Abstract
The Irp9 protein of Yersinia enterocolitica participates in the synthesis of salicylate, the precursor of the siderophore yersiniabactin. In Pseudomonas species, salicylate synthesis is mediated by two enzymes: isochorismate synthase and isochorismate pyruvate-lyase. Both enzymes are required for complementation of a Yersinia irp9 mutant. However, irp9 is not able to complement Escherichia coli entC for the production of enterobactin, which requires isochorismate as a precursor. These results suggest that Irp9 directly converts chorismate into salicylate.
The pathogenicity of Yersinia pestis (causing bubonic plague), Yersinia pseudotuberculosis, and Yersinia enterocolitica biogroup 1B (enteropathogens) is determined by a common virulence plasmid (pYV) and a high-pathogenicity island (HPI) which is inserted into the tRNA gene asnT (1, 3, 5, 20). The HPI of Y. enterocolitica carries two gene clusters, the cluster irp2, irp1, irp3, irp4, and irp5 and the cluster irp6, irp7, irp8, and irp9 (Fig. 1a), which are involved in the biosynthesis and transport of the siderophore yersiniabactin (Ybt), and the fyuA gene, which encodes the outer membrane receptor FyuA for Ybt (17, 20). Targeted disruption of irp genes (excluding irp8, whose function is unknown) results in an attenuation of virulence (1, 2, 6). Previously, it has been demonstrated that salicylate is the precursor of Ybt biosynthesis and that the irp9 gene homolog of Y. pestis ybtS is required for synthesis of this precursor (11).
FIG. 1.
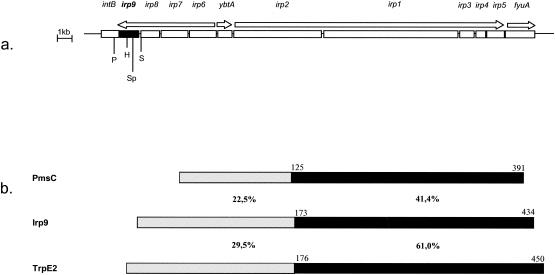
(a) Structure of the HPI core in Y. enterocolitica. Bars indicate genes, and arrows indicate gene clusters and the direction of transcription (20). intB, gene for P4-like integrase (see also text); H, HincII; P, PstI; Sp, SphI; S, SalI. (b) Alignment of chorismate binding proteins PmsC (ICS of P. fluorescens), Irp9 (Y. enterocolitica), and TrpE2/MtbI (putative anthranilate synthase of M. tuberculosis). Black bars indicate the regions of high homology comprising the predicted chorismate binding sites. Similarities for Irp9/PmsC and Irp9/TrpE2 are indicated.
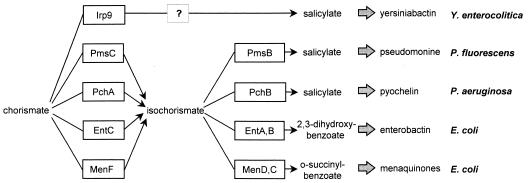
The predicted amino acid sequence of Irp9/YbtS demonstrates a close similarity to the sequence of anthranilate synthase component I (TrpE of Enterobacteriaceae and TrpE2/MbtI of Mycobacterium tuberculosis) and a lower degree of similarity to the sequences of the isochorismate synthases (ICSs) of Pseudomonas aeruginosa (PchA), Pseudomonas fluorescens (PmsC), and Escherichia coli (EntC) (11) (Fig. 1b). The chorismatebinding domain is localized between amino acids 173 and 428 (according to an NCBI conserved domain search). The anthranilate synthase converts chorismate to anthranilate (the amino analog of salicylate) by using glutamine as the nitrogen source. ICSs convert chorismate into isochorismate, which is needed for the synthesis of the siderophores pseudomonine (P. fluorescens) (13), pyochelin (P. aeruginosa) (22), and enterobactin (E. coli) (16) (Fig. 2). The precursor of Ybt, pyochelin, and pseudomonine is salicylate, which can be generated in a second step from isochorismate by isochorismate pyruvate lyases (IPLs) (Fig. 2). P. aeruginosa and P. fluorescens carry pchB and pmsB, respectively, which have been shown to encode IPL (9, 13, 22). A similar pathway has been described previously for the salicylate biosynthesis (the precursor of mycobactin) of M. tuberculosis (7, 18). On the other hand, it has been suggested that TrpE2/MbtI may directly convert chorismate into salicylate and thus may function like anthranilate synthase (7, 18).
FIG. 2.
Pathways of siderophore and menaquinone synthesis in Y. enterocolitica, E. coli, P. fluorescens, and P. aeruginosa. MenF (E. coli), EntC (E. coli), PmsC (P. fluorescens) and PchA (P. aeruginosa) are ICSs which convert chorismate into salicylate. In E. coli, isochorismate is converted into o-succinylbenzoate by MenD and MenC and into 2,3-dihydroxybenzoate by EntA and EntB. Pseudomonas converts isochorismate into salicylate by PmsB/PchB. Irp9 shows similarities to ICSs but shows a higher degree of similarity to anthranilate synthase (see Fig. 1b).
Strikingly, YbtS/Irp9 does not carry related sequences to PchB or PmsB, which suggests that there is an IPL-encoding gene outside of the HPI on the chromosome. However, sequence analysis of the available genome sequence of Y. pestis or Y. enterocolitica (http://www.sanger.ac.uk/Projects/Y_pestis and http://www.sanger.ac.uk/Projects/Y_enterocolitica) failed to identify a PchB/PmsB homolog. Therefore, the question whether YbtS/Irp9 is a bifunctional enzyme with ICS and IPL activity (two-step salicylate synthesis) or converts chorismate directly into salicylate remains open.
To address this issue, we disrupted the irp9 gene of Y. enterocolitica and introduced pmsC and pmsB of P. fluorescens into the irp9 mutant for restoration of Ybt biosynthesis. Moreover, for restoration of ICS activity or salicylate production, we introduced irp9, pmsC, and pmsB into an E. coli entC mutant. Our results support the assumption that Irp9/YbtS functions as salicylate synthase by converting chorismate directly into salicylate.
Inactivation of irp9 in Y. enterocolitica leads to a chrome azurol S (CAS)-negative phenotype.
For a first approach, we disrupted the irp9 gene of Y. enterocolitica O:8 strain WA-CS. A 580-bp HpaI/SalI fragment from cosmid 12H2 (2, 19) (Table 1) harboring a SphI site in gene irp9 (Fig. 1a) was ligated into the EcoRV/SalI site of plasmid pKS (Stratagene). A HindII-cut kanamycin cassette from plasmid pSB315, which lacks a transcriptional terminator (10), was inserted into the blunted SphI site of irp9. The fragment carrying the kanamycin cassette was inserted into suicide vector pKAS32 (24) by means of the KpnI and SacI sites of the pKS vector. The resulting construct, pSVIrp9, was transformed into S17-1 λ pir+ tra+ (14, 23) and mobilized into WA-CS. Mutants were selected on agar plates containing kanamycin (40 μg/ml), streptomycin (100 μg/ml), and nalidixin (100 μg/ml), and results were confirmed by Southern blotting.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype and/or phenotype | Reference or source |
|---|---|---|
| Y. enterocolitica | ||
| WA-C | Derivative of Y. enterocolitica, BG1B, serotype O:8, WA-314, pYV plasmid cured; Nalr | 12 |
| WA-CS | Sm resistant derivative of WA-C; Nalr Smr | 17 |
| WA-CSirp1::Kanr | irp1 mutant of WA-CS, Ybt deficient strain; Smr Kanr Nalr | 17 |
| WA-CSirp9::Kanr | WA-CS carrying a kanamycin cassette without transcriptional terminator in gene irp9; Smr Kanr Nalr | This study |
| WA-CSG | WA-CS carrying reporter construct pCJFY5G3; Nalr Cmr | This study |
| WA-CSirp1::KanrG | WA-CSirp1::Kanr carrying reporter construct pCJFY5G3; Smr Kanr Nalr Cmr | This study |
| WA-CSirp9::KanrG | WA-CSirp9::Kanr carrying reporter construct pCJFY5G3; Smr Kanr Nalr Cmr | This study |
| E. coli | ||
| S17-1 λpir | pir+tra+ | 14, 23 |
| PBB7 | entC mutant, enterobactin deficient, CAS-negative phenotype | 15 |
| BL21(DE3) | Enterobactin positive carrying IPTG-inducible T7 polymerase gene | Stratagene |
| Plasmids | ||
| pSB315 | Containing kanamycin cassette without transcriptional terminator; Ampr Kanr | 10 |
| 12H2 | pLAFR2 carrying a DNA insert of WA-C comprising irp6-9 operon, ybtA, and irp2; Tcr | 2, 19 |
| pKAS32 | Suicide vector with rpsL gene; Ampr | 24 |
| pSVIrp9 | pKAS32 carrying a SalI/HpaI DNA-fragment of the HPI with, a kanamycin cassette (without transcriptional terminator) integrated into the SphI site of irp9; Ampr Kanr | This study |
| pGP1-2 | Vector containing heat-inducible T7 polymerase; Kanr | 25 |
| pT7-5 | Cloning vector; Ampr | 25 |
| pTlrp9 | pT7-5 vector carrying irp9 (SalI/PstI fragment of cosmid 12H2) upstream of T7 promoter; Ampr | This study |
| pCJFY5G3 | Reporter construct, pACYC184 carrying 153 bp of fyuA promoter region fused to gfp:Cmr | 2 |
| pE3R | pGEM-3Z carrying the pmsCEAB operon of P. fluorescens WCS374; Ampr | 13 |
| pE3 | pGEM-3Z carrying the T7 promoter upstream of the pmsCEAB operon of P. fluorescens WCS374; Ampr | 13 |
| pPmsCB | pGEM-3Z carrying pmsC and pmsB genes (ApaI/SacII fragment of pE3R) of P. fluorescens WCS374; Ampr | This study |
| pPmsC | pGEM-3Z carrying pmsC gene (ApaI/NdeI fragment of pE3R) of P. fluorescens WCS374; Ampr | This study |
| pPmsB | pGEM-3Z carrying pmsB gene (StuI/SacII fragment of pE3R) of P. fluorescens WCS374; Ampr | This study |
| pUC57 | Cloning vector; Ampr | MBI Fermentas |
| pUCEntC | pUC57 carrying entC of E. coli; Ampr | This study |
The resulting mutant strain, WA-CSirp9::Kanr, was tested on CAS agar, a siderophore indicator. The CAS assay relies on the color change from a green-blue CAS-iron complex to orange desferrated CAS around siderophore-producing colonies (producing a CAS halo, or CAS positive) (12, 21). WA-CSirp9::Kanr expressed no CAS halo, indicating the loss of Ybt synthesis. The CAS-positive phenotype was restored after the introduction of pTIrp9 (Table 2). The plasmid harbors the 2.8-kb SalI/PstI fragment of cosmid 12H2 carrying irp9 downstream of the T7 promoter in the pT7-5 backbone (Table 1). Complementation of WA-CSirp9::Kanr was possible without plasmid pGP1-2 (carrying the T7 polymerase), indicating that there was sufficient expression of the irp9 gene for Ybt biosynthesis even in the absence of the T7 polymerase.
TABLE 2.
Ybt production of WA-CSirp9::Kanr carrying different plasmids as determined by CAS agar or a cross-inducer reporter assay
| Strain | Plasmid | CAS phenotype | Fluorescence intensity of reporter strain WA-CSirp1::KanrGa |
|---|---|---|---|
| WA-CS | + | 42 ± 14 | |
| WA-CSirp9::Kanr | − | 6.5 ± 0.8 | |
| pTlrp9 | + | NDb | |
| pPmsCB | + | 37 ± 7 | |
| pPmsC | − | 8 ± 1 | |
| pPmsB | − | 25 ± 5 |
Arbitrary units (au) of fluorescence intensities and standard deviations of three experiments. Note that NBD induces 5.5 ± 0.5 au (background level).
ND, not determined.
Salicylate feeding restores Ybt production of an irp9 mutant.
Assuming that Irp9 converts chorismate into salicylate, we examined Ybt production of WA-CSirp9::Kanr after feeding the bacteria with chorismate or salicylate by a Ybt cross-inducer reporter assay and the conventional cross-feeding assay (2, 17). The cross-inducer reporter assay is based on the observation that Ybt-containing culture supernatant is able to induce the expression of the fyuA gene in yersinia mutants with disrupted Ybt biosynthesis genes (e.g., irp1 or irp9). Reporter strains WA-CSirp9::KanrG and WA-CSirp1::KanrG carrying plasmid pCJFY5G3 (translational fusion between a fyuA promoter and green fluorescent protein [GFP] reporter gene gfp) (Table 1) show enhanced GFP fluorescence under iron limitation in response to Ybt-containing culture supernatants of overnight cultures of tester strains. Fluorescence intensity was determined by cytofluorometry (fluorescence-activated cell sorting) on a single-cell level for 12,000 bacteria after 24 h of growth at 28°C in stoppered vessels (2). The culture supernatants of WA-CSirp9::Kanr grown overnight in (i) iron-limited nutrient broth (NBD; negative control), (ii) NBD plus chorismate, (iii) NBD plus salicylate, and (iv) the supernatant of WA-CS grown in NBD (positive control) were examined for the presence of Ybt. The supernatants were collected, sterile filtered, mixed with fresh NBD medium (1:3, vol/vol), and inoculated with overnight cultures of the indicated reporter strains in nutrient broth (1:50, vol/vol).
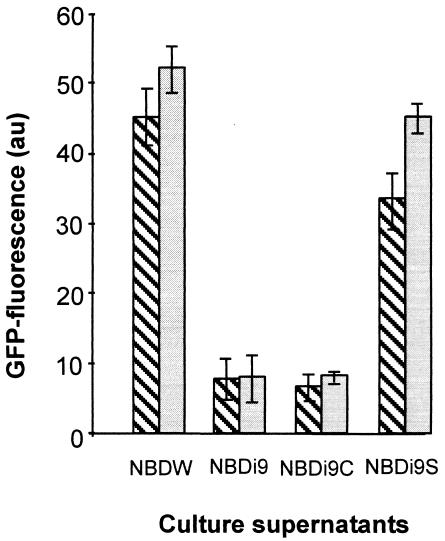
As expected, the supernatant of WA-CSirp9::Kanr cultured in NBD plus salicylate was nearly as efficient as the WA-CS supernatant in inducing the fyuA-gfp reporter gene of the irp9 and irp1 mutants, indicating the presence of Ybt (Fig. 3). The supernatant collected from an irp9 mutant grown in NBD or NBD plus chorismate showed no significant Ybt-inducing effect.
FIG. 3.
Ybt cross-inducer assay performed with two reporter strains. Reporter strains (WA-CSirp1::KanrG [hatched bars] and WA-CSirp9::KanrG [shaded bars]) were grown in NBD medium containing (i) sterile filtered supernatant of WA-CS grown in NBD (NBDW), (ii) WA-CSirp9::Kanr grown in NBD (NBDi9), (iii) WA-CSirp9::Kanr grown in NBD plus 300 μM chorismate (NBDi9C), and (iv) WA-CSirp9::Kanr grown in NBD plus 300 μM salicylate (NBDi9S). Bars represent GFP fluorescence of the strains. Standard deviations (for three experiments) are indicated by error bars. au, arbitrary units.
A cross-feeding assay confirmed the results of cross-induction (results not shown). Filter tips soaked with 12 μl of NBD containing 3 μM purified Ybt, 300 μM chorismate, 300 μM salicylate, or tester culture supernatants (see above) were placed on the agar layer. For the indicator strain, we used WA-CSirp1::Kanr, which grows only poorly in iron-limited CDM-H (chemically defined medium with Hefe [0.25% yeast extract] and containing 40 μM EDDHA [ethylenediamine di-o-hydroxy-phenylacetic acid], an iron chelator) (8). The indicator strain showed no growth in the presence of salicylate or chorismate, unlike in the presence of Ybt. Tips soaked with sterile filtered supernatants of the irp9 mutant grown in (i) NBD, (ii) NBD containing chorismate, and (iii) NBD containing salicylate revealed growth of the seeded indicator strain only around the tip that had been soaked with the supernatant of WA-CSirp9::Kanr grown in NBD with the addition of salicylate; this result indicates that Ybt synthesis of the irp9 mutant is salicylate dependent, as has been demonstrated with the cross-inducer assay.
In summary, these results clearly demonstrate that irp9 inactivation may be complemented by the addition of salicylate in the culture medium, which confirms that Irp9 is involved in salicylate synthesis.
Both pmsC and pmsB are required for complementation of WA-CSirp9::Kanr.
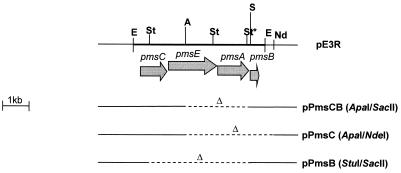
Irp9 has sequences homologous to the sequences of ICSs (Fig. 1b), but neither irp9 nor any other gene of the HPI encodes a protein with significant homology to PchB or PmsB, the IPLs of the Pseudomonas species. This raises the question whether Irp9 functions as ICS in cooperation with an IPL-encoding gene unrelated to pchB or pmsB elsewhere on the chromosome. As a first step, we subcloned the genes pmsC and pmsB as ApaI/SacII fragments of pE3R (carrying the pmsCEAB operon of P. fluorescens) (Fig. 4). Blunt ends were generated with T4 DNA polymerase (MBI Fermentas) and religated. The resulting plasmid, pPmsCB, restored the CAS-positive phenotype of WA-CSirp9::Kanr (Table 2). In contrast, WA-CSirp9::Kanr carrying only pmsC (pPmsC, cut with ApaI/NdeI) or pmsB (pPmsB, cut with StuI/SacII) did not produce a halo on CAS agar. As CAS agar might not be sensitive enough for detection of low Ybt concentrations, we analyzed the NBD supernatants of WA-CSirp9::Kanr carrying plasmid pPmsCB, pPmsB, or pPmsC for the presence of Ybt by using the WA-CSirp1:KanrG reporter strain (Ybt cross-inducer assay). As expected, a strong induction of the fyuA-gfp reporter gene was detected after the addition of the supernatant from WA-CSirp9::Kanr(pPmsCB) cultivated in NBD medium (which was comparable to the supernatant of the parental strain WA-CS) (Table 2). However, a significant induction of fyuA-gfp was also detected after the addition of the culture supernatant from WA-CSirp9::Kanr(pPmsB), although plasmid pPmsB expressing an IPL cannot restore the CAS-positive phenotype of the irp9 mutant. These results indicate that both the pmsC and pmsB genes are necessary for the restoration of the CAS-positive phenotype of the irp9 mutant but that pmsB is sufficient to partially restore Ybt production under liquid culture conditions (in stoppered vessels).
FIG. 4.
Plasmid pE3R carrying the pmsCEAB operon of P. fluorescens. Plasmid pE3R was cut with different restriction enzymes and religated. E, EcoRI; A, ApaI; S, SacII; St, StuI; St*, methylated StuI cutting site. Arrows indicate genes and the direction of transcription. Dashed lines denote generated deletions. The resulting plasmids were designated pPmsCB (cut with ApaI/SacII, harboring pmsCB), pPmsC (cut with ApaI/NdeI, harboring pmsC), and pPmsB (cut with StuI/SacII, harboring pmsB).
These apparently contrary results suggest that growth in liquid culture results in a level of isochorismate production sufficient for conversion by PmsB to salicylate and subsequently to Ybt. Is there a plausible explanation for this? First, we have to consider that yersiniae grow aerobically on CAS agar but microaerobically in the stoppered liquid culture vessels used in the cross-inducer assay. Second, for E. coli it is known that about 2% of the isochorismate produced by MenF (an isochorismate synthase of the menaquinone pathway, expressed predominantly under anaerobic conditions) (Fig. 2) can channel into the enterobactin pathway (4, 26). As Y. enterocolitica harbors a gene encoding a protein with 66% similarity to MenF of E. coli (http://www.sanger.ac.uk/Projects/Y_enterocolitica), it is very likely that a sufficient level of isochorismate is produced by WA-CSirp9::Kanr(pPmsB) in liquid culture and is then converted to salicylate by PmsB and subsequently to Ybt.
irp9 leads to salicylate synthesis in an E. coli entC mutant.
To differentiate between one-step and two-step conversion of chorismate into salicylate by Irp9, we introduced pmsC, pmsCEAB of P. fluorescens, irp9 of Y. enterocolitica, and, as a control, entC of E. coli into the E. coli entC mutant PBB7. The entC gene and its promoter region were amplified by using primers EntC70 (AATCCGTCCCCTCGCCTTTG) and EntC1637 (TGCGTCAGAATGTCGGTCAG). The PCR product was subcloned into vector pUC57 (MBI Fermentas), yielding pUCEntC (Table 1). As a first step, the resulting transformants were tested for enterobactin synthesis on CAS agar (Table 3). As expected, introduction of entC (pUCEntC) restored siderophore production (CAS positive) in PBB7. Transfer of pPmsC led also to a CAS-positive phenotype of E. coli entC, indicating that entC and pmsC are interchangeable. In contrast, transfer of pTIrp9 did not restore siderophore production in PBB7 even though pGP1-2 (carrying the T7 polymerase) was present and induced (30 min at 42°C). Obviously, Irp9 does not produce isochorismate as an available intermediate for enterobactin biosynthesis in the E. coli entC mutant.
TABLE 3.
Salicylate synthesis and enterobactin production (CAS agar) of strain E. coli BL21(DE3) and E. coli entC mutant PBB7(pGP1-2) carrying different plasmids
| Strain | Plasmid | Salicylate concn (mean ± SD) (μM)a | CAS phenotype |
|---|---|---|---|
| BL21 (DE3) | pT7-5 | 0 | + |
| pPmsB | 114 ± 24 | NDb | |
| pTIrp9 | 439 ± 81 | ND | |
| PBB7(pGP1-2) | pT7-5 | 0 | − |
| pE3 | 34 ± 7 | ND | |
| pPmsC | ND | + | |
| pUCEntC | ND | + | |
| pTIrp9 | 57 ± 13 | − |
Results are means for three experiments.
ND, not determined.
To verify Irp9-mediated salicylate synthesis in E. coli strains, the plasmids pT7-5 (negative control), pE3 or pPmsB (positive control), and pTIrp9 were transferred into PBB7 (pGP1-2) and into the enterobactin-positive E. coli strain BL21(DE3), as indicated in Table 3. After induction at an optical density of 0.5 [PBB7 (pGP1-2) at 42°C for 30 min and BL21(DE3) in 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside)], strains were grown for 15 h at 37°C. Salicylate was detected as described by Mercado-Blanco et al. (13). The purple iron-salicylate complex in the aqueous phase was quantified photometrically at 527 nm. PBB7 and BL21(DE3) produced detectable amounts of salicylate when harboring pTIrp9 (Table 3). Similar results were obtained with PBB7 carrying pE3 and BL21(DE3) carrying pPmsB. Thus, E. coli becomes a salicylate producer after receiving the corresponding Yersinia or Pseudomonas genes.
In conclusion, these results demonstrate that Yersinia Irp9 functions as a salicylate synthase by the conversion of chorismate into salicylate (probably in one step), in contrast to Pseudomonas species, which require the two enzymes ICS and IPL.
Considering the high degree of similarity between Irp9 and TrpE2/MbtI and that both corresponding genes are located within the gene cluster of yersiniabactin and mycobactin biosynthesis, respectively, we suggest that Irp9 and TrpE2/MbtI function similarly to anthranilate synthase component I by channelling a hydroxyl residue to the active site for salicylate production instead of an amino residue for anthranilate production.
Acknowledgments
Daniela Brem and Cosima Pelludat contributed equally to this work.
We are indebted to Peter Bakker and Eckhard Leistner for kindly providing plasmids (pE3 and pE3R) and strain PBB7, respectively, and to Alexander Rakin for the alignment of predicted amino acid sequences.
This study was supported by a grant (HE1297/8) from the Deutsche Forschungsgemeinschaft to J.H.
REFERENCES
- 1.Bearden, S. W., J. D. Fetherston, and R. D. Perry. 1997. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brem, D., C. Pelludat, A. Rakin, C. A. Jacobi, and J. Heesemann. 2001. Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica. Microbiology 147:1115-1127. [DOI] [PubMed] [Google Scholar]
- 3.Buchrieser, C., R. Brosch, S. Bach, A. Guiyoule, and E. Carniel. 1998. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol. Microbiol. 30:965-978. [DOI] [PubMed] [Google Scholar]
- 4.Buss, K., R. Muller, C. Dahm, N. Gaitatzis, E. Skrzypczak-Pietraszek, S. Lohmann, M. Gassen, and E. Leistner. 2001. Clustering of isochorismate synthase genes menF and entC and channeling of isochorismate in Escherichia coli. Biochim. Biophys. Acta 1522:151-157. [DOI] [PubMed] [Google Scholar]
- 5.Carniel, E., I. Guilvout, and M. Prentice. 1996. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J. Bacteriol. 178:6743-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carniel, E., A. Guiyoule, I. Guilvout, and O. Mercereau-Puijalon. 1992. Molecular cloning, iron-regulation and mutagenesis of the irp2 gene encoding HMWP2, a protein specific for the highly pathogenic Yersinia. Mol. Microbiol. 6:379-388. [DOI] [PubMed] [Google Scholar]
- 7.De Voss, J. J., K. Rutter, B. G. Schroeder, and C. E. Barry III. 1999. Iron acquisition and metabolism by mycobacteria. J. Bacteriol. 181:4443-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flossmann, K. D., C. Grajetzki, and H. Rosner. 1985. Nachweis von Eisen-Transport-Aktivität in Pasteurella multocida-Kulturen. J. Basic Microbiol. 25:559-567. [DOI] [PubMed] [Google Scholar]
- 9.Gaille, C., P. Kast, and D. Haas. 2002. Salicylate biosynthesis in Pseudomonas aeruginosa: purification and characterization of PchB, a novel bifunctional enzyme displaying isochorismate pyruvate-lyase and chorismate mutase activities. J. Biol. Chem. 277:21768-21775. [DOI] [PubMed] [Google Scholar]
- 10.Galán, J. E., C. Ginocchio, and P. Costeas. 1992. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J. Bacteriol. 174:4338-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gehring, A. M., E. DeMoll, J. D. Fetherston, I. Mori, G. F. Mayhew, F. R. Blattner, C. T. Walsh, and R. D. Perry. 1998. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5:573-586. [DOI] [PubMed] [Google Scholar]
- 12.Heesemann, J. 1987. Chromosomal-encoded siderophores are required for mouse virulence of enteropathogenic Yersinia species. FEMS Microbiol. Lett. 48:229-233. [Google Scholar]
- 13.Mercado-Blanco, J., K. M. G. M. van der Drift, P. E. Olsson, J. E. Thomas-Oates, L. C. van Loon, and P. A. H. M. Bakker. 2001. Analysis of the pmsCEAB gene cluster involved in biosynthesis of salicylic acid and the siderophore pseudomonine in the biocontrol strain Pseudomonas fluorescens WCS374. J. Bacteriol. 183:1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller, R., C. Dahm, G. Schulte, and E. Leistner. 1996. An isochorismate hydroxymutase isogene in Escherichia coli. FEBS Lett. 378:131-134. [DOI] [PubMed] [Google Scholar]
- 16.Ozenberger, B. A., T. J. Brickman, and M. A. McIntosh. 1989. Nucleotide sequence of Escherichia coli isochorismate synthetase gene entC and evolutionary relationship of isochorismate synthetase and other chorismate-utilizing enzymes. J. Bacteriol. 171:775-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pelludat, C., A. Rakin, C. A. Jacobi, S. Schubert, and J. Heesemann. 1998. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J. Bacteriol. 180:538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quadri, L. E., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 19.Rakin, A., E. Saken, D. Harmsen, and J. Heesemann. 1994. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol. Microbiol. 13:253-263. [DOI] [PubMed] [Google Scholar]
- 20.Rakin, A., C. Noelting, S. Schubert, and J. Heesemann. 1999. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect. Immun. 67:5265-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 22.Serino, L., C. Reimmann, H. Baur, M. Beyeler, P. Visca, and D. Haas. 1995. Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol. Gen. Genet. 249:217-228. [DOI] [PubMed] [Google Scholar]
- 23.Simon, R., U. Priefer, and A. Pühler. 1988. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-785. [Google Scholar]
- 24.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 25.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young, I. G., T. J. Batterham, and F. Gibson. 1969. The isolation, identification and properties of isochorismic acid: an intermediate in the biosynthesis of 2, 3-dihydroxybenzoic acid. Biochim. Biophys. Acta 177:389-400. [DOI] [PubMed] [Google Scholar]