Abstract
The nitrogen-fixing symbiosis between Sinorhizobium meliloti and Medicago sativa requires complex physiological adaptation by both partners. One method by which bacteria coordinately control physiological adaptation is the stringent response, which is triggered by the presence of the nucleotide guanosine tetraphosphate (ppGpp). ppGpp, produced by the RelA enzyme, is thought to bind to and alter the ability of RNA polymerase (RNAP) to initiate and elongate transcription and affect the affinity of the core enzyme for various sigma factors. An S. meliloti relA mutant which cannot produce ppGpp was previously shown to be defective in the ability to form nodules. This mutant also overproduces a symbiotically necessary exopolysaccharide called succinoglycan (38). The work presented here encompasses the analysis of suppressor mutants, isolated from host plants, that suppress the symbiotic defects of the relA mutant. All suppressor mutations are extragenic and map to either rpoB or rpoC, which encode the β and β′ subunits of RNAP. Phenotypic, structural, and gene expression analyses reveal that suppressor mutants can be divided into two classes; one is specific in its effect on stringent response-regulated genes and shares striking similarity with suppressor mutants of Escherichia coli strains that lack ppGpp, and another reduces transcription of all genes tested in comparison to that in the relA parent strain. Our findings indicate that the ability to successfully establish symbiosis is tightly coupled with the bacteria's ability to undergo global physiological adjustment via the stringent response.
Successful nitrogen-fixing symbiosis between Sinorhizobium meliloti and Medicago sativa (alfalfa) requires developmental and physiological adaptation by both partners. Alfalfa secretes a signaling molecule, luteolin, recognized by the bacteria, which in turn produce Nod factor, a compound responsible for the initiation of nodule development on plant roots (10, 35). During the morphological maturation of nodules, bacteria enter into the plant from root hair cells via infection threads, the initiation and development of which require the bacteria to produce an exopolysaccharide called succinoglycan (7). Bacteria are ultimately released from infection threads into nodule cortical cells, where they differentiate into large, branched bacteroids, which provide fixed nitrogen for the plant in exchange for carbon (31). Thus, to establish symbiosis, the bacteria must undergo complex signal exchange with the host to make the adaptation from the free-living to the symbiotically active bacteroid state.
A major determinant of physiological adaptation in bacteria is the stringent response, which is initiated when bacteria are subjected to nutrient-limiting conditions. Specifically, uncharged tRNA molecules are thought to trigger ribosome-associated RelA to convert GTP into guanosine pentaphosphate (pppGpp), which is subsequently hydrolyzed to guanosine tetraphosphate, or ppGpp (5). In Escherichia coli, a second highly homologous enzyme, SpoT, also can produce pppGpp but is thought to be primarily involved in the hydrolysis of ppGpp. Although most gram-negative bacteria have two (p)ppGpp synthetase enzymes, RelA and SpoT (27), a number of alpha proteobacteria, like S. meliloti, have a single enzyme with dual function. While this contrasts with the general paradigm for gram-negative bacteria, dual-function enzymes, known as Rel, are common to gram-positive bacteria. In all bacteria, however, ppGpp elicits a pleiotropic response that causes down-regulation of stable RNA species, resulting in global metabolic slowing. ppGpp has additionally been shown to activate amino acid biosynthetic genes and transcription factors involved in stress responses, such as heat shock and the transition to stationary phase (5). Recently, studies of the stringent response in bacteria which interact with eukaryotic organisms have led to insights into the role of ppGpp in virulence and persistence within hosts (6).
The pleiotropic effects of ppGpp are thought to occur specifically by modification of the behavior of RNA polymerase (RNAP). Within bacteria, core RNAP consists of five subunits (α2, β, β′, and ω) and is responsible for all transcription. The alternative binding of various σ subunits to core RNAP forms holoenzymes with different promoter specificities. Recent studies suggest that binding of ppGpp to RNAP occurs primarily within the β′ subunit of RNAP, encoded by rpoC (33). Prototrophic suppressors of E. coli ppGpp0 strains (which lack both relA and spoT and are auxotrophic) harbor mutations exclusively in rpoB, which encodes the β subunit of RNAP; rpoC; or rpoD, which encodes σ70 (3, 11, 14, 40). The σ70 mutants are thought to have decreased ability to associate with RNAP, thereby giving alternative σ factors such as σS (rpoS) greater likelihood of encountering and forming a holoenzyme with RNAP (14). This interpretation is supported by recent work showing that underproduction of σ70 in a ppGpp0 strain mimics the stringent response (23), and indeed ppGpp directly reduces the competitiveness of σ70 with σ32 (rpoH) for core binding (19). In addition to affecting the affinity of the core enzyme for various sigma factors, ppGpp has also been shown to affect both transcription initiation and elongation: ppGpp specifically decreases the half-life of open complex formation between ribosomal promoters and RNAP holoenzyme (2) and causes a significant decrease in the rate of transcription elongation (36).
Further understanding of the effect of ppGpp on RNAP has come from RNAP core and holoenzyme structures from thermophilic bacteria (28, 29, 34). The RNAP core enzyme is shaped like a claw, comprised of the β and β′ subunits, which form a channel that the DNA-RNA hybrid (the transcription bubble) passes through during transcription elongation (8, 15). In E. coli, ppGpp0 suppressor mutations that map to rpoB and rpoC have effects on transcription initiation similar to those of ppGpp; both decrease the half-life of open complex formation of rrnB P1 (1, 2, 40). Further, these mutations map exclusively within the region corresponding to the claw of RNAP (1), indicating that this region is critical for regulation of transcription during the stringent response. Although these mutations have been analyzed only with regard to transcription initiation, it is also possible that they affect other aspects of transcription that are controlled by ppGpp, such as elongation and affinity of core RNAP for σ.
To better understand the mechanism of physiological adjustment and adaptation by S. meliloti during symbiosis, a relA mutant strain that is unable to produce ppGpp was generated (38). This strain is auxotrophic, fails to form nodules on alfalfa, and overproduces the symbiotically necessary exopolysaccharide succinoglycan, suggesting a major role for the stringent response in the early stages of symbiosis. On rare occasions, however, the relA mutant does form nodules on alfalfa. All strains reisolated from such nodules are suppressor mutants, which can be divided into two phenotypic classes. Class I suppressors are able to grow on minimal medium, while class II strains remain largely auxotrophic (38).
In the following study, we present a detailed characterization of four relA suppressor mutants. During free-living growth, class I suppressors generally suppress the defects of the relA mutant more thoroughly than class II suppressors, which have more modest effects. Identification of the mutations revealed that all map to either rpoB or rpoC, and subsequent alignment and molecular modeling of the affected amino acids revealed that three of the four lie in conserved regions of either β or β′. Class I mutations map to a region of rpoB where suppressor mutations in E. coli ppGpp0 strains are found and most likely function by causing RNAP to mimic its behavior under stringent conditions. The class II suppressors reduce expression of all genes tested in a similar manner and therefore may have a nonspecific effect on transcription. The characterization of S. meliloti suppressor mutations has revealed that metabolic adjustment by the bacteria, whether to signals from the environment or from the host plant, is critical throughout the process of symbiosis.
MATERIALS AND METHODS
Growth conditions and media.
All S. meliloti strains, derivatives of Rm1021, were grown at 30°C in either Luria broth (LB), M9 sucrose (26), or TY-MGAS (38). Calcofluor white M2R (Sigma) was filter sterilized and added to autoclaved medium at a final concentration of 0.02% (21). E. coli strains were grown at 37°C in LB medium. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 50 μg/ml; neomycin, 50 μg/ml; and streptomycin, 500 μg/ml. Nodulation assays with alfalfa (M. sativa AS13R) were performed as described by Oke and Long (30).
Mapping suppressor mutations.
The TnphoA transposon on pRK609 (22) was introduced into DW362 by conjugal transfer. Approximately 10,000 insertions were isolated and pooled to generate an N3 phage lysate (24). All insertions were then moved into the relA background (DW186) by transduction, and neomycin-resistant individuals carrying the suppressor mutation were isolated based on decreased fluorescence on LB plates with calcofluor and the slight growth advantage that the suppressor mutation confers over relA, probably due to high salt concentration in the medium. Lysates were made from 15 isolates and transduced again into the relA background to calculate linkage. Arbitrary PCR (4) was performed to identify the locations of transposons in three strains that showed 94, 96, and 98% linkage to the suppressor mutation in DW362. The first round of arbitrary PCR was performed by using an oligonucleotide primer within the inverted repeat of TnphoA (and directed outward) and one of three arbitrary primers with known 20-base sequences at the 5′ ends followed by 10 random nucleotides and distinct anchors of five bases with various G+C contents. The second round of arbitrary PCR involved a nested primer, also within the inverted repeat of TnphoA, and a primer identical to the known 20 bases of the arbitrary primers. Positive PCR products were subcloned into the TOPO vector (Invitrogen Life Technologies), purified, and sequenced by using standard techniques.
Identifying mutations and molecular modeling.
To generate merodiploid strains, large segments of genomic DNA (2 to 4 kb) were amplified by PCR, sequenced, and cloned into the XbaI restriction site of pVO155. These plasmids were in turn introduced into either the relA mutant or DW362 by triparental mating (9, 12), and gain or loss of suppressor phenotype, respectively, was screened on LB calcofluor plates as described above. Analysis of the region beyond nusG (see Fig. 3B), containing rpoB and rpoC, was continued by PCR amplification of slightly overlapping 500-bp segments, TOPO cloning, and sequencing. All sequences in DW362 in which differences were observed were resequenced and checked against those of Rm1021 and the parent relA mutant by using Sequencher software (Gene Codes Corporation). The mutations in all other suppressor strains proved to be linked to rpoB or rpoC by transduction of a nearby marker used for generating merodiploids (see above) and screening either on LB calcofluor or minimal medium for restoration of the relA phenotype. The changes in suppressor strains are as follows: DW362, CTG to GTG in rpoC, resulting in L792V in β′; DW364, GAC to TAC in rpoB, resulting in D453Y in β; DW366, TTC to GTC in rpoC, resulting in F620V in β′; DW371, deletion of TCGGCAACC in rpoB, resulting in ΔLGN458-460 in β (Table 1).
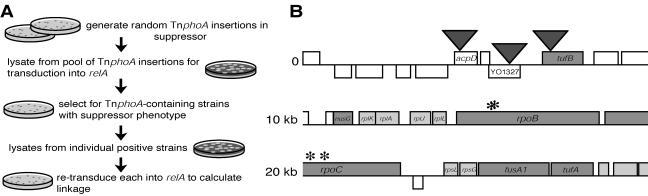
FIG. 3.
Flow chart of cloning strategy and map of genomic locations of suppressor mutations. (A) A Tn5 derivative transposon was used to mark the suppressor mutation. Transduction of linked markers by using N3 phage was used to genetically map the mutation. (B) The map of the region carrying the suppressor mutations was redrawn from a contig at http://sequence.toulouse.inra.fr/meliloti.html. Bars above and below lines represent open reading frames directed from left to right and vice versa, respectively. Arrowheads show the locations of the most closely linked transposons. Approximate locations of suppressor mutations in rpoB and rpoC are marked by asterisks.
TABLE 1.
Mutations in suppressor strains
| Strain (class) | Gene (RNAP subunit) | Mutation |
|---|---|---|
| DW362 (II) | rpoC (β′) | L792V |
| DW364 (I) | rpoB (β) | D453Y |
| DW366 (II) | rpoC (β′) | F620V |
| DW371 (I) | rpoB (β) | Δ458-460 (LGN) |
The 2.6-Å structure of RNAP holoenzyme from Thermus thermophilus (National Center for Biotechnology Information pdb: 1IW7) was used to map the location of amino acids affected by changes in the suppressor strains. Amino acids in β and β′ from T. thermophilus, homologous to those changed by the S. meliloti suppressor mutations, were identified by sequence alignments generated using the Lasergene/DNAstar software. The pdb file was viewed and modified in Swiss Pdb Viewer, and final rendering of the holoenzyme, with colored subunits and marked amino acids, was performed by using the Persistence of Vision ray-tracing shareware program (POVray; http://www.povray.org).
Gene expression.
To generate a transcriptional fusion between the rrnC promoter and the β-glucuronidase gene (uidA or gus), the upstream regulatory region was amplified by PCR, confirmed by sequencing, and cloned into the SpeI and XhoI sites of pVO155 (30), resulting in plasmid pDW187 (Table 2). The plasmid bearing the nodF reporter fusion, pDW110, was generated as previously described (37). The fusion to SMb21243 was generated by insertion into the SalI and XbaI sites of pVO155, yielding pDKR50, except that an internal fragment of the gene was used, which results in both a transcriptional fusion and a deletion (D. H. Keating, unpublished data). All plasmids were introduced into Rm1021 by triparental mating, and strains positive for the insertion were identified by resistance to neomycin and the ability to generate a PCR product with primers internal to uidA and external to the PCR-amplified segment used in the initial cloning. N3 phage lysates from these strains were transduced into Rm1021, DW186, and all suppressor strains (the lysate from Rm1021 with integrated pDKR50 was provided by D. H. Keating). All reporter strains were diluted from overnight cultures and grown for 3 to 4 h to an optical density at 600 nm (OD600) of 1.0 (to gauge growth stage, in this case late exponential phase) in TY-MGAS. In the case of nodF strains, luteolin, at a concentration of 3 μM, was added upon subculturing. Expression assays were performed as described by Jefferson et al. (16), except that CFU were used instead of OD600, to correct for the significant variability in viable cell counts at a given OD between relA mutants, suppressors, and the wild type (data not shown).
TABLE 2.
Bacterial strains and plasmids used in this study
| Plasmid or strain | Relevant characteristics | Source or reference |
|---|---|---|
| Plasmids | ||
| pRK600 | Conjugal transfer helper, Cm | 9 |
| pRK609 | pRK600 with TnphoA, Cm, Nm | 22 |
| pVO155 | Vector used for transcriptional fusions to uidA; Ap Nm | 30 |
| pDKR50 | pVO155 derivative, SMb21243 | D. H. Keating (unpublished data) |
| pDW110 | pVO155 derivative, nodF | 37 |
| pDW187 | pVO155 derivative, rrnC | This work |
| Strains | ||
| Rm1021 | S. meliloti wild type, Str | 25 |
| DW186 | ΔrelA Str | 38 |
| DW362 | ΔrelA (L792V) rpoC Str | This work |
| DW364 | ΔrelA (D453Y) rpoB Str | This work |
| DW366 | ΔrelA (F620V) rpoC Str | This work |
| DW371 | ΔrelA Δ458-460 (LGN) rpoB Str | This work |
| DW386 | Rm1021, integrated pDW110, Str Nm | 37 |
| DW387 | DW186, integrated pDW110, Str Nm | This work |
| DW527 | Rm1021, integrated pDKR50, Str Nm | This work |
| DW528 | DW186, integrated pDKR50, Str Nm | This work |
| DW529 | DW362, integrated pDKR50, Str Nm | This work |
| DW530 | DW364, integrated pDKR50, Str Nm | This work |
| DW531 | DW366, integrated pDKR50, Str Nm | This work |
| DW532 | DW371, integrated pDKR50, Str Nm | This work |
| DW564 | Rm1021, integrated pDW187, Str Nm | This work |
| DW565 | DW186, integrated pDW187, Str Nm | This work |
| DW566 | DW362, integrated pDW187, Str Nm | This work |
| DW567 | DW364, integrated pDW187, Str Nm | This work |
| DW568 | DW366, integrated pDW187, Str Nm | This work |
| DW569 | DW371, integrated pDW187, Str Nm | This work |
| DW576 | DW362, integrated pDW110, Str Nm | This work |
| DW577 | DW364, integrated pDW110, Str Nm | This work |
| DW578 | DW366, integrated pDW110, Str Nm | This work |
| DW579 | DW371, integrated pDW110, Str Nm | This work |
RESULTS
Suppressor mutants rescue the symbiotic defects of a relA mutant.
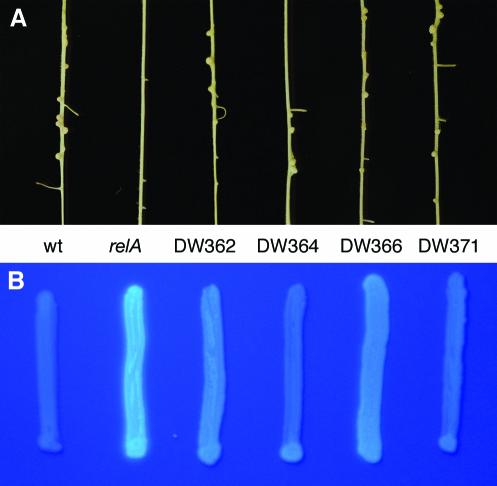
To examine the role of the stringent response in symbiosis, we characterized the ability of the suppressors to restore symbiotic competence to the relA mutant. We chose DW364 and DW371 as representative class I strains and DW362 and DW366 as representative class II strains. Figure 1A shows the nodulation phenotypes of these strains in comparison to those of the wild type, Rm1021, and the relA mutant, DW186. At 2 weeks after inoculation, the wild-type and suppressor strains formed nodules on alfalfa, whereas the relA mutant did not. The protrusions observed on the relA mutant-inoculated plant are emerging lateral roots. All four suppressor strains restored nodule formation equally well, although they generally formed fewer nodules than the wild type. Figure 1B shows the levels of succinoglycan production of all strains. By growing the bacteria on rich medium plates containing calcofluor, which binds specifically to succinoglycan and fluoresces under long-wave UV light, it is possible to qualitatively determine how much exopolysaccharide is produced. In this assay, the relA mutant overproduced succinoglycan compared to the wild type. While all four suppressor mutants produced less succinoglycan than the relA mutant, strains DW364 and DW371 displayed levels closer to that of the wild type than did strains DW362 and DW366, which were attenuated to a lesser degree (Fig. 1B).
FIG. 1.
Nodulation and succinoglycan production phenotypes of relA and suppressor mutants. (A) Alfalfa plants 2 weeks postinoculation with wild-type (wt), relA, and the four reisolated suppressor strains. (B) Bacteria patched onto rich TY-MGAS medium containing 0.02% calcofluor. Fluorescence was visualized under long-wave UV light. (Class I strains, DW364 and DW371; class II strains, DW362 and DW366.)
The suppressor mutants exhibit different growth phenotypes.
In addition to suppression of symbiotic defects of the relA parent strain, the two classes of suppressor mutants show an effect on free-living growth. On rich medium, the class II suppressors formed colonies of sizes equivalent to those of both wild-type and relA strain colonies. In contrast, the class I suppressors formed smaller colonies, which may reflect a reduction of growth rate in rich medium (data not shown). On minimal medium, the relA strain was unable to grow compared to wild-type and class I suppressor strains, both of which formed large single colonies. Although the class II strains were severely impaired in growth on minimal medium, they eventually formed single colonies (data not shown), perhaps due to an absence of competition from other cells, but the colonies were significantly smaller than those of the prototrophic wild-type and class I strains.
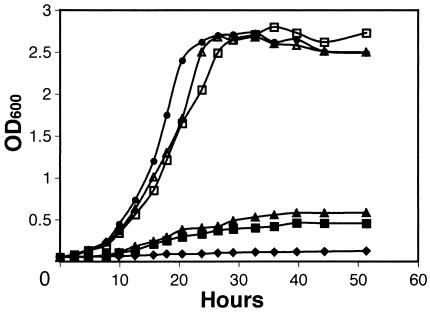
Figure 2 shows the growth curves of these strains after transfer and dilution from rich to minimal medium. In this experiment, the relA mutant increased in OD600 by slightly more than twofold. This most likely reflects the completion of cell division events which were already under way upon transfer, followed by inhibition of growth as remaining nutrients were depleted. Consistent with the growth phenotype on solid minimal medium, the class II suppressor strains showed only a modest increase in OD600 in this assay. Although these strains grew slightly better in minimal medium than the relA strain, they remained significantly impaired for growth under these conditions and were thus originally characterized as auxotrophic. The class I strains, on the other hand, grew similarly to the wild type in minimal medium (Fig. 2). While both class I strains reached the same final OD600 as the wild type, they did show a modest delay in growth during log phase.
FIG. 2.
Growth curves of relA and suppressor mutants in minimal medium. Growth in M9 liquid medium was measured as OD600 of the bacterial culture versus time. Strains assayed were as follows: wild type, closed circles; relA mutant, closed diamonds; DW362 (class II), closed squares; DW364 (class I), open triangles; DW366 (class II), closed triangles; DW371 (class I), open squares.
Cloning and identifying suppressor mutations.
Based on the observation that class II suppressor mutants rescued both nodule formation and succinoglycan production yet remained significantly auxotrophic, we predicted that these strains might permit identification of symbiotic genes that are regulated by the stringent response. We therefore chose DW362 for further analysis. Figure 3A shows a flow chart of the strategy used to identify the mutation in DW362. Since all suppressors arose spontaneously, we introduced a Tn5 derivative transposon, TnphoA, into DW362 to mark the mutation with neomycin resistance. These individual insertion strains were then pooled for transduction into the relA mutant (by using N3 phage). We next isolated relA strains carrying the suppressor mutation, which we scored by calcofluor and growth phenotypes, and retransduced them into the relA mutant to calculate genetic linkage (see Materials and Methods for details). After isolation of individual insertion strains that showed the highest degree of linkage with the suppressor mutation, we identified the locations of transposons by arbitrary PCR and DNA sequencing. Arrowheads in Fig. 3B show the locations of transposons in three strains displaying the highest degrees of linkage to the suppressor mutation.
To determine the precise nature of the DW362 mutation, we generated merodiploids and carried out DNA sequencing. From the most closely linked transposon (in tufB) (Fig. 3B), large segments of DNA from both the suppressor strain and the relA mutant, containing intact genes and their promoters, were amplified by PCR and cloned into pVO155 (30). The plasmids from DW362 were in turn introduced into the relA strain (to account for dominant mutations), those from relA were introduced into DW362, and either the acquisition of the suppressor phenotype or the loss thereof, respectively, was assayed as described above (see Materials and Methods). This procedure was used to “walk” along the chromosome to the end of nusG. Surprisingly, despite the high degree of transductional linkage to TnphoA, the mutation did not map to this region. We could not continue this procedure with the region beyond nusG, which encodes four ribosomal proteins and contains rpoBC (Fig. 3B), due to either lethality in E. coli or difficulty in generating a sufficiently large PCR product. Therefore, we identified the DW362 mutation by PCR amplification and DNA sequencing of 500-bp segments downstream of nusG.
The mutation in DW362 proved to be a single-base substitution that results in a leucine-to-valine change at position 792 of the β′ subunit of RNAP, corresponding to rpoC (Table 1). Although this mutation is over 12 kb away from the closest transposon (98% linkage would correlate with approximately 1.5 kb of distance, according to estimates of N3 transducing particle size), the discrepancy can most likely be explained by either suppression of recombination in this region or the low survival rate of the relA mutant compared to that of the suppressors during the transduction process. We subsequently determined that the other suppressor strains were linked to the region containing rpoB and rpoC (see Materials and Methods). The other class II mutation, in strain DW366, results in a phenylalanine-to-valine change at position 620 of β′. The mutation in DW364 yields an aspartic acid-to-tyrosine change at residue 453 of β, and DW371 carries a deletion of LGN at positions 458 to 460 of β (Table 1). To confirm that these mutations are solely responsible for suppression of relA, and since we could not identify any plasmid bearing this region in existing libraries, we introduced a suicide vector containing wild-type DNA that spans the deletion into strain DW371. Recombinants that restored the deletion in DW371 and hence the relA phenotype, which contained intact rpoB as confirmed by sequencing, were recovered at a rate of 1 in 18,000 (data not shown). This indicates that the nine-base deletion in rpoB is responsible for suppression of the defects of the relA mutant.
Altered amino acids are located in different regions of RNAP.
To identify the regions of RNAP to which the suppressor mutations map, we projected the affected amino acids onto the 2.6-Å crystal structure from T. thermophilus (34). Figure 4A shows a comparison of the primary sequences of β and β′ from S. meliloti and T. thermophilus. The β subunits are shown in light aqua, the β′ subunits are shown in yellow, and the locations of regions which are highly conserved among all RNAPs (from prokaryotic to eukaryotic organisms) are shown in green. Although the primary sequences of the β subunits from S. meliloti and T. thermophilus share only 56% similarity and 42% identity, the altered amino acids in class I suppressor strains DW364 (D453) and DW371 (LGN458-460) are both clearly in conserved region C of β (8, 32). These amino acids are highlighted in red in the alignment shown and correspond, respectively, to residues D323 and LGN328-330 from T. thermophilus. The altered amino acid in the class II suppressor strain DW362 (L792) (Table 1) is shown in orange and corresponds to L1094 from T. thermophilus in conserved region F of the β′ subunit (8, 20). The primary sequences of the β′ subunits from the two organisms share 55% similarity and 40% identity. The altered amino acid in the other class II strain, DW366 (F620), did not align with any conserved regions of β′, nor was there a homologous phenylalanine residue in β′ from T. thermophilus.
FIG. 4.
Predicted location of amino acids altered by suppressor mutations (within the crystal structure of a model RNAP). (A) Primary sequences of the β (light aqua) and β′ (yellow) subunits of RNAP from S. meliloti (Sm) and T. thermophilus (Tt) and evolutionarily conserved segments (green). Amino acid numbering is shown above the bars. The first alignment shows the location of amino acids altered by class I suppressor mutations (in red) in conserved region C of β. The second alignment shows the amino acid altered by the class II mutation in DW362 (in orange) in conserved region F of β′. (B) Amino acids affected by suppressor mutations projected onto the crystal structure of RNAP holoenzyme from T. thermophilus (National Center for Biotechnology Information pdb: 1IW7). Colors are as follows: α2, blue and light blue; β, light aqua; β′, yellow; σ, green; ω, dark aqua. The altered amino acid in DW362 (L792) is shown in orange in the β′ subunit (rpoC). The altered amino acids in DW364 and DW371 (D453 and LGN458-460) are shown in red in the β subunit (rpoB).
Figure 4B shows the locations of the altered amino acids on the three-dimensional structure of RNAP from T. thermophilus. The subunits are colored as follows: the two α subunits are blue and light blue, the β subunit is light aqua, β′ is yellow, σ is green, and ω is dark aqua. From this perspective, DNA enters RNAP from the bottom right and exits behind the molecule and into the page. In the middle of the enzyme, DNA is unwound, and transcription elongation occurs within a channel of the claw of RNAP formed by β and β′. At the center and base of the claw is the active site of the enzyme. The inset of Fig. 4B shows an entirely space-filled rendering of RNAP, and the arrows show the overall direction of transcription, towards the 5′ end of the template DNA strand (downstream). The large RNAP structure in Fig. 4B shows the location in β′ of the altered amino acid in class II strain DW362. This residue, L792, is close to where β′ meets β near the α subunits. The altered amino acids in the class I strains, in the β subunit, are near the active site within the claw of RNAP. It is directly through this region that the unwound DNA template is read during transcription. Further, mutations in E. coli in precisely this region have been shown to suppress auxotrophy of ppGpp0 strains and are thought to cause RNAP to mimic its behavior in the presence of ppGpp (1).
Expression studies indicate the specificity of RNAP mutations.
To determine the overall effect of RNAP mutations on expression of genes controlled by the stringent response, we generated transcriptional fusions between promoters of target genes and promoterless uidA (β-glucuronidase) as a reporter. As a representative of genes that are positively regulated by the stringent response, we used a gene (SMb21243) that encodes a putative sulfotransferase (see http://sequence.toulouse.inra.fr/meliloti.html). SMb21243, which shows a significant rise in expression levels at the transition to stationary phase (M. G. Müller and D. H. Keating, unpublished data), was used because of lack of expression data for more traditional positively regulated genes in S. meliloti, such as those that control amino acid biosynthesis. We generated a transcriptional fusion to the promoter of rrnC to observe the effect of suppressors on the expression of negatively regulated genes. This promoter lies upstream of the 16S rRNA gene of one of three rRNA operons in S. meliloti (13). To determine the overall specificity of these mutations on transcription, we generated a transcriptional fusion to the promoter of nodF (37), a Nod factor biosynthetic gene, which proved not to be regulated by the stringent response (see Table 2 for a detailed description of strains used in this assay). To minimize the effects of growth rate differences between the strains, all were assayed in rich medium at an OD600 of approximately 1.0. At this point in the growth curve, all strains are in late exponential phase (data not shown).
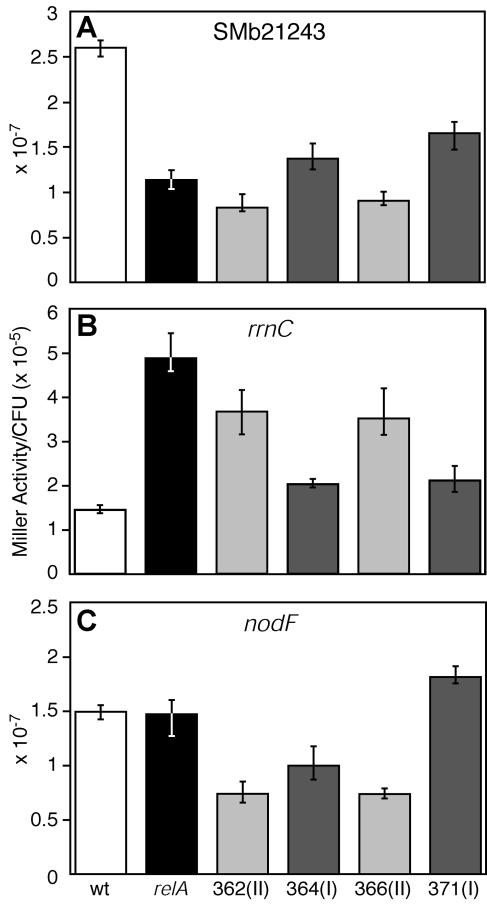
Figure 5A shows that the promoter activity of SMb21243 was significantly decreased in late exponential phase in the relA mutant compared to that in the wild type. Class I strains showed a moderate increase in activity, towards wild-type levels, whereas both class II strains surprisingly had even lower levels than the relA mutant. As shown in Fig. 5B, the promoter activity of rrnC in class I strains was shifted from the levels observed in the relA mutant towards those of the wild-type strain, similar to the moderate restorative effect represented in Fig. 5A. The class II strains also showed a decrease in promoter activity of rrnC compared to that in the relA parent strain, but the decrease was less than that observed in class I strains. As shown in Fig. 5C, promoter activity of nodF in the relA mutant was indistinguishable from that in the wild type. In comparison to the relA mutant (and the wild type), both class II strains displayed a decrease in activity of nodF, a trend similar to that exhibited in Fig. 5A and B. Furthermore, while promoter activity of nodF was decreased in one class I suppressor strain, DW364, it was increased in the other, DW371. These results indicate that lack of nod gene expression is not the cause of the inability of the relA mutant to form nodules, because (i) relA mutants did not show decreased nod gene expression compared to the wild type and (ii) three of four suppressor strains expressed nodF at lower levels than the relA mutant yet were symbiotically competent.
FIG. 5.
The effect of suppressor mutations on gene expression. (A) Levels of expression of a gene (SMb21243) that is positively regulated by the stringent response. Levels of expression were measured by using a transcriptional fusion to uidA (or gus). Strains were grown in rich medium, and all were harvested in late exponential phase at an OD600 of ∼1.0. Values are shown as Miller units per CFU of bacteria. Background activity in wild-type S. meliloti (which lacks β-glucuronidase) is zero. Average expression values (10−7) for SMb21243 were as follows: wild type (wt), 2.599; relA mutant, 1.133; DW362, 0.827; DW364, 1.371; DW366, 0.904; DW371, 1.653. Error bars represent minimum and maximum values. (B) Levels of expression of rrnC, which is negatively regulated by the stringent response. Levels of expression were measured as described for panel A. Average expression values (10−5) for rrnC were as follows: wild type, 1.450; relA mutant, 4.882; DW362, 3.671; DW364, 2.033; DW366, 3.513; DW371, 2.113. (C) Levels of expression of nodF, which is not regulated by the stringent response. Average expression values (10−7) for nodF were as follows: wild type, 1.495; relA mutant, 1.472; DW362, 0.738; DW364, 0.997; DW366, 0.737; DW371, 1.814. All experiments were repeated at least three times. Suppressor class is shown in parentheses after the strain name.
DISCUSSION
The detailed characterization of the two classes of relA suppressor mutants has revealed several insights into the role of the stringent response in symbiosis. Class I mutations suppress all defects in the relA mutant strain. The class I mutants form nodules on alfalfa, produce less succinoglycan, and are prototrophic (Fig. 1 and 2). Both class I mutations are in rpoB (Table 1), which encodes the β subunit of RNAP, and are predicted by modeling to affect amino acids that lie within the claw of the enzyme (Fig. 4), close to where transcription occurs. In E. coli, suppressor mutations in a similar region of rpoB, which restore prototrophy to ppGpp0 mutants (3, 11), are thought to cause RNAP to mimic its overall behavior in the presence of ppGpp (1). The evidence presented here supports this idea, since in comparison to those in the relA mutant, we observed an increase in expression levels of a positively regulated gene and a decrease in expression levels of a negatively regulated gene in both class I strains (Fig. 5A and B).
The class I mutations may therefore mimic the effects of the stringent response by altering transcription at the level of initiation (1, 40), elongation (36), or the ability of core RNAP to bind various σ factors (19, 36). It is of note that the region in which these mutations are located in RNAP has been implicated directly in binding to σ factors (39). Regardless of the specific means by which suppressor mutations affect transcription, the slow growth of class I strains on rich medium (data not shown) provides further evidence that RNAP in these strains functions as if it were under stringent conditions. If RNAP in these strains is locked in the stringent state, then growth rate may be slowed towards that of the wild type during the stringent response. However, despite the overall ability of these mutations to restore the relA mutant to a state closer to that of the wild type, it is striking that the class I strains showed different effects on expression of a gene that is not regulated by the stringent response: DW364 showed levels of nodF expression that were lower than those observed in wild-type and relA strains, while DW371 showed levels that were higher (Fig. 5C). It is therefore possible that although these mutations may have similar effects on stringent response-related phenotypes, they may differ significantly in their effects on the expression of genes that are not controlled by ppGpp.
The ability of class II mutants to restore nodulation without fully restoring prototrophy to the relA mutant (Fig. 1 and 2) suggests that these suppressors could specify stringently controlled genes that are necessary for symbiosis. However, further results indicate that these mutations may affect transcription in a nonspecific manner. Both mutations map to rpoC (Table 1), a gene that encodes the β′ subunit of RNAP. Since F620 is not within a conserved region of β′, it is not likely to be in a region critical for RNAP function. In addition, L792 lies within a region of RNAP that is not likely to alter any of the known aspects of transcription affected by the stringent response, because σ binding, initiation, and elongation all occur on the opposite face of the β′ subunit (8, 34).
Expression data from all genes assayed in class II strains point to a global decrease in efficiency of transcription in comparison to levels observed in the relA parent strain. Both class II mutants displayed expression levels of a positively regulated gene, SMb21243, that were even lower than those seen with the relA mutant (Fig. 5A). This trend was observed in the expression of rrnC as well (Fig. 5B); however, a decrease in the expression levels of ribosomal promoters from those observed in the relA mutant gives the class II strains a profile more similar to that of the wild type. This effect alone may allow for the weak ability of class II strains to grow on minimal medium, since these mutations may decrease the metabolic load on the relA mutant.
Finally, expression of a gene that is not regulated by the stringent response, nodF, was also lower in class II strains (Fig. 5C). This result suggests not only that the effects of class II suppressors are nonspecific but also that the inability of the relA mutant to form nodules is not due to low levels of nod gene expression, since strains that restore nodulation have even lower levels of nodF promoter activity. Furthermore, the relA mutant has been shown to induce calcium oscillations on host plants (38), which indicates that the inability to produce Nod factor is not the primary cause of this symbiotic defect.
To further investigate the nature of the suppressor mutations, we have conducted preliminary whole-genome expression analysis of relA and suppressor strains by using Affymetrix GeneChips. Evidence thus far suggests that both types of suppressor mutations function to restore wild-type expression levels of the majority of the aberrantly expressed genes in the relA parent strain. Class I suppressor mutations restored expression to almost 90% of all genes misregulated in the relA parent, and surprisingly, class II mutations restored expression to nearly 80% of genes. In addition, the array data confirm the succinoglycan phenotype shown in Fig. 1 (by expression levels of various exo genes) and identify numerous putative targets for further study of the role of the stringent response in symbiosis. Although these data will be presented in detail elsewhere, they show, at first approximation, that both suppressors function to broadly attenuate aberrant gene expression in the relA background and that neither specifies a distinct or small set of genes that are clearly involved in the ability of S. meliloti to establish a functional symbiosis.
The preliminary array data indicate that suppressor mutations function to broadly affect genes misregulated in the relA parent strain, yet determining their specific effect on different aspects of transcription requires the development of an S. meliloti in vitro transcription system, which is currently unavailable. Since the lack of in vitro transcription prevents our experimentally testing putative phenotypes of suppressors, we cannot yet determine precisely whether class I mutations cause RNAP to functionally mimic its behavior in the presence of ppGpp (by measuring half-lives of open complex formation, for example) or how class II mutations result in the decrease in expression levels of all genes assayed in comparison to the levels observed in the relA mutant.
Although all other suppressors that we isolated (nine additional strains) are linked to rpoBC, it remains possible that there exists a single gene or small set of genes, downstream of the stringent response, which regulates symbiotic traits such as the ability to form nodules. While further extensive mutagenesis is necessary to answer this question, it is interesting that the relA mutation does not affect expression of a biosynthetic nod gene, the product of which is the primary determinant for nodule formation by the bacteria. This suggests that there may not be a single gene downstream of the stringent response that regulates symbiosis (which would be a likely target for suppression) and that the only means of restoring nodule formation by a relA mutant is a major alteration in cellular metabolism. This is further supported by the array data, which suggest that suppressor mutations function to globally and not specifically affect gene expression. The decrease in transcription observed with all genes assayed in class II suppressors suggests that overall metabolic demand on bacteria which cannot produce ppGpp may be the very cause of the inability to form nodules.
The analysis of suppressor mutations that attenuate the defects of an S. meliloti relA mutant establishes the tight coupling between bacterial physiological state and symbiosis. Indeed, it is striking that suppressor strains that suppress the symbiotic defects of an S. meliloti relA mutant show such strong similarities with suppressors of E. coli ppGpp0 mutations, which were isolated based on the ability to either restore growth on minimal medium (1, 3, 11) or confer resistance to rifampin (3, 11, 17, 18, 40). This indicates that the ability of S. meliloti to form nodules requires significant metabolic adjustment which fundamentally involves the stringent response. Although we did not identify specific symbiotic genes that are regulated by the stringent response, suppressor mutants, in particular class I strains, ultimately will help to identify genes required for symbiosis by restoring their expression from levels in the relA mutant to those in the wild type. Future analysis of these suppressor mutants with regard to global gene expression, by using the DNA microarray analysis mentioned above, for example, will allow us to identify the entire cast of genes that coordinate ppGpp-regulated symbiosis.
Acknowledgments
We thank David Keating for helpful discussions and advice throughout the course of this study and for providing the N3 lysate of Rm1021 with integrated pDKR50. We additionally thank Robert Fisher and Erin Gaynor for critical readings of the manuscript and V. J. Hernandez for helpful advice. We thank Marvin Whiteley for design of arbitrary PCR primers and Tom Purcell for assistance with the POVray software.
This work was supported by the NIH predoctoral training grant T32 G07276 and NIH grant GM30962 to S.R.L.
REFERENCES
- 1.Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305:689-702. [DOI] [PubMed] [Google Scholar]
- 2.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett, M. S., T. Gaal, W. Ross, and R. L. Gourse. 1998. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J. Mol. Biol. 279:331-345. [DOI] [PubMed] [Google Scholar]
- 4.Caetano-Anolles, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85-94. [DOI] [PubMed] [Google Scholar]
- 5.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella. ASM Press, Washington, D.C.
- 6.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darst, S. A. 2001. Bacterial RNA polymerase. Curr. Opin. Struct. Biol. 11:155-162. [DOI] [PubMed] [Google Scholar]
- 9.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher, R. F., and S. R. Long. 1992. Rhizobium-plant signal exchange. Nature 357:655-660. [DOI] [PubMed] [Google Scholar]
- 11.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278:2092-2097. [DOI] [PubMed] [Google Scholar]
- 12.Glazebrook, J., and G. C. Walker. 1991. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 204:398-418. [DOI] [PubMed] [Google Scholar]
- 13.Gustafson, A. M., K. P. O'Connell, and M. F. Thomashow. 2002. Regulation of Sinorhizobium meliloti 1021 rrnA-reporter gene fusions in response to cold shock. Can. J. Microbiol. 48:821-830. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez, V. J., and M. Cashel. 1995. Changes in conserved region 3 of Escherichia coli sigma 70 mediate ppGpp-dependent functions in vivo. J. Mol. Biol. 252:536-549. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, L. M. 2002. Open season on RNA polymerase. Nat. Struct. Biol. 9:502-504. [DOI] [PubMed] [Google Scholar]
- 16.Jefferson, R. A., S. M. Burgess, and D. Hirsh. 1986. Beta-glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83:8447-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin, D. J., and C. A. Gross. 1989. Characterization of the pleiotropic phenotypes of rifampin-resistant rpoB mutants of Escherichia coli. J. Bacteriol. 171:5229-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin, D. J., and C. A. Gross. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 202:45-58. [DOI] [PubMed] [Google Scholar]
- 19.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jokerst, R. S., J. R. Weeks, W. A. Zehring, and A. L. Greenleaf. 1989. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol. Gen. Genet. 215:266-275. [DOI] [PubMed] [Google Scholar]
- 21.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 82:6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long, S., S. McCune, and G. C. Walker. 1988. Symbiotic loci of Rhizobium meliloti identified by random TnphoA mutagenesis. J. Bacteriol. 170:4257-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magnusson, L. U., T. Nystrom, and A. Farewell. 2003. Underproduction of sigma 70 mimics a stringent response. A proteome approach. J. Biol. Chem. 278:968-973. [DOI] [PubMed] [Google Scholar]
- 24.Martin, M. O., and S. R. Long. 1984. Generalized transduction in Rhizobium meliloti. J. Bacteriol. 159:125-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meade, H. M., and E. R. Signer. 1977. Genetic mapping of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 74:2076-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mittenhuber, G. 2001. Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J. Mol. Microbiol. Biotechnol. 3:585-600. [PubMed] [Google Scholar]
- 28.Murakami, K. S., S. Masuda, E. A. Campbell, O. Muzzin, and S. A. Darst. 2002. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296:1285-1290. [DOI] [PubMed] [Google Scholar]
- 29.Murakami, K. S., S. Masuda, and S. A. Darst. 2002. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science 296:1280-1284. [DOI] [PubMed] [Google Scholar]
- 30.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 31.Oke, V., and S. R. Long. 1999. Bacteroid formation in the Rhizobium-legume symbiosis. Curr. Opin. Microbiol. 2:641-646. [DOI] [PubMed] [Google Scholar]
- 32.Sweetser, D., M. Nonet, and R. A. Young. 1987. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc. Natl. Acad. Sci. USA 84:1192-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toulokhonov, I. I., I. Shulgina, and V. J. Hernandez. 2001. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta′-subunit. J. Biol. Chem. 276:1220-1225. [DOI] [PubMed] [Google Scholar]
- 34.Vassylyev, D. G., S. Sekine, O. Laptenko, J. Lee, M. N. Vassylyeva, S. Borukhov, and S. Yokoyama. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 417:712-719. [DOI] [PubMed] [Google Scholar]
- 35.Vijn, I., L. das Nevas, A. van Kammen, H. Franssen, and T. Bisseling. 1993. Nod factors and nodulation in plants. Science 260:1764-1765. [DOI] [PubMed] [Google Scholar]
- 36.Vogel, U., M. Sorensen, S. Pedersen, K. F. Jensen, and M. Kilstrup. 1992. Decreasing transcription elongation rate in Escherichia coli exposed to amino acid starvation. Mol. Microbiol. 6:2191-2200. [DOI] [PubMed] [Google Scholar]
- 37.Wais, R. J., D. H. Wells, and S. R. Long. 2002. Analysis of differences between Sinorhizobium meliloti 1021 and 2011 strains using the host calcium spiking response. Mol. Plant-Microbe Interact. 15:1245-1252. [DOI] [PubMed] [Google Scholar]
- 38.Wells, D. H., and S. R. Long. 2002. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol. Microbiol. 43:1115-1127. [DOI] [PubMed] [Google Scholar]
- 39.Wigneshweraraj, S. R., S. Nechaev, K. Severinov, and M. Buck. 2002. Beta subunit residues 186-433 and 436-445 are commonly used by Esigma54 and Esigma70 RNA polymerase for open promoter complex formation. J. Mol. Biol. 319:1067-1083. [DOI] [PubMed] [Google Scholar]
- 40.Zhou, Y. N., and D. J. Jin. 1998. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2908-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]