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geyer, R. K., Z. K. Yu, and C. G. Maki. 2000. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2:569-573. [DOI] [PubMed] [Google Scholar]
- 10.Giaccia, A. J., and M. B. Kastan. 1998. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 12:2973-2983. [DOI] [PubMed] [Google Scholar]
- 11.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 12.Henderson, B., and A. Eleftheriou. 2001. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 13.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 14.Honda, R., and H. Yasuda. 1999. Association of p19ARF with MDM2 inhibits ubiquitin ligase activity of MDM2 for tumor suppressor p53. EMBO J. 18:22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubbutat, M. H. G., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 16.Li, M., D. Chen, A. Shiloh, J. Luo, A. Y. Nikolaev, J. Qin, and W. Gu. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416:648-653. [DOI] [PubMed] [Google Scholar]
- 17.Liang, S.-H., and M. F. Clarke. 2001. A bipartite nuclear localization signal is required for p53 nuclear import regulated by a carboxyl-Terminal domain. J. Biochem. 274:32699-32703. [DOI] [PubMed] [Google Scholar]
- 18.Meek, D. W. 1999. Mechanisms of switching on p53: a role for covalent modification? Oncogene 18:7666-7675. [DOI] [PubMed] [Google Scholar]
- 19.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 20.Nigg, E. A. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779-787. [DOI] [PubMed] [Google Scholar]
- 21.Pomerantz, J., N. Schreiber-Agus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, and R. A. DePinho. 1998. The INK4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92:713-723. [DOI] [PubMed] [Google Scholar]
- 22.Roth, J., M. Dobbelstein, D. A. Freedman, T. Shenk, and A. J. Levine. 1998. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17:554-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan, K. M., A. C. Phillips, and K. H. Vousden. 2001. Regulation and function of the p53 tumor suppressor protein. Curr. Opin. Cell Biol. 13:332-337. [DOI] [PubMed] [Google Scholar]
- 24.Sherr, C. J., and J. D. Weber. 2000. The ARF-p53 pathway. Curr. Opin. Genet. Dev. 10:94-99. [DOI] [PubMed] [Google Scholar]
- 25.Shieh, S.-Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 26.Shirangi, T. R., A. Zaika, and U. M. Moll. 2002. Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J. 16:420-422. [DOI] [PubMed] [Google Scholar]
- 27.Stommel, J. M., N. D. Marchenko, G. S. Jimenez, U. M. Moll, T. J. Hope, and G. M. Wahl. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18:1660-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao, W., and A. J. Levine. 1999. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc. Natl. Acad. Sci. USA 96:3077-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao, W., and A. J. Levine. 1999. p19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. USA 96:6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thut, C. J., J. A. Goodrich, and R. Tjian. 1997. Repression of p53-mediated transcription by MDM2, a dual mechanism. Genes Dev. 11:1974-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 32.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 33.Wu, X., J. H. Bayle, D. Olson, and A. J. Levine. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7:1126-1132. [DOI] [PubMed] [Google Scholar]
- 34.Xirodimas, D. P., C. W. Stephen, and D. P. Lane. 2001. Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp. Cell Res. 270:66-77. [DOI] [PubMed] [Google Scholar]
- 35.Yu, Z. K., R. K. Geyer, and C. G. Maki. 2000. MDM2-dependent ubiquitination of nuclear and cytoplasmic p53. Oncogene 19:5892-5897. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Y., and Y. Xiong. 2001. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 12:175. [PubMed] [Google Scholar]
- 37.Zhang, Y., and Y. Xiong. 1999. Mutation in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell 3:579-591. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, Y., and Y. Xiong. 2001. A p53 Amino terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 292:1910. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725-734. [DOI] [PubMed] [Google Scholar]