Abstract
Lipopolysaccharide (LPS) O polysaccharide was identified as the principle factor impeding intercellular formation of intact thin aggregative fimbriae (Tafi) in Salmonella enterica serovar Enteritidis. The extracellular nucleation-precipitation assembly pathway for these organelles was investigated by quantifying fimbrial formation between ΔagfA (AgfA recipient) and ΔagfB (AgfA donor) cells harboring mutations in LPS (galE::Tn10) and/or cellulose (ΔbcsA) synthesis. Intercellular complementation could be detected between ΔagfA and ΔagfB strains only when both possessed the galE mutation. LPS O polysaccharide appears to be an impenetrable barrier to AgfA assembly between cells but not within individual cells. The presence of cellulose did not restrict Tafi formation between cells. Transmission electron microscopy of w+ S. enterica serovar Enteritidis 3b cells revealed diffuse Tafi networks without discernible fine structure. In the absence of cellulose, however, individual Tafi fibers were clearly visible, appeared to be occasionally branched, and showed the generally distinctive appearance described for Escherichia coli K-12 curli. A third extracellular matrix component closely associated with cellulose and Tafi was detected on Western blots by using immune serum raised to whole, purified Tafi aggregates. Cellulose was required to tightly link this material to cells. Antigenically similar material was also detected in S. enterica serovar Typhimurium and one diarrheagenic E. coli isolate. Preliminary analysis indicated that this material represented an anionic, extracellular polysaccharide that was distinct from colanic acid. Therefore, Tafi in their native state appear to exist as a complex with cellulose and at least one other component.
Thin aggregative fimbriae (Tafi) (formerly SEF17) are produced by most Salmonella strains and isolates of Escherichia coli, in which these organelles have been termed curli. In Salmonella, the divergently transcribed agfDEFG and agfBAC operons are required for production of functional Tafi (10, 35). Tafi are comprised primarily of AgfA subunits, and thus far there is evidence for the presence of only one other protein in the fiber, AgfB (7, 46). Tafi and curli fibers are remarkably stable, requiring treatment with 90% formic acid to depolymerize (3, 12). These fibers bind the hydrophobic dye Congo red (CR) (13), presumably due to the similar β-strand structures of AgfA and AgfB (31, 46).
AgfA and AgfB are interesting sequence homologues that are likely the result of several gene duplications; they are nearly identical in size and share many structural features. Despite their similarities, they have very different biochemical properties (46). Both proteins are required for fimbrial polymerization, with AgfA substantially predominating in the fimbriae. AgfEFG proteins participate in fiber formation and somehow ensure the fidelity of assembly. Deletion of agfE or agfF leads to the production of fimbriae with altered biochemical properties (9), whereas deletion of agfG disrupts fimbrial formation altogether (28). AgfD is a positive transcriptional regulator of the agf operon (19) and is required for Tafi biosynthesis (36).
There is growing interest in the relationship between Tafi (or curli) production in Salmonella (or E. coli) and the formation of biofilms (4, 24, 32, 39). Tafi (curli) production by many S. enterica and E. coli strains is tightly regulated by growth conditions, normally occurring at temperatures below 30°C (17, 37). In Salmonella enterica serovar Enteritidis 3b, Tafi are produced under less stringent conditions at both 28 and 37°C (12). These differences in regulation are likely due to differences in the agfD promoter sequence (37, 44). Recently, AgfD was shown to regulate production of another extracellular substance, cellulose (36, 48). Tafi and cellulose form a highly resistant extracellular matrix which may be important for ensuring bacterial survival in harsh environments (39). Capsular polysaccharides, such as colanic acid (CA), have also been implicated in the formation of Tafi- and curli-associated biofilms (33). These substances appear to be important for maintenance of space between cells and development of complex three-dimensional architectures (14).
Assembly of Tafi or curli fibers has been proposed to occur extracellularly, perhaps in the midst of other components. The current assembly hypothesis allows for fimbrial growth by addition of subunits principally to the distal end of the growing fibers (20). This is unique among fimbriae and has been termed the extracellular nucleation-precipitation pathway (40). The hypothesis is based upon the intercellular complementation of an E. coli MC4100 csgA mutant grown in close proximity to a csgB mutant (CsgA donor) on solid medium (20). Complementation yielded cells which possessed assembled curli and as a result were able to bind the dye CR. However, these experiments could not be reproduced in S. enterica serovar Enteritidis 3b with precisely defined agfA and agfB mutant strains (46). For this study we devised an intercellular complementation strategy applicable to enteric strains more native than E. coli MC4100.
S. enterica serovar Enteritidis 3b possesses smooth lipopolysaccharide (LPS) O polysaccharide and produces cellulose in concert with Tafi (48). In contrast, E. coli MC4100 produces neither LPS O polysaccharide (27) nor cellulose (48). To allow for these differences, S. enterica serovar Enteritidis 3b ΔagfA and ΔagfB strains were rendered LPS O-polysaccharide deficient by introducing a galE::Tn10 mutation and cellulose deficient by deleting bcsA, coding for cellulose synthase (34). Intercellular complementation between strains did not readily occur and did so only when the donor and acceptor were LPS O-polysaccharide deficient. Thus, LPS O polysaccharide substantially interferes with Tafi formation between cells under normal conditions, indicating that Tafi assembly does not normally occur intercellularly. Additional experiments with ΔbcsA strains revealed that Tafi in their native state exist as a complex with cellulose and at least one other extracellular polysaccharide (EPS).
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
S. enterica serovar Enteritidis 27655 strain 3b and its ΔwcaJ mutant, S. enterica serovar Typhimurium SL7207 (22), and E. coli Vietnam I/1 (13) were routinely grown in T broth (12) at 28 or 37°C for 24 or 48 h, with shaking at 200 rpm, or on T agar (T) or T agar plus CR (100 μg/ml) indicator plates (TCR agar) as previously described (11).
Generation of ΔbcsA and ΔwcaJ strains.
An in-frame deletion removing 1,998 bp in bcsA (encoding amino acids 165 to 828 in BcsA) was generated as previously described (48) with some modifications. Primer YHJ05 was modified to contain an EcoRI site instead of a BamHI site, and a new YHJ07 primer (CCACTGCAGATTCGCGCCGCCTTCAGTAA [a PstI site is underlined]) was generated since S. enterica serovar Typhimurium bcsA contains a unique EcoRI site corresponding to amino acids 828 to 829; primers YHJ06 and YHJ08 were used as described previously (48). An in-frame deletion removing amino 1,206 bp in wcaJ (encoding amino acids 34 to 436 in WaaJ) was generated similarly by using PCR primers wcaJ01 (GGTGAATTCAAAAACCGGGCACGC [an EcoRI site is underlined]), wcaJ02 (GAACTGCAGCCCGCCAAACA [a PstI site is underlined]), wcaJ03 (GACCTGCAGTAAATCCGCGA [a PstI site is underlined]), and wcaJ04 (GTAAAGCTTGGTCAGCTCCA [a HindIII site is underlined]). For each construct, the two PCR fragments generated were directionally cloned into EcoRI-HindIII-cut pTZ18R (Amersham Biosciences) and subcloned into pHSG415 (21). The ΔbcsA or ΔwcaJ mutation was introduced into the chromosomes of S. enterica serovar Enteritidis strains according to procedures described previously (47) with modifications: Apr plasmid-cointegrate colonies were selected after growth for 18 h at 42°C, and Aps ΔbcsA mutants were selected after two or three 18-h passages at 28°C. Mutants were identified by PCR screening with primers bcsAko1 (CGGCCCGTTACCTCATTCAG), bcsAko2 (TTCAGCACCGCTTTCGACGC), bcsAwt1 (CAGAAACGAGCGTGTCGGCA), wcaJko1 (CGCTGCTGAATCAGTAACGT), wcaJko2 (TTAGCGCCGCTGATCGTTTT), and wcaJwt1 (GCGACGCCAGCACGATATCT), based on the S. enterica serovar Typhimurium DNA sequences (GenBank accession numbers AJ315770 and AF285085).
Generation of galE::Tn10 mutants of S. enterica serovar Enteritidis 3b ΔagfA and ΔagfB strains.
Phage stocks prepared after growth of transducing phage P22 int3 HT 12/4 on S. enterica serovar Typhimurium LT2-TN1117 were used to infect S. enterica serovar Enteritidis 3b, ΔagfA, and ΔagfB cells at multiplicities of infection ranging from 0.01 to 5. Transductants were selected on Luria-Bertani plates containing tetracycline (16 μg/ml) and were rendered phage free by passage on Green plates (8). Potential galE::Tn10 transductants were screened with indicator plates specific for galactose fermentation (23). galE phenotypes were confirmed by the absence of LPS O polysaccharide (O antigen) after growth on nutrient agar without galactose (23). galE::Tn10 mutants grown on T agar (see Fig. 1 to 3) were severely depleted in O antigen as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and LPS-silver staining, but trace amounts (<10% of wild-type [w+] levels) were detected.
FIG. 1.
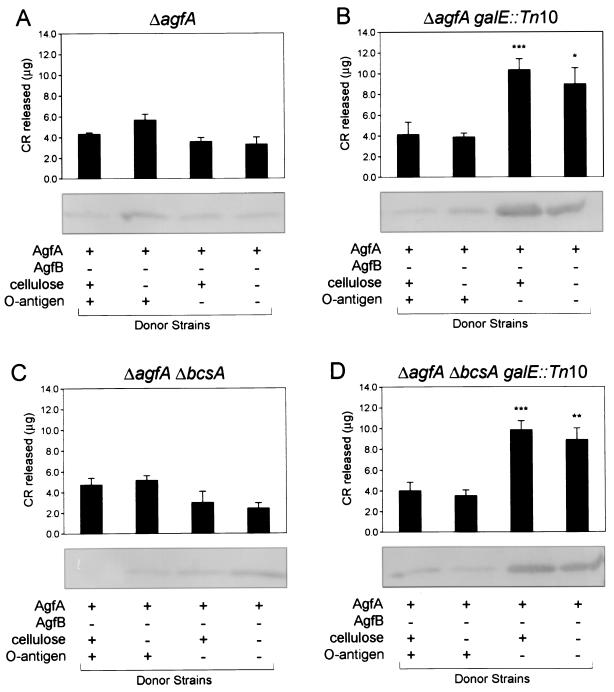
Intercellular complementation of Tafi between S. enterica serovar Enteritidis 3b ΔagfB donor and ΔagfA recipient strains. CR binding of different ΔagfA recipient strains grown together with ΔagfB donor strains is shown. Recipient strain genotypes are indicated above each graph, while ΔagfB donor strain phenotypes are listed at the bottom. CR binding values for each combination of mutants were normalized by subtracting values for the corresponding ΔagfB or ΔagfA strains grown individually. Each bar shows the averages and standard errors from four separate experiments, and asterisks indicate significant differences within each group (*, P < 0.05; **, P < 0.01; ***, P < 0.001 [Tukey-Kramer multiple comparisons test]). The panels below the bar graphs show immunoblot analysis performed with immune serum raised to purified Tafi. The amount of AgfA fimbrial material associated with recipient and donor cells scraped off the TCR agar is noted for each recipient-donor combination.
FIG. 3.
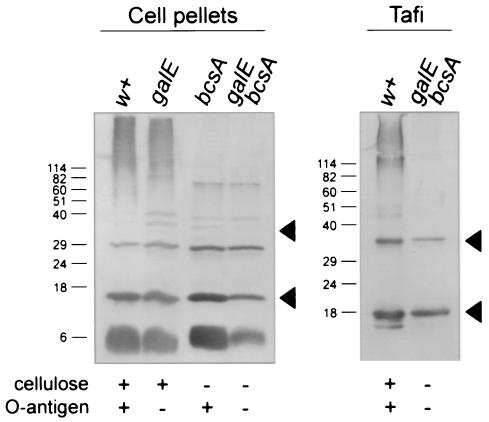
Immunoblot analysis of AgfA and associated material produced by S. enterica serovar Enteritidis 3b and related strains. Proteins in the debris left over after boiling cells in SDS-PAGE sample buffer (cell pellets) or purified fimbrial material (Tafi) from S. enterica serovar Enteritidis 3b (w+) the galE::Tn10 strain (galE), the ΔbcsA strain (bcsA), or the galE::Tn10 ΔbcsA strain (galE bcsA) were loaded as indicated. Strain phenotypes are listed below each immunoblot. All samples were treated with 90% formic acid prior to loading on SDS-PAGE. AgfA and associated material were detected by using immune serum raised to whole Tafi; the lower and upper arrowheads indicate monomeric and dimeric AgfA, respectively. Molecular mass markers (in kilodaltons) are indicated on the left..
Purification of Tafi from S. enterica serovar Enteritidis ΔbcsA galE::Tn10.
Fimbriae were isolated and purified from plate-grown cells as previously described (12). SDS-PAGE sample buffer-insoluble material which did not enter the stacking gel was recovered, washed three times in distilled water (dH2O), and lyophilized. This material was resuspended in dH2O and treated with 90% formic acid before SDS-PAGE and Western blotting.
Intercellular complementation.
S. enterica serovar Enteritidis 3b and the ΔagfA, ΔagfB, and related mutants were grown overnight at 28°C in T broth; cultures were normalized to an optical density at 600 nm (OD600) of 1, and sterile swabs were used to streak different ΔagfA and ΔagfB combinations immediately adjacent to each other on TCR agar. Any differences in colony color due to variable CR binding were recorded after growth for 24 or 48 h at 37°C. For coplating experiments, 500-μl portions (OD600 of 0.5) of two different ΔagfA and ΔagfB cultures were combined, and a 100-μl aliquot was plated onto TCR agar. For each control strain, 100 μl of the original cell suspension at an OD600 of 1 was plated directly. Following growth for 24 h at 37°C, cells were scraped off the agar and resuspended in 1 ml of 10 mM Tris (pH 7.2). For SDS-PAGE and immunoblotting, cell samples at an OD600 of 4 were aliquoted, buffer was removed, and cells were treated as described below. For the CR binding assay (45), cells were normalized to an OD600 of 10 per ml and left to equilibrate for 1 h at room temperature (RT). After removal of cells by centrifugation, the amount of CR released into the buffer was determined by measuring the absorbance at 480 nm.
SDS-PAGE and Western blot analyses.
Cell pellets from 4 OD600 units of cells were resuspended in 100 μl of SDS-PAGE sample buffer (26) and boiled for 10 min, and cellular debris was pelleted by centrifugation (15,600 × g, 5 min). These samples were washed with 500 μl of dH2O, resuspended in 250 μl of 90% formic acid, frozen, and lyophilized. For analysis of proteins secreted into the agar, cells were removed and plugs of ∼8 mm in diameter were taken (9), resuspended in 500 μl of dH2O or 98% formic acid, frozen, and lyophilized. Samples were resuspended in SDS-PAGE sample buffer before separation by SDS-PAGE (26). Proteins and other materials were electrophoretically transferred to nitrocellulose in a glycine (192 mM)-Tris (25 mM, pH 8.3)-methanol (20%) solution by using a Mini-Protean II apparatus (Bio-Rad Laboratories). Proteins and associated material were detected by using immune serum raised to purified whole Tafi followed by goat anti-rabbit immunoglobulin G-alkaline phosphatase secondary antibody (Cedarlane Laboratories Ltd.) and nitroblue tetrazolium and BCIP (5-bromo-4-chloro-3-indolylphosphate) substrates (12).
Electron microscopy.
S. enterica serovar Enteritidis 3b strains grown on T agar were resuspended in 10 mM Tris (pH 8) and deposited on 0.5% Formvar-coated nickel grids. The immunogold labeling procedure was as follows: 40 min at RT in 1% bovine serum albumin-0.15% NaCl-10 mM Tris (pH 8), 45 min at RT with AgfA-specific monoclonal antibody ascites 3A-12 diluted 1:1,000 in 0.1% bovine serum albumin-Tris-NaCl (diluent), and 30 min at RT with goat anti-mouse immunoglobulin G-10-nm-diameter gold (Cedarlane Laboratories Inc.) diluted 1:20 in diluent. Washes between steps were performed with Tris-NaCl. For negative staining only, cells were deposited on 0.5% Formvar-coated copper grids, rinsed in dH2O, and stained with 1 to 2% uranyl acetate (pH 7). All samples were visualized with a Hitachi H7600 transmission electron microscope under HC-zoom mode at 100 kV.
RESULTS
Morphological characterization of S. enterica serovar Enteritidis ΔbcsA and galE mutants.
Strains with ΔbcsA and/or galE::Tn10 mutations were generated from S. enterica serovar Enteritidis 3b or the ΔagfA or ΔagfB mutant. The phenotypes of these strains were compared after growth on TCR agar (Table 1). All agfBA+ strains exhibited an aggregative colony morphology and high CR binding, whereas all ΔagfA or ΔagfB strains were nonaggregative, with a much lower CR binding (Table 1). Introduction of the bcsA deletion caused a color change in all strains and a slight drop in CR binding in ΔagfA or ΔagfB strains (Table 1). These phenotypes matched those recently reported for similar ΔbcsA strains of S. enterica serovar Typhimurium (48). Introduction of the galE mutation resulted in different effects. In agfBA+ strains a drop in aggregative morphology was observed, but minimal changes in CR binding were measured (Table 1). In ΔagfA or ΔagfB strains, a color change was observed, and a small increase in CR binding was measured (Table 1). These results confirm that AgfA, AgfB, and subsequent Tafi fibers represent the major CR binding component produced by S. enterica serovar Enteritidis 3b (11), while cellulose and LPS O polysaccharide and have relatively minor effects.
TABLE 1.
Morphological characterization of S. enterica serovar enteritidis 3b and isogenic agfA or agfB mutants
| Straina | Colony colorb | Morphologyc | S. enterica serovar Typhimurium morphologyd | CR bound (μg)e |
|---|---|---|---|---|
| 3b (agfBA+) | Red | Ag(++) | rdar | 35.8 ± 9.9 |
| ΔbcsA | Red-brown | Ag(++) | bdar | 44.1 ± 12.3 |
| galE::Tn10 | Orange-brown | Ag(+) | N/Af | 34.7 ± 6.1 |
| ΔbcsA galE::Tn10 | Orange-brown | Ag(+) | N/A | 34.5 ± 2.9 |
| ΔagfA mutant | Pink | NAg | pdar | 5.6 ± 0.6 |
| ΔbcsA | White | NAg | saw | 3.7 ± 0.4 |
| galE::Tn10 | Pink-orange | NAg | N/A | 8.4 ± 1.5 |
| ΔbcsA galE::Tn10 | Pink | NAg | N/A | 7.3 ± 1.5 |
| ΔagfB mutant | Pink | NAg | pdar | 4.6 ± 0.6 |
| ΔbcsA | White | NAg | saw | 3.6 ± 0.6 |
| galE::Tn10 | Pink-orange | NAg | N/A | 11.2 ± 2.2 |
| ΔbcsA galE::Tn10 | Pink | NAg | N/A | 10.6 ± 1.1 |
S. enteritidis strains as described in Materials and Methods.
Color of colonies judged after growth on TCR agar for 24 h at 37°C.
The morphology of colonies was compared to that of the 3b parent strain Ag(++), most aggregative (colonies dry and difficult to break apart); Ag(+), aggregative (colonies dry but easier to break apart); NAg, nonaggregative (colonies mucoid).
Colony phenotypes of analogous S. enterica serovar Typhimurium strains grown on Luria agar without salt (containing 40 μg of CR per ml and 20 μg of Coomassie brilliant blue per ml) rdar, red, dry, and rough; bdar, brown, dry, and rough; pdar, pink, dry, and rough; saw, smooth and white. Phenotypes are as reported previously (35, 36, 48).
Amount of CR eluted from 10 A600 units of cells ± standard error of the mean from four independent experiments.
N/A, not applicable.
Intercellular complementation of Tafi between S. enterica serovar Enteritidis 3b ΔagfB and ΔagfA strains.
To effect intercellular formation of Tafi between cells, S. enterica serovar Enteritidis ΔagfA and ΔagfB mutant strains depleted of LPS O polysaccharide (O antigen) or devoid of cellulose were grown closely side by side on TCR agar at 28 or 37°C for 24 or 48 h. No regions of CR binding or other changes in colony color were apparent for any the ΔagfA colonies grown beside ΔagfB colonies, indicating that Tafi were not being formed between cells. Cross-streaking the different strains together, as recently demonstrated in E. coli (9), also did not enable complementation. No colony-associated color changes were seen, indicating that Tafi were not being formed between the cells (data not shown).
To enhance complementation, liquid cultures of each of the four S. enterica serovar Enteritidis ΔagfB (AgfA donor) and ΔagfA (AgfA recipient) strains (Table 1) were mixed and grown together on TCR agar. After 24 h of growth, cells from the 16 different combinations were harvested and the relative CR binding was measured (Fig. 1, upper panels). Significant increases in CR binding were observed for the four combinations in which both donor and recipient strains carried the galE::Tn10 mutation (Fig. 1B and D, last two bars). No significant differences in CR binding were measured in the remaining groups, in which at least one strain was galE+ (Fig. 1A and C and Fig. 1B and D, first two bars).
Different levels of CR binding were correlated with cell-associated AgfA detected in the various donor-recipient combinations (Fig. 1, lower panels). Larger amounts of cell-associated AgfA were detected in combinations in which both donor and recipient strains contained galE::Tn10 (Fig. 1B and D, last two lanes). This was in contrast to the other 12 combinations, in which only a trace AgfA band was detectable for the majority of combinations (Fig. 1A and C and Fig. 1B and D, first two lanes). There was some experimental variability, and the levels of cell-associated AgfA did not always correlate with CR binding values (Fig. 1C, compare second and fourth lanes). However, the overall trend indicated that more cell-associated AgfA correlated with increased CR binding. This follows previous findings that increased AgfA polymerization or fimbrial formation correlates with increased levels of CR binding (6, 7, 15, 45).
Characterization of S. enterica serovar Enteritidis 3b ΔagfA and ΔagfB strains and detection of Tafi-associated material.
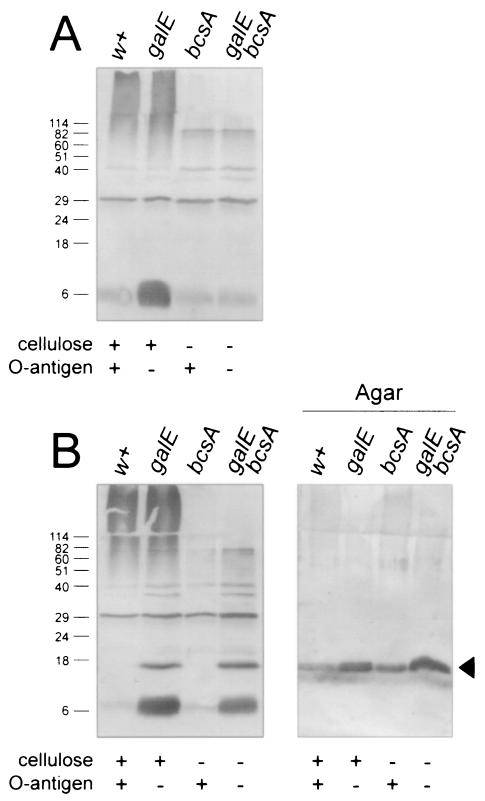
S. enterica serovar Enteritidis 3b ΔagfA and ΔagfB strains (Table 1) were analyzed for the presence of AgfA on Western blots to see if differences existed between the different strains. As expected, AgfA was not detected in any ΔagfA strains (Fig. 2A). Cell pellet samples from ΔagfB and ΔagfB ΔbcsA strains were also devoid of AgfA (Fig. 2B, lanes w+ and bcsA). Unexpectedly, residual cell-associated AgfA was detected in ΔagfB strains carrying the galE::Tn10 mutation (Fig. 2, lanes galE and galE bcsA),. This also correlated with an increase in CR binding in these strains (Table 1).
FIG. 2.
Western blot analysis of S. enterica serovar Enteritidis 3b ΔagfB donor and ΔagfA recipient strains for the production of AgfA. S. enterica serovar Enteritidis 3b ΔagfA (A) and ΔagfB (B) strains were analyzed for production of AgfA after growth on T agar. Samples from strains were loaded as indicated, and strain phenotypes are listed below each lane. Panel A and the first four lanes of panel B represent proteins in the debris left over after boiling cells in SDS-PAGE sample buffer, whereas agar samples (B) represent proteins present in agar plugs after cells were removed. All samples were treated with formic acid prior to loading on SDS-PAGE. AgfA and associated material were detected by using immune serum raised to purified Tafi; the arrowhead in panel B indicates monomeric AgfA. Molecular mass markers (in kilodaltons) are indicated on the left.
To test for secretion of AgfA by the ΔagfB mutants, cell-free agar plug samples were also analyzed. AgfA was detected in samples corresponding to each of the four S. enterica serovar Enteritidis 3b ΔagfB strains (Fig. 2B, right panel). Similar amounts of AgfA were detected without formic acid-induced depolymerization, indicating that the AgfA monomers were in an unpolymerized soluble form (data not shown). More AgfA was detected for galE::Tn10 strains (Fig. 2, lanes galE and galE bcsA), but the difference between strains was small. Thus, removal of cellulose or O antigen did not have a large effect on secretion of AgfA in the ΔagfB strains.
High-molecular-weight (high-MW) immunoreactive material was detected in two of the four ΔagfA and ΔagfB cell pellet samples (Fig. 2, lanes w+ and galE). In contrast, the high-MW material was not detected in cell pellet samples from ΔbcsA strains (Fig. 2, lanes bcsA and galE bcsA). This indicated that the material was associated with the presence of cellulose. However, pure cellulose was not immunoreactive with Tafi-specific immune serum (data not shown). Therefore, the high-MW material was distinct from AgfA, AgfB, and cellulose.
Immunological characterization of S. enterica serovar Enteritidis 3b agfBA+ strains.
To investigate Tafi production by agfBA+ S. enterica serovar Enteritidis 3b strains (Table 1), cell pellet samples were analyzed for levels of AgfA (Fig. 3). Overall, the levels of AgfA detected in each strain were very similar (Fig. 3, left panel), which correlates with high levels of CR binding for each strain (Table 1). Thus, depleting O polysaccharide or removing cellulose had little or no effect on overall Tafi production within individual cells.
Like for the ΔagfB or ΔagfA strains, high-MW immunoreactive material was detected in cell pellets from bcsA+ strains (Fig. 3, lanes w+ and galE). Again, this immunoreactive material was not found in cell pellets from ΔbcsA strains (Fig. 3, lanes bcsA and galE bcsA). The high-MW material appeared to be tightly cell associated only when cellulose was produced.
To test whether Tafi could be separated from this high-MW material, we purified fimbrial material from S. enterica serovar Enteritidis 3b ΔbcsA galE::Tn10 by the procedure outlined by Collinson et al. (12). Western blot analysis of these purified Tafi showed that only AgfA and its associated dimer were detected (Fig. 3, lanes galE bcsA) and not the high-MW immunoreactive material; however, it could be seen along with AgfA in Tafi purified from S. enterica serovar Enteritidis 3b (Fig. 3, lanes w+). Therefore, Tafi could be separated from this high-MW material in S. enterica serovar Enteritidis 3b, but only when cellulose was not produced,.
Electron microscopy of S. enterica serovar Enteritidis 3b and Tafi with or without cellulose.
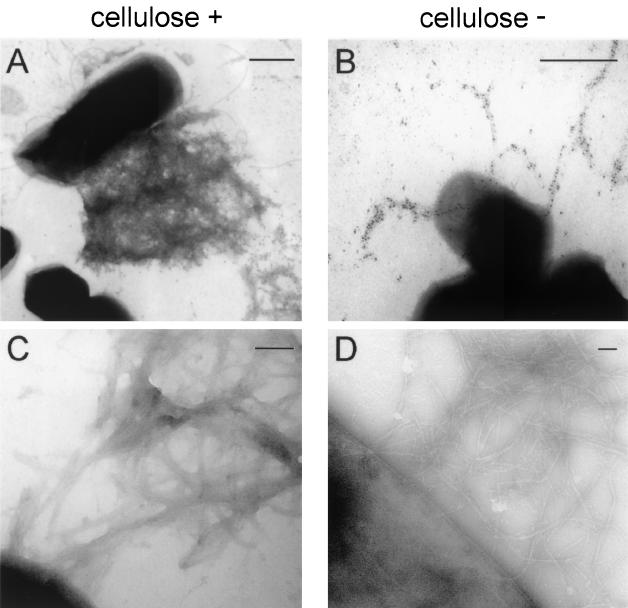
The appearances of Tafi produced by S. enterica serovar Enteritidis 3b bcsA+ and ΔbcsA strains were compared by using electron microscopy (Fig. 4). Immunogold labeling of S. enterica serovar Enteritidis 3b cells with an AgfA-specific monoclonal antibody most often showed Tafi as a diffuse fimbrial mat extending from the cell surface (Fig. 4A). Some cells were devoid of labeled fimbrial material, while the majority were intermediate in Tafi production (data not shown). In dramatic contrast, Tafi produced by S. enterica serovar Enteritidis 3b ΔbcsA were visualized as distinct, curling fibers with precise antibody labeling (Fig. 4B). In general, less fimbrial material was labeled on these cells.
FIG. 4.
Transmission electron microscopy of Tafi fimbriae. Fimbriae produced by S. enterica serovar Enteritidis strain 3b (cellulose positive) (A and C) or the ΔbcsA mutant (cellulose negative) (B and D) were immunogold labeled with AgfA-specific monoclonal antibody 3A-12 ascites followed by goat anti-mouse immunoglobulin-10-nm-diameter gold (A and B) or simply negatively stained with uranyl acetate (C and D). Bars, 500 nm (A and B) or 100 nm (C and D).
For observation of fimbrial material at higher magnifications, cell samples were negatively stained without antibody labeling (Fig. 4C and D). When S. enterica serovar Enteritidis 3b Tafi networks were visualized under higher magnifications, no fine detail or individual fibers could be resolved (Fig. 4C). The fimbrial material was quite diffuse. Tafi produced by S. enterica serovar Enteritidis 3b ΔbcsA were easily resolved into distinct individual fibers (Fig. 4D), although some fibers appeared to be occasionally branched. These observations indicated that S. enterica serovar Enteritidis 3b Tafi are closely associated with cellulose at the cell surface.
Detection of Tafi-associated material in S. enterica serovar Typhimurium and E. coli strains.
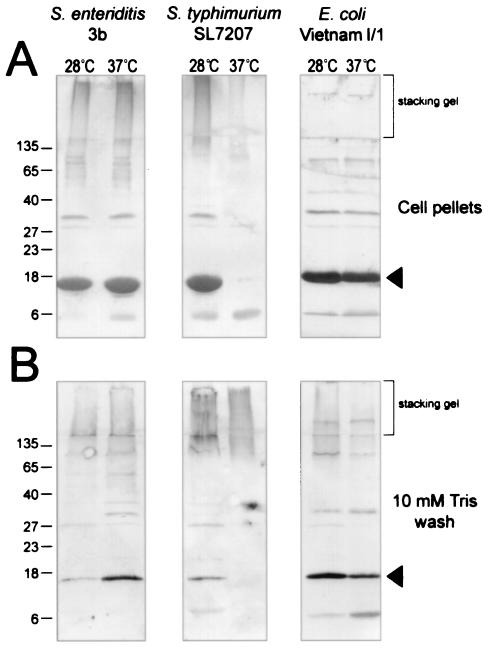
S. enterica serovar Enteritidis 3b, S. enterica serovar Typhimurium SL7207, and E. coli Vietnam I/1 were tested for production of Tafi and high-MW material after growth at 28 or 37°C (Fig. 5). AgfA (CsgA) was detected in cell pellets from each strain grown at 28°C but was detected only in S. enterica serovar Enteritidis 3b and E. coli Vietnam I/1 at 37°C (Fig. 5A). CsgA produced by E. coli Vietnam I/1 (Fig. 5A) migrated more slowly than AgfA, as reported previously (13).
FIG. 5.
Detection of immunoreactive Tafi-associated material in different enterobacterial species. Proteins in the debris left over after boiling cells in SDS-PAGE sample buffer (cell pellets) or acetone-precipitated proteins from a 10 mM Tris (pH 8) wash of whole cells of S. enterica serovar Enteritidis 3b, S. enterica serovar Typhimurium SL7207 or E. coli Vietnam I/1 after growth on T agar at 28 or 37°C were loaded as indicated. The brackets show the regions of immunoblots corresponding to the stacking gel from SDS-PAGE. AgfA and associated material were detected by using immune serum raised to whole Tafi; arrowheads indicate monomeric AgfA (Salmonella) or CsgA (E. coli). Molecular mass markers (in kilodaltons) are indicated on the left.
High-MW immunoreactive material found in the stacking gel was detected in cell pellet samples from S. enterica serovar Enteritidis 3b at 28 and 37°C and in S. enterica serovar Typhimurium SL7207 at 28°C (Fig. 5A, first three lanes, stacking gel). The E. coli samples were devoid of this material, and only traces were detected for S. enterica serovar Typhimurium SL7207 grown at 37°C (Fig. 5A, last three lanes). However, the high-MW material was detected in a pH 8, 10 mM Tris wash of these same cells (Fig. 5B, last three lanes, stacking gel). The material appeared to wash completely off the cells, suggesting that these cells were devoid of cellulose. The high-MW material was also detected in washes of S. enterica serovar Enteritidis 3b grown at 28 and 37°C and in washes of S. enterica serovar Typhimurium SL7207 grown at 28°C (Fig. 5B, first three lanes), indicating that it was not entirely associated with the cell pellet. However, small amounts of AgfA were detected in these lanes (Fig. 5B), and the material may have been associated with Tafi that washed off. The immunoreactive high-MW material likely represents a substance which is commonly found with Tafi (curli) in Salmonella and E. coli.
The high-MW material associated with cellulose and Tafi is a polysaccharide but not CA.
CA is an anionic EPS that has been detected in association with curli (Tafi) in E. coli (14, 33). Purified CA (25) has a high-MW smearing migration pattern on SDS-PAGE, and its composition is relatively conserved between S. enterica serovar Enteritidis, S. enterica serovar Typhimurium, and E. coli (16, 18). To investigate whether the high-MW material detected by Tafi-specific immune serum represented CA, an isogenic S. enterica serovar Enteritidis 3b CA mutant strain (ΔwcaJ mutant) was generated. The wcaJ gene encodes the initiating undecaprenylphosphate glucose phosphotransferase (41) that is required for CA production in E. coli (1).
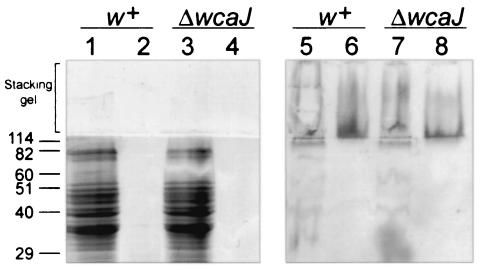
Immunoreactive high-MW material was detected on Western blots of cell pellet samples from both the 3b parent and ΔwcaJ strains (Fig. 6, lanes 5 and 7), indicating that the material did not represent CA. Furthermore, no differences in colony morphology were observed between the ΔwcaJ strain and S. enterica serovar Enteritidis 3b after growth at 28 or 37°C on TCR agar (data not shown). The high-MW material was resistant to digestion with proteinase K (Fig. 6, lanes 6 and 8) and did not stain with GelCode blue (Fig. 6, lanes 1 to 4), indicating that it was not proteinaceous. Small amounts of purified material tested positive for the presence of uronic acids (data not shown). These data, together with the migration pattern on SDS-PAGE, suggest that the high-MW material represents an uncharacterized anionic polysaccharide that is distinct from CA. This material may be a common component of the Tafi-cellulose extracellular matrix.
FIG. 6.
Detection of high-MW immunoreactive material in S. enterica serovar Enteritidis 3b and ΔwcaJ strains. SDS-PAGE (lanes 1 to 4) or immunoblot analysis (lanes 5 to 8) of whole cells of the S. enterica serovar Enteritidis 3b (w+) and ΔwcaJ strains boiled in SDS-PAGE sample buffer (lanes 1, 3, 5, and 7) and digested with 0.5 mg of proteinase K per ml for 1 h at 65°C (lanes 2, 4, 6, and 8) is shown. Proteins were detected with GelCode staining (Pierce) (lanes 1 to 4). High-MW material was detected by using immune serum raised to purified Tafi (lanes 5 to 8). The bracket shows the stacking gel region of SDS-PAGE and the corresponding immunoblot. Molecular mass markers (in kilodaltons) are indicated on the left.
DISCUSSION
We discovered that LPS O polysaccharide is the principle factor impeding intercellular Tafi formation in S. enterica serovar Enteritidis 3b. Experiments to achieve intercellular formation of Tafi between ΔagfA (AgfA recipient) and ΔagfB (AgfA donor) strains of S. enterica serovar Enteritidis 3b were repeatedly unsuccessful. Increases in CR binding occurred only when ΔagfA and ΔagfB strains were grown together and only when both donor and recipient cells carried a galE::Tn10 mutation interfering with LPS O-polysaccharide biosynthesis. Previously we observed that neither AgfA nor AgfB was freely diffusible in S. enterica serovar Enteritidis 3b (46), unlike CsgA and CsgB in E. coli MC4100 (20). In E. coli MC4100, CsgA subunits secreted by ΔcsgB colonies readily polymerize onto the cell surface of adjacent cells in ΔcsgA colonies, resulting in an increase in CR binding (20). Consistent with our findings, E. coli MC4100 is a K-12 derivative and does not produce LPS O polysaccharide (27).
It is apparent, then, that an intact LPS integument physically precludes access of exogenous AgfA monomers to the nucleating activity of AgfB at the recipient outer membrane surface. When galE+ ΔagfA recipient cells were used, AgfA subunits secreted from adjacent ΔagfB cells were unable to polymerize into intact fimbriae. All ΔagfB donor strains were able to secrete extracellular AgfA, although AgfA subunits were somewhat restricted from diffusing away from the ΔagfB cell surface when galE+ ΔagfB donor cells were used, presumably due to the LPS barrier effect, but this barrier appears to be less restrictive toward AgfA exit than toward AgfA entrance. Introduction of the galE mutation did not affect fimbrial formation or subsequent CR binding in wild-type agfBA+ cells. In addition, all agfBA+ strains had aggregation characteristics similar to those of the S. enterica serovar Enteritidis 3b parent. These findings seem to question the role of the fimbrial assembly pathway with respect to the significance of intercellular participation in fimbrial assembly with w+ cells, colonies, and biofilms. This is particularly evident with w+ S. enterica serovar Enteritidis 3b, where there is normally no escape of soluble AgfA monomers to adjacent cells and no assembly into intact fimbriae even when AgfA escape is forced via an AgfB deficiency, unless, of course, access is permitted through a disruption in the LPS integument. Thus, the nucleation-precipitation pathway (20) for assembly of these fimbriae would a priori be restricted to operating principally at the immediate cell surface for each w+ cell. The data here seem to confirm AgfB-mediated assembly of AgfA monomers at the cell surface. However, this raises the intriguing question as to how Tafi or curli then undergo entropically driven assembly at the distal fimbrial ends, as envisioned in the original model, considerably distant from the LPS integument in the apparent absence of extracellular soluble AgfA monomers.
Interestingly, the presence of cellulose did not interfere with intercellular Tafi formation. This was surprising, since cellulose is produced at the cell surface (34) and forms part of an extracellular matrix with Tafi (39, 48). Although Tafi and cellulose are coregulated under most conditions (39, 48), this could reflect differences in temporal production.
The appearance of Tafi, viewed by electron microscopy, changed drastically when cellulose was not produced. In S. enterica serovar Enteritidis 3b (bcsA+), Tafi fibers appeared as a tangled amorphous matrix, as reported previously (12, 36). The ultrastructure of the fibers was not discernible at higher magnifications. In comparison, individual Tafi fibers formed by S. enterica serovar Enteritidis 3b ΔbcsA were easily resolved and had a distinctive curling appearance. At higher magnifications, the ultrastructure was more distinguishable and what appeared to be branch points were observed. These observations help explain other differences between S. enterica serovar Enteritidis 3b and E. coli MC4100. While S. enterica serovar Enteritidis Tafi fibers were unresolved and sometimes linear in appearance (12, 45), E. coli curli fibers appeared to be clearly defined with a curling appearance (9, 20). It can be safely assumed that these differences were due to the presence and absence of cellulose, respectively. It was recently demonstrated that S. enterica serovar Enteritidis 3b produces cellulose, while E. coli MC4100 does not (48). These observations indicate that cellulose forms a very tight association with Tafi in S. enterica serovar Enteritidis 3b.
We have detected another component as part of the extracellular matrix formed by Tafi and cellulose in S. enterica serovar Enteritidis 3b. This component migrates as a high-MW smear on SDS-PAGE and is recognized by immune serum raised to purified Tafi. In all prior analyses, it was assumed that the immunoreactive high-MW material represented different levels of Tafi depolymerization. In this study, by comparing cellulose-deficient and -sufficient S. enterica serovar Enteritidis strains on Western blots, it was apparent that this high-MW material was quite distinct from AgfA, AgfB, and cellulose. However, cellulose was required to tightly link this material to cells. As confirmation of this, purification of Tafi without this material was successful in a ΔbcsA strain. Preliminary analysis showed that the high-MW material is not proteinaceous and is slightly anionic. The material does not appear to be CA, since it is still produced in an S. enterica serovar Enteritidis ΔwcaJ mutant strain.
Cross-reactive high-MW material was also detected in S. enterica serovar Enteritidis, S. enterica serovar Typhimurium, and E. coli. The high-MW material was detected in S. enterica serovar Typhimurium SL7207 under conditions in which Tafi and presumably cellulose were not produced. This indicates that production of the unknown material is not regulated by AgfD. However, extracellular polysaccharide production is often induced in the presence of excess carbon when nitrogen or phosphates are limiting (43), conditions in which agfD transcription is also up-regulated (17). We hypothesize that the high-MW material represents an uncharacterized EPS that is sometimes produced in association with Tafi and cellulose. Structural characterization to test this hypothesis is in progress.
We have examined the extracellular components associated with Tafi in order to understand the native state of Tafi (curli) found on Salmonella and E. coli cell surfaces. The genes coding for Tafi (curli) and cellulose are highly conserved between Salmonella spp. and E. coli and have been detected in most strains tested so far (5, 15, 34, 41, 42). Production of these components is more limited, to between 60 and 70% of strains tested (13, 15, 18, 30, 34, 39). Production of Tafi (curli) and cellulose has not been directly linked to virulence. Highly invasive E. coli strains do not appear to produce curli (30, 38), and csg genes coding for E. coli curli are almost universally disrupted in closely related Shigella spp. (38). This suggests there is a selection pressure against Tafi (curli) production during host invasion, especially since the majority of Tafi (curli)-expressing strains produce fimbriae only at temperatures of below 30°C (15, 30, 37). Cellulose-deficient mutants of S. enterica serovar Enteritidis showed no difference in virulence in both the BALB/c mouse and 1-day-old chick infection models (39). Again, there is evidence of selective pressure against cellulose production in highly invasive strains (34).
Tafi and curli do not exist alone as protein fibers under normal conditions. In association with cellulose, they are thought to form an important cellular coating associated with biofilm formation and cell-cell attachment (4, 33, 36, 39), which may be analogous to the aggregative physiological state, the Rugose phenotype (2). Production of a more complex matrix consisting of Tafi, cellulose, and EPS would seem to be particularly advantageous for environmental persistence of Salmonella spp. and may even aid in survival in the mammalian gut. Cellulose and EPS have been shown to protect cells against acid and heat (29) and chlorine treatment (39) and presumably would prevent desiccation of cells, aid in nutrient trapping, and contribute to buffering (29, 43). Thus, the evidence presented here suggests that Tafi are an integral component of a stable and protective, complex protein-polysaccharide matrix.
Acknowledgments
We thank Ian Sutherland for the generous gift of purified CA samples from E. coli and Enterobacter cloacae and for helpful discussions. We also thank R. Beecroft and T. Otto for production of AgfA-specific monoclonal antibodies, J. Halverson and C. Rajotte for excellent technical assistance, and E. E. Ishiguro, C. Whitfield, A. Boraston, and W. Kim for helpful discussions.
Funding for this work was provided by grants to W. W. Kay from the National Sciences and Engineering Research Council and the Canadian Bacterial Diseases Network.
REFERENCES
- 1.Aguilar, A., S. Merino, M. M. Nogueras, M. Regue, and J. M. Tomas. 1999. Two genes from the capsule of Aeromonas hydrophila (serogroup O:34) confer serum resistance to Escherichia coli K12 strains. Res. Microbiol. 150:395-402. [DOI] [PubMed] [Google Scholar]
- 2.Anriany, Y. A., R. M. Weiner, J. A. Johnson, C. E. De Rezende, and S. W. Joseph. 2001. Salmonella enterica serovar Typhimurium DT104 displays a rugose phenotype. Appl. Environ. Microbiol. 67:4048-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnqvist, A., A. Olsen, J. Pfeifer, D. G. Russell, and S. Normark. 1992. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol. Microbiol. 6:2443-2452. [DOI] [PubMed] [Google Scholar]
- 4.Austin, J. W., G. Sanders, W. W. Kay, and S. K. Collinson. 1998. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 162:295-301. [DOI] [PubMed] [Google Scholar]
- 5.Baumler, A. J., A. J. Gilde, R. M. Tsolis, A. W. van der Velden, B. M. Ahmer, and F. Heffron. 1997. Contribution of horizontal gene transfer and deletion events to development of distinctive patterns of fimbrial operons during evolution of Salmonella serotypes. J. Bacteriol. 179:317-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bian, Z., A. Brauner, Y. Li, and S. Normark. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 181:602-612. [DOI] [PubMed] [Google Scholar]
- 7.Bian, Z., and S. Normark. 1997. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 16:5827-5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, R. K., D. Botstein, T. Watanabe, and Y. Ogata. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology 50:883-898. [DOI] [PubMed] [Google Scholar]
- 9.Chapman, M. R., L. S. Robinson, J. S. Pinkner, R. Roth, J. Heuser, M. Hammar, S. Normark, and S. J. Hultgren. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collinson, S. K., S. C. Clouthier, J. L. Doran, P. A. Banser, and W. W. Kay. 1996. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J. Bacteriol. 178:662-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collinson, S. K., P. C. Doig, J. L. Doran, S. Clouthier, T. J. Trust, and W. W. Kay. 1993. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J. Bacteriol. 175:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collinson, S. K., L. Emody, K. H. Muller, T. J. Trust, and W. W. Kay. 1991. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173:4773-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collinson, S. K., L. Emody, T. J. Trust, and W. W. Kay. 1992. Thin aggregative fimbriae from diarrheagenic Escherichia coli. J. Bacteriol. 174:4490-4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doran, J. L., S. K. Collinson, J. Burian, G. Sarlos, E. C. Todd, C. K. Munro, C. M. Kay, P. A. Banser, P. I. Peterkin, and W. W. Kay. 1993. DNA-based diagnostic tests for Salmonella species targeting agfA, the structural gene for thin, aggregative fimbriae. J. Clin. Microbiol. 31:2263-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garegg, P. J., B. Lindberg, T. Onn, and I. W. Sutherland. 1971. Comparative structural studies on the M-antigen from Salmonella typhimurium, Escherichia coli and Aerobacter cloacae. Acta Chem. Scand. 25:2103-2108. [DOI] [PubMed] [Google Scholar]
- 17.Gerstel, U., and U. Romling. 2001. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ. Microbiol. 3:638-648. [DOI] [PubMed] [Google Scholar]
- 18.Grant, W. D., I. W. Sutherland, and J. F. Wilkinson. 1969. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. J. Bacteriol. 100:1187-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammar, M., A. Arnqvist, Z. Bian, A. Olsen, and S. Normark. 1995. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661-670. [DOI] [PubMed] [Google Scholar]
- 20.Hammar, M., Z. Bian, and S. Normark. 1996. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:6562-6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashimoto-Gotoh, T., F. C. Franklin, A. Nordheim, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene 16:227-235. [DOI] [PubMed] [Google Scholar]
- 22.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 23.Hone, D., R. Morona, S. Attridge, and J. Hackett. 1987. Construction of defined galE mutants of Salmonella for use as vaccines. J. Infect. Dis. 156:167-174. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Junkins, A. D., and M. P. Doyle. 1992. Demonstration of exopolysaccharide production by enterohemorrhagic Escherichia coli. Curr. Microbiol. 25:9-17. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Liu, D., and P. R. Reeves. 1994. Escherichia coli K12 regains its O antigen. Microbiology 140:49-57. [DOI] [PubMed] [Google Scholar]
- 28.Loferer, H., M. Hammar, and S. Normark. 1997. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol. Microbiol. 26:11-23. [DOI] [PubMed] [Google Scholar]
- 29.Mao, Y., M. P. Doyle, and J. Chen. 2001. Insertion mutagenesis of wca reduces acid and heat tolerance of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:3811-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsen, A., A. Arnqvist, M. Hammar, S. Sukupolvi, and S. Normark. 1993. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol. Microbiol. 7:523-536. [DOI] [PubMed] [Google Scholar]
- 31.Olsen, A., H. Herwald, M. Wikstrom, K. Persson, E. Mattsson, and L. Bjorck. 2002. Identification of two protein-binding and functional regions of curli, a surface organelle and virulence determinant of Escherichia coli. J. Biol. Chem. 277:34568-34572. [DOI] [PubMed] [Google Scholar]
- 32.Prigent-Combaret, C., E. Brombacher, O. Vidal, A. Ambert, P. Lejeune, P. Landini, and C. Dorel. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183:7213-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450-464. [DOI] [PubMed] [Google Scholar]
- 34.Romling, U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153:205-212. [DOI] [PubMed] [Google Scholar]
- 35.Romling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romling, U., M. Rohde, A. Olsen, S. Normark, and J. Reinkoster. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10-23. [DOI] [PubMed] [Google Scholar]
- 37.Romling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 38.Sakellaris, H., N. K. Hannink, K. Rajakumar, D. Bulach, M. Hunt, C. Sasakawa, and B. Adler. 2000. Curli loci of Shigella spp. Infect. Immun. 68:3780-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 40.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson, G., R. Lan, and P. R. Reeves. 2000. The colanic acid gene cluster of Salmonella enterica has a complex history. FEMS Microbiol. Lett. 191:11-16. [DOI] [PubMed] [Google Scholar]
- 43.Sutherland, I. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3-9. [DOI] [PubMed] [Google Scholar]
- 44.Uhlich, G. A., J. E. Keen, and R. O. Elder. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:2367-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White, A. P., S. K. Collinson, P. A. Banser, D. J. Dolhaine, and W. W. Kay. 2000. Salmonella enteritidis fimbriae displaying a heterologous epitope reveal a uniquely flexible structure and assembly mechanism. J. Mol. Biol. 296:361-372. [DOI] [PubMed] [Google Scholar]
- 46.White, A. P., S. K. Collinson, P. A. Banser, D. L. Gibson, M. Paetzel, N. C. Strynadka, and W. W. Kay. 2001. Structure and characterization of AgfB from Salmonella enteritidis thin aggregative fimbriae. J. Mol. Biol. 311:735-749. [DOI] [PubMed] [Google Scholar]
- 47.White, A. P., S. K. Collinson, J. Burian, S. C. Clouthier, P. A. Banser, and W. W. Kay. 1999. High efficiency gene replacement in Salmonella enteritidis: chimeric fimbrins containing a T-cell epitope from Leishmania major. Vaccine 17:2150-2161. [DOI] [PubMed] [Google Scholar]
- 48.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]