Abstract
We report the first study of tRNA modification in psychrotolerant archaea, specifically in the archaeon Methanococcoides burtonii grown at 4 and 23°C. For comparison, unfractionated tRNA from the archaeal hyperthermophile Stetteria hydrogenophila cultured at 93°C was examined. Analysis of modified nucleosides using liquid chromatography-electrospray ionization mass spectrometry revealed striking differences in levels and identities of tRNA modifications between the two organisms. Although the modification levels in M. burtonii tRNA are the lowest in any organism of which we are aware, it contains more than one residue per tRNA molecule of dihydrouridine, a molecule associated with maintenance of polynucleotide flexibility at low temperatures. No differences in either identities or levels of modifications, including dihydrouridine, as a function of culture temperature were observed, in contrast to selected tRNA modifications previously reported for archaeal hyperthermophiles. By contrast, S. hydrogenophila tRNA was found to contain a remarkable structural diversity of 31 modified nucleosides, including nine methylated guanosines, with eight different nucleoside species methylated at O-2′ of ribose, known to be an effective stabilizing motif in RNA. These results show that some aspects of tRNA modification in archaea are strongly associated with environmental temperature and support the thesis that posttranscriptional modification is a universal natural mechanism for control of RNA molecular structure that operates across a wide temperature range in archaea as well as bacteria.
The posttranscriptional processing of tRNA produces a diverse wealth of modified nucleotides (29, 30, 41), most of which occur at conserved RNA sequence locations in all three phylogenetic domains (4, 45). Many of the functional roles of these modifications, in addition to other factors, such as G-C and metal ion content, are associated with their influence on secondary and tertiary structures in RNA (1, 14, 43). Thus, RNA modifications offer an important means of mediation of RNA structure across the entire temperature range of natural habitats for microorganisms: in low-temperature organisms, a degree of conformational flexibility in tRNA must be maintained during translation, while in the case of thermophiles, protection against environmental temperatures which may exceed the melting point of unmodified base-paired stems is required (27, 50). For example, it has been shown with increases in growth temperature for a single species (2, 27, 51) or through comparison of closely related organisms growing optimally at different temperatures (32) that selected stabilizing tRNA modifications are associated with increased culture temperature. By contrast, in bacterial psychrophiles, low levels of modification have been reported, with the exception of dihydrouridine (10), a modified tRNA nucleoside which is associated with enhancement and maintenance of molecular flexibility at low temperatures (12).
From a phylogenetic perspective, it is interesting that the nucleoside structural motifs used for RNA stabilization at higher temperatures are known to be different in bacterial thermophiles (13, 24) and archaeal thermophiles (17), but tRNA modifications in low-temperature archaea have not previously been examined. We report here a detailed study of the identities and levels of nucleoside modifications in unfractionated tRNA from the psychrotolerant archaeon Methanococcoides burtonii (20). Cultures were grown at 23°C (the optimum growth temperature) and at 4°C (closer to the natural habitat temperature of 1 to 2°C [20]) in order to examine the overall level of tRNA modifications in comparison with that in bacteria, as well as recently studied mesophilic and thermophilic methanococci (32), and to assess the influence of a significant reduction in culture temperature upon modification. For contrast within the archaeal domain, we examined tRNA from the hyperthermophile Stetteria hydrogenophila (25) cultured at 93°C, near the growth optimum of 95°C. In both cases nucleoside modifications were measured by analysis of total enzymatic digests of tRNA using combined LC/MS, a definitive method for structural identification of RNA nucleoside modifications (7, 39).
MATERIALS AND METHODS
Abbreviations and symbols used.
Systematic names and structures for each nucleoside can be found on the World Wide Web at http://medlib.med.utah.edu/RNAmods. Abbreviations and symbols used are as follows: D, dihydrouridine; Ψ, pseudouridine; Um, 2′-O-methyluridine; m1Ψ, 1-methylpseudouridine; s2U, 2-thiouridine; s4U, 4-thiouridine; mnm5s2U, 5-methylaminomethyl-2-thiouridine; m5C, 5-methylcytidine; m5Cm, 5,2′-O-dimethylcytidine; Cm, 2′-O-methylcytidine;  Cm, N4,N4,2′-O-trimethylcytidine; ac4C, N4-acetylcytidine; ac4Cm, N4-acetyl-2′-O-methylcytidine; m1A, 1-methyladenosine; m2A, 2-methyladenosine; m6A, N6-methyladenosine;
Cm, N4,N4,2′-O-trimethylcytidine; ac4C, N4-acetylcytidine; ac4Cm, N4-acetyl-2′-O-methylcytidine; m1A, 1-methyladenosine; m2A, 2-methyladenosine; m6A, N6-methyladenosine; 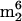 A, N6,N6-dimethyladenosine; t6A, N6-threonylcarbamoyladenosine; m6t6A, N6-methyl-N6-threonylcarbamoyladenosine; hn6A, N6-hydroxynorvalylcarbamoyladenosine; hn6A + CH3, monomethyl derivative of hn6A; ms2t6A, 2-methylthio-N6-threonylcarbamoyladenosine; ms2hn6A, 2-methylthio-N6-hydroxynorvalylcarbamoyladenosine; Am, 2′-O-methyladenosine; I, inosine; m1I, 1-methylinosine; m1G, 1-methylguanosine; m2G, N2-methylguanosine; m7G, 7-methylguanosine; Gm, 2′-O-methylguanosine; m22G, N2,N2-dimethylguanosine; m2Gm, N2,2′-O-dimethylguanosine;
A, N6,N6-dimethyladenosine; t6A, N6-threonylcarbamoyladenosine; m6t6A, N6-methyl-N6-threonylcarbamoyladenosine; hn6A, N6-hydroxynorvalylcarbamoyladenosine; hn6A + CH3, monomethyl derivative of hn6A; ms2t6A, 2-methylthio-N6-threonylcarbamoyladenosine; ms2hn6A, 2-methylthio-N6-hydroxynorvalylcarbamoyladenosine; Am, 2′-O-methyladenosine; I, inosine; m1I, 1-methylinosine; m1G, 1-methylguanosine; m2G, N2-methylguanosine; m7G, 7-methylguanosine; Gm, 2′-O-methylguanosine; m22G, N2,N2-dimethylguanosine; m2Gm, N2,2′-O-dimethylguanosine; 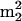 Gm, N2,N2,2′-O-trimethylguanosine; m3G, trimethylguanosine; m2,7Gm, N2,7,2′-O-trimethylguanosine; o8G, 8-oxoguanosine; imG, wyosine; imG2, isomer of imG; imG*, demethyl isomer of wyosine; mimG, methylwyosine; G+, archaeosine; MH+, molecular ion, corresponding to the protonated molecule;
Gm, N2,N2,2′-O-trimethylguanosine; m3G, trimethylguanosine; m2,7Gm, N2,7,2′-O-trimethylguanosine; o8G, 8-oxoguanosine; imG, wyosine; imG2, isomer of imG; imG*, demethyl isomer of wyosine; mimG, methylwyosine; G+, archaeosine; MH+, molecular ion, corresponding to the protonated molecule; 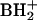 , base fragment ion, corresponding to the protonated free base of a nucleoside; SME medium, synthetic marine medium; HPLC, high-performance liquid chromatography; LC/MS, liquid chromatography-electrospray ionization mass spectrometry; Tm, melting temperature.
, base fragment ion, corresponding to the protonated free base of a nucleoside; SME medium, synthetic marine medium; HPLC, high-performance liquid chromatography; LC/MS, liquid chromatography-electrospray ionization mass spectrometry; Tm, melting temperature.
Cell sources and culture conditions.
M. burtonii (DSM 6242) was isolated from Ace Lake in Antarctica, where the in situ temperature is annually 1 to 2°C (20). It has an optimal growth temperature of 23°C and an upper growth temperature limit of approximately 28°C (20). To prepare biomass for tRNA extraction, cells were grown anaerobically in a modified methanogen growth medium (47) at 4 and and 23°C, the cells were harvested by centrifugation (10,000 × g), and the pellet was lyophilized.
S. hydrogenophila (DSM 11227) was cultivated in half-strength SME medium as described previously (25). Mass cultures were grown in a 100-liter enamel-coated fermentor at 93°C at pH 6.0. The agitation rate was 100 rpm, and the gassing rate was 2 liters/min (H2/CO2 = 80:20).
Isolation and enzymatic digestion of tRNA.
tRNAs were isolated as described previously (5) and totally digested to nucleosides using nuclease P1, phosphodiesterase I, and bacterial alkaline phosphatase (8), usually on a scale of several tens of micrograms of tRNA.
Analysis of tRNA digests by LC/MS.
Nucleoside mixtures produced by enzymatic hydrolysis of tRNA were analyzed using an LC/MS system consisting of a Quattro II (Micromass) triple-quadrupole mass spectrometer with a Z-spray ion source interfaced to an HP 1090 (Hewlett-Packard) liquid chromatograph with a photodiode array detector, with both instruments under the control of MassLynx 3.4 (Micromass) software. Under typical operating conditions, mass spectra of quality suitable for nucleoside identification using this system can be obtained at limits of detection of approximately one modified nucleoside per 5 × 103 total nucleosides in a digest of unfractionated tRNA, a ratio constrained largely by HPLC column loading limits.
Chromatographic separation of nucleosides (see Fig. 1 and 2) was carried out with a Luna C18 (Phenomenex) reversed-phase column (2.0 by 250 mm), thermostatted at 40°C, with a buffer gradient system composed of 5 mM ammonium acetate (pH 5.3) (buffer A) and acetonitrile-water (40:60, vol/vol) (buffer B) at a flow rate of 0.3 ml/min, essentially as described previously (19). Analysis of dihydrouridine in M. burtonii tRNAs (see Fig. 3) was made with a Develosil RP-Aqueous (C30) (Phenomenex) reversed-phase column (250 by 2 mm), with the same buffer system as above but using a multilinear gradient from 100% A to 100% B over 24 min. The procedures for the LC/MS experiments and the interpretation of resulting data for identification of nucleosides were similar to those detailed earlier for a thermospray, rather than electrospray, ionization-based protocol (39).
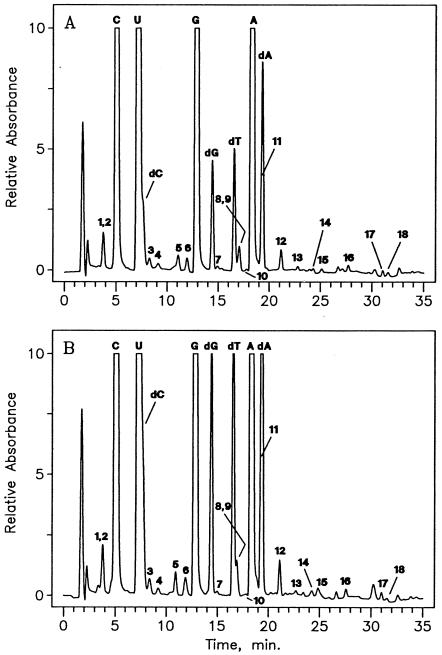
FIG. 1.
LC/MS analysis of nucleosides from unfractionated tRNA of M. burtonii cultured at 4°C (A) or 23°C (B). Components: 1, D; 2, Ψ; 3, m1Ψ; 4, m1A; 5, m7G; 6, Cm; 7, s4U; 8, m1G; 9, Gm; 10, m2G; 11, G+; 12, 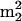 G; 13, t6A; 14, m6A; 15, imG*; 16, hn6A; 17,
G; 13, t6A; 14, m6A; 15, imG*; 16, hn6A; 17, 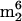 A (non-tRNA nucleoside; see the text); 18, ms2hn6A.
A (non-tRNA nucleoside; see the text); 18, ms2hn6A.
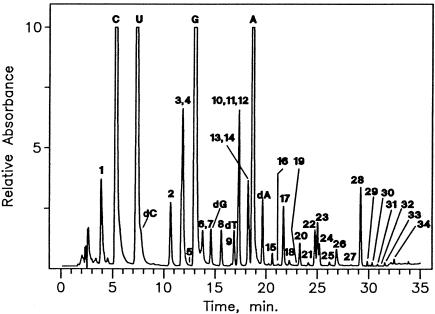
FIG. 2.
LC/MS analysis of nucleosides from unfractionated tRNA of S. hydrogenophila cultured at 90°C. Components: 1, Ψ; 2, m5C; 3, m1A; 4, Cm; 5, I; 6, m7G; 7, o8G (non-tRNA nucleoside; see the text); 8, Um; 9, m5Cm; 10, m1I; 11, m1G; 12, Gm; 13, ac4C; 14, m2G; 15, t6A; 16, m3G isomer of unknown structure; 17, 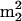 G; 18, Am; 19, m24Cm (non-tRNA nucleoside, see the text); 20, ac4Cm; 21, m2A; 22, m6A; 23, m2Gm; 24, hn6A; 25, ms2t6A; 26, m2,7Gm; 27, imG; 28,
G; 18, Am; 19, m24Cm (non-tRNA nucleoside, see the text); 20, ac4Cm; 21, m2A; 22, m6A; 23, m2Gm; 24, hn6A; 25, ms2t6A; 26, m2,7Gm; 27, imG; 28, 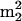 Gm; 29, hn6A + CH3; ms2hn6A; 30, 31, imG isomer of unknown structure; 32, unknown nucleoside N415; 33,
Gm; 29, hn6A + CH3; ms2hn6A; 30, 31, imG isomer of unknown structure; 32, unknown nucleoside N415; 33, 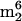 A (non-tRNA nucleoside, see text); 34, mimG.
A (non-tRNA nucleoside, see text); 34, mimG.
FIG. 3.
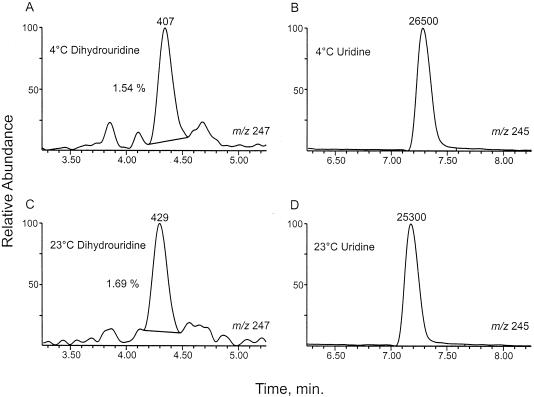
LC/MS detection of D in tRNA from M. burtonii cultured at 4°C (A) and 23°C (C), based on response using the MH+ ion, m/z 247. Relative peak areas (407 and 429 units) compared with corresponding responses from uridine (26,500 and 25,300 units) (B and D) show relative levels of D of 1.54% at 4°C and 1.69% at 23°C. The baselines used for area integrations are indicated in panels A and C.
Some LC/MS experiments were carried out by using deuterated chromatographic mobile phases, such as D2O and ND4COCH3, to allow complete exchange of labile hydrogen (protium) atoms in nucleosides by deuterium during chromatography (18). Labeled HPLC solvents were prepared by closely following an earlier protocol (39), or for small volumes and limited use by simply diluting concentrated unlabeled reagents with D2O. The mass shifts resulting from deuterium exchange, when unambiguous values were obtained, were used to verify nucleoside structure assignments and to distinguish isomers differing in the number of exchangeable hydrogen atoms.
Calculation of relative tRNA modification levels.
Crude estimates of overall modification levels for each tRNA were calculated from HPLC peak areas from UV detection at 260 nm, using MassLynx chromatography software routines. Based on total peak areas of 100% (including unmodified A, U, G, and C), the sum of modified nucleoside peak areas for each organism is presented as a portion of the total. Although individual nucleosides exhibit different molar absorptivities, for example in purines versus pyrimidines, the aggregate absorptivities and molar ratios of purines and pyrimidines are considered sufficiently similar to allow crude estimates of modification levels to be derived in this fashion. Responses from deoxynucleosides resulting from residual DNA impurities, readily recognized from their mass spectra, were excluded from the calculations.
Estimates of the average numbers of residues of modified nucleosides m2G, m2Gm, 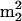 G, and
G, and 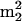 Gm per tRNA molecule were calculated as follows. Percents A, U, G, and C were calculated from respective HPLC peak areas using the following relative 260 nm UV response values, which were established from an equimolar nucleoside digest that had been analyzed by HPLC using the same buffered gradient as for LC/MS measurements: A, 1.0; U, 0.685; G, 0.80; and C, 0.49. Individual nucleoside abundances were then referenced via HPLC peak areas to percent G, assuming a molar absorptivity (ɛ) ratio of 0.86 for
Gm per tRNA molecule were calculated as follows. Percents A, U, G, and C were calculated from respective HPLC peak areas using the following relative 260 nm UV response values, which were established from an equimolar nucleoside digest that had been analyzed by HPLC using the same buffered gradient as for LC/MS measurements: A, 1.0; U, 0.685; G, 0.80; and C, 0.49. Individual nucleoside abundances were then referenced via HPLC peak areas to percent G, assuming a molar absorptivity (ɛ) ratio of 0.86 for 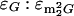 (22) (and therefore for
(22) (and therefore for 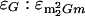 as well), and a molar absorptivity ratio of 1.22 for ɛG:ɛm2G (and therefore for
as well), and a molar absorptivity ratio of 1.22 for ɛG:ɛm2G (and therefore for 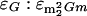 ), all at 260 nm (22). The residue-per-tRNA abundance values were calculated assuming 76 nucleotides per tRNA molecule.
), all at 260 nm (22). The residue-per-tRNA abundance values were calculated assuming 76 nucleotides per tRNA molecule.
RESULTS
Modified nucleosides identified by LC/MS analysis are summarized in Table 1, representing assignments shown in Fig. 1 and 2. In selected cases (see below), UV spectra of HPLC effluents on line, and mass spectra of deuterium-exchanged nucleosides, were used to distinguish isomers. All nucleoside assignments shown were made primarily on the basis of their mass spectra and relative HPLC retention times (39) and are considered definitive. Eighteen ribonucleosides were detected in digested tRNA isolates from M. burtonii and 34 were detected in tRNA from S. hydrogenophila, of which one in M. burtonii (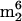 A) and three in S. hydrogenophila (o8G,
A) and three in S. hydrogenophila (o8G, 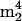 Cm, and
Cm, and 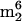 A) were judged not to be native nucleosides from tRNA (see below). Therefore, a total of 17 nucleosides from M. burtonii tRNA and 31 nucleosides from S. hydrogenophila tRNA were characterized. All isolates were found to contain 2′-deoxyribonucleosides resulting from DNA impurities, which were readily identified from their characteristic mass spectra and relative retention times (39).
A) were judged not to be native nucleosides from tRNA (see below). Therefore, a total of 17 nucleosides from M. burtonii tRNA and 31 nucleosides from S. hydrogenophila tRNA were characterized. All isolates were found to contain 2′-deoxyribonucleosides resulting from DNA impurities, which were readily identified from their characteristic mass spectra and relative retention times (39).
TABLE 1.
Summary of ribonucleosides identified in unfractionated tRNA
| Nucleoside | Result for organism at growth temp
|
|||
|---|---|---|---|---|
| Methanoc- coides burtonii (4°C) | Methanoc- coides burtonii (23°C) | Stetteria hydro- genophila (90°C) | Methano- coccus igneus (85°C)a | |
| Uridines | ||||
| D | + | + | ||
| Ψ | + | + | + | + |
| Um | + | + | ||
| m1Ψ | + | + | tr | |
| s4U | + | + | + | |
| Cytidines | ||||
| s2C | + | |||
| m5C | + | + | ||
| m5Cm | + | |||
| Cm | + | + | + | + |
| ac4C | + | |||
| ac4Cm | + | |||
| Adenosines | ||||
| m1A | + | + | + | + |
| m2A | + | |||
| m6A | + | + | + | + |
| t6A | + | + | + | + |
| hn6A | + | + | + | + |
| hn6A + CH3b | + | |||
| ms2t6A | + | + | ||
| ms2hn6A | + | + | + | |
| Am | + | + | ||
| I | + | + | ||
| m1I | + | + | ||
| Guanosines | ||||
| m1G | + | + | + | + |
| m2G | + | + | + | + |
| m7G | + | + | + | |
| Gm | + | + | + | + |
| m22G | + | + | + | + |
| m2Gm | + | + | ||
| m22Gm | + | + | ||
| m3Gb | + | |||
| m2,7Gm | + | |||
| imG | + | + | ||
| imG2b | + | |||
| imG*b | + | + | + | |
| mimG | + | |||
| Other nucleosides | ||||
| G+ | + | + | + | |
| Unknown N415b | + | |||
From reference 32.
Complete structure unknown.
M. burtonii nucleosides.
No qualitative difference in identities of nucleosides between the cultures grown at 4 and 23°C was observed. Relative modification levels from 4 and 23°C cultures were measured as 1.9 and 2.6%, respectively. In consideration of the low levels of modification involved, the difference between these values is not considered significant. Likewise, no differences in the modification patterns exhibited in Fig. 1A and 1B are apparent. Ion current responses from dihydrouridine and uridine at the two culture temperatures are shown in Fig. 3. The relative D levels of 1.5% (4°C) and 1.7% (23°C) are experimentally indistinguishable.
The detection of D, a modified nucleoside having essentially no UV-absorbing chromophore and thus not detectable with a conventional UV detector, was based on mass analysis using the MH+ ion, m/z 247 (Fig. 3A and C). For this purpose a C30 reversed-phase HPLC column was used in which D (Mr, 246) separates completely from Ψ (Mr, 244). Use of this column avoids overlapping (in time) signals from D and the second isotope peak of Ψ (both m/z 247), as found with the more commonly used C18 reversed-phase system (39). The D/uridine ion current ratios shown in Fig. 3 reflect an average D level of between one and two residues per tRNA molecule. This estimate is based on comparison with D and uridine ion current signals from unfractionated Escherichia coli tRNA in which the amounts of the two nucleosides had been accurately determined by a stable-isotope dilution method (11) and the assumption that uridine contents of E. coli and M. burtonii tRNAs are similar.
s4U (component 7) was distinguished from the 2-thio isomer by its UV spectrum: λmax, 331 nm for component 7; the values in the literature (21) are 331 nm for s4U and 275 nm for s2U. The 17-min HPLC peak in Fig. 1A and B is composed of two nucleoside isomers (components 8 and 9) with the same molecular mass (Mr, 297) corresponding to monomethyl guanosines and having essentially the same retention times (39). They are recognized and assigned as m1G and Gm, as indicated by presence of both protonated base ions m/z 166 and 152, respectively, occurring within approximately the same elution profile (7). Similarly, the hypermodified nucleoside G+ (component 11 in Fig. 1) elutes in the leading edge of a much larger amount of 2-deoxyadenosine from DNA impurity but is easily mass detected by using its unusual molecular ion (MH+, m/z 325) and base ion (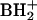 , m/z 193) mass values. The complex amino acid-containing nucleoside hn6A (component 16 in Fig. 1) was distinguished from an isomer known in bacterial tRNA (m6t6A), which differs from hn6A by the placement of a methyl group in the amino acid side chain (40). Differentiation between the two isomers was based on further fragmentation of the base ion (
, m/z 193) mass values. The complex amino acid-containing nucleoside hn6A (component 16 in Fig. 1) was distinguished from an isomer known in bacterial tRNA (m6t6A), which differs from hn6A by the placement of a methyl group in the amino acid side chain (40). Differentiation between the two isomers was based on further fragmentation of the base ion (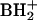 , m/z 295), whose mass is common to both possibilities. Formation of m/z 136 from adenine and m/z 134 from protonated hydroxynorvaline, rather than m/z 150 (methyladenine) and m/z 120 (threonine), as would be required for the isomer m6t6A, indicated the identity of component 16 as hn6A. One nucleoside with a partially known structure was encountered in M. burtonii tRNA, designated imG* in Fig. 1 and Table 1. This nucleoside (Mr, 321), previously found in tRNAs from five methanococci (32), is a member of the imG tricyclic nucleoside family, as shown by its characteristic UV spectrum (26) (UV λmax, 228 and 282 nm). The molecular mass corresponds to a nucleoside having two fewer methyl groups than the archaeal nucleoside mimG (Mr, 349), which has three methyls (31). The structure of nucleoside imG* is under investigation. Component 17 in Fig. 1,
, m/z 295), whose mass is common to both possibilities. Formation of m/z 136 from adenine and m/z 134 from protonated hydroxynorvaline, rather than m/z 150 (methyladenine) and m/z 120 (threonine), as would be required for the isomer m6t6A, indicated the identity of component 16 as hn6A. One nucleoside with a partially known structure was encountered in M. burtonii tRNA, designated imG* in Fig. 1 and Table 1. This nucleoside (Mr, 321), previously found in tRNAs from five methanococci (32), is a member of the imG tricyclic nucleoside family, as shown by its characteristic UV spectrum (26) (UV λmax, 228 and 282 nm). The molecular mass corresponds to a nucleoside having two fewer methyl groups than the archaeal nucleoside mimG (Mr, 349), which has three methyls (31). The structure of nucleoside imG* is under investigation. Component 17 in Fig. 1, 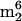 A, is a common minor product in tRNA preparations, arising as an easily detected impurity derived from 16S rRNA, in which its presence is unique and ubiquitous in highly conserved sequences (48, 49). Unannotated peaks in Fig. 1 were shown from their mass spectra not to be nucleosides.
A, is a common minor product in tRNA preparations, arising as an easily detected impurity derived from 16S rRNA, in which its presence is unique and ubiquitous in highly conserved sequences (48, 49). Unannotated peaks in Fig. 1 were shown from their mass spectra not to be nucleosides.
S. hydrogenophila nucleosides.
Nucleosides released by hydrolysis of S. hydrogenophila tRNA are represented in a chromatogram of unusual complexity (Fig. 2). o8G (component 7 in Fig. 2) coelutes with m7G (MH+, m/z 298; BH2+, m/z 168) and was identified following deuterium labeling (m/z 300 shift to 308; m/z 168 shift to 174), which showed seven exchangeable hydrogen atoms in the neutral molecule, one more than in guanosine. Component 7 therefore exhibits 16 Da of modification in the base compared with guanosine and no modification in ribose. Assignment of the likely structure as o8G was confirmed by rigorous comparison against authentic material (gift of T. Hashizume, University of Utah): HPLC retention time, 13.7 min; MS (MH+, m/z 300; 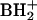 , m/z 168). This unexpected component is viewed as a likely product of adventitious oxidation of guanosine, as might result from xenobiotic or metabolic oxidation processes (for discussion and leading references to the sources of this nucleoside in RNA, see references 23 and 52). Support for what we view as the unlikely authenticity of o8G as being native to tRNA would require at a minimum its localization to specific tRNA sequence sites, which was not undertaken. Similar observations and conclusions regarding trace amounts of o8G in some archaeal hyperthermophile tRNAs have been previously made (J. A. McCloskey and K. O. Stetter, unpublished results).
, m/z 168). This unexpected component is viewed as a likely product of adventitious oxidation of guanosine, as might result from xenobiotic or metabolic oxidation processes (for discussion and leading references to the sources of this nucleoside in RNA, see references 23 and 52). Support for what we view as the unlikely authenticity of o8G as being native to tRNA would require at a minimum its localization to specific tRNA sequence sites, which was not undertaken. Similar observations and conclusions regarding trace amounts of o8G in some archaeal hyperthermophile tRNAs have been previously made (J. A. McCloskey and K. O. Stetter, unpublished results).
The assignments of site of cytosine methylation in components 2 and 9 as position C-5 (i.e., m5C and m5Cm) rather than as the likewise biologically plausible N4- or N-3 methyl isomers, were made by deuterium labeling. Component 2 showed five exchangeable hydrogens in the neutral molecule and three in the neutral base, while component 9 showed four and three, respectively. Those values mandate methylation at C-5. Substitution at heteroatom positions (N4-or N-3) would result in one fewer exchangeable hydrogen in each case.
The unusual diversity of modifications exhibited by S. hydrogenophila tRNA includes m2A, not previously reported for any type of archaeal RNA (34). Deuterium exchange experiments were used to confirm the assignments shown for the methyladenosine isomers m2A and m6A (Fig. 2) made initially by relative retention times and mass spectra. Their isotopic exchange patterns likewise differ due to C versus N alkylation sites, respectively: m2A contains five exchangeable hydrogens in the neutral molecule (component 21), while m6A contains four (component 22).
The presence of both methylguanosine isomers m1G and Gm (components 11 and 12 in Fig. 2) was established by using the base ions as in the case of M. burtonii (Fig. 1) described previously. All other assignments of multiple components eluting in unresolved HPLC peaks were made on the basis of molecular ion and base ion mass values and relative retention times, as appropriate (39).
Three additional nucleosides with partially known structures and one with an unknown structure were found in the S. hydrogenophila digest. In all four cases, the characteristic 132- mass-unit spacing between MH+ and 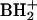 ions (due to loss of ribose) was used to identify each component as a ribonucleoside, without a substitution in ribose. Component 16 (Fig. 1) (UV λmax, 261.5 nm) is judged from its molecular mass (Mr, 325) and base mass as likely to be a base-trimethylated derivative of guanosine. The potential mass isomer acetylguanosine (not a known RNA component) cannot be excluded, although its elution position would likely be much later. Component 29 is assigned as a methyl homolog of the complex amino acid-containing nucleoside hn6A (40), based on a molecular mass of 440, which is 14 higher than that of hn6A (Mr, 426). Upon deuterium exchange the MH+ ion m/z 441 shifted to m/z 449, requiring seven exchangeable hydrogens in the neutral molecule. This shift value is the same as for nucleoside hn6A, consistent with substitution by the additional methyl group to be on carbon rather than a heteroatom (N or O).
ions (due to loss of ribose) was used to identify each component as a ribonucleoside, without a substitution in ribose. Component 16 (Fig. 1) (UV λmax, 261.5 nm) is judged from its molecular mass (Mr, 325) and base mass as likely to be a base-trimethylated derivative of guanosine. The potential mass isomer acetylguanosine (not a known RNA component) cannot be excluded, although its elution position would likely be much later. Component 29 is assigned as a methyl homolog of the complex amino acid-containing nucleoside hn6A (40), based on a molecular mass of 440, which is 14 higher than that of hn6A (Mr, 426). Upon deuterium exchange the MH+ ion m/z 441 shifted to m/z 449, requiring seven exchangeable hydrogens in the neutral molecule. This shift value is the same as for nucleoside hn6A, consistent with substitution by the additional methyl group to be on carbon rather than a heteroatom (N or O).
Two nucleosides observed are considered likely to arise as minor impurities from small-subunit rRNA: 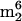 A and
A and 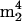 Cm (components 33 and 19 in Fig. 2). The arguments concerning the origin of
Cm (components 33 and 19 in Fig. 2). The arguments concerning the origin of 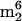 A are presented above. The novel trimethylcytidine
A are presented above. The novel trimethylcytidine 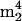 Cm was recently discovered and characterized in trace amounts in a tRNA isolate from the hyperthermophilic archaeon Aeropyrum pernix; it was concluded to arise as an impurity from 16S rRNA, where it was found in greater abundance (R. Van Wagoner, E. Bruenger, G. M. Caballero, N. Nomura, Y. Sako, and J. A. McCloskey, unpublished data), and is therefore similarly assigned in the present case to rRNA.
Cm was recently discovered and characterized in trace amounts in a tRNA isolate from the hyperthermophilic archaeon Aeropyrum pernix; it was concluded to arise as an impurity from 16S rRNA, where it was found in greater abundance (R. Van Wagoner, E. Bruenger, G. M. Caballero, N. Nomura, Y. Sako, and J. A. McCloskey, unpublished data), and is therefore similarly assigned in the present case to rRNA.
DISCUSSION
The overall differences found between tRNA modifications in the two archaea studied are dramatic, particularly in three respects. First, the modification level in the psychrotolerant archaeon M. burtonii of approximately 2% is the lowest of which we are aware in any organism, including the bacterial psychrophile ANT-300 (Table 2). This comparison also considers Mycoplasma capricolum tRNAs, which are often considered undermodified, based on the complete set of published tRNA sequences (3). Within the domain Archaea, and based on the same measurement criteria, modification of M. burtonii tRNA is significantly lower than in the mesophilic archaeal methanogen Methanococcus maripaludis (32). We conclude that tRNA adaptation to cold in archaea parallels, and could be more pronounced than, that observed for cold-adapted bacteria (10), but additional organisms should be studied. (Comments on the differences between two culture temperatures in M. burtonii are presented below.)
TABLE 2.
Structural diversity and modification levels of nucleosides in selected unfractionated tRNAs
| Organism | Culture temp (°C) | No. of modified tRNA nucleosides detected | % Modifi- cationa | Reference |
|---|---|---|---|---|
| Methanococcoides burtonii | 4 | 17b | 1.9 | This study |
| Methanococcoides burtonii | 23 | 17b | 2.6 | This study |
| Stetteria hydrogenophila | 90 | 31c | 13 | This study |
| Methanococcus maripaludis | 37 | 23 | 9.3 | 32 |
| Methanococcus igneus | 85 | 24 | 9.6 | 32 |
| Escherichia coli | 37 | 26 | 5.1 | 21; this study |
| ANT-300d | 5 | 13 | ∼6.6e | 10 |
Crude estimate based on HPLC peak areas; see the text.
One additional nucleoside was detected (m26A) and judged to be not native to tRNA; see the text.
Three additional nucleosides were detected and judged to be not native to tRNA (o8G, m24Cm, and m26A); see the text.
Estimate calculated by using unpublished HPLC chromatograms associated with an earlier report (10).
By contrast, the 13% tRNA modification level found in the hyperthermophile S. hydrogenophila is exceptional, compared for example to that in the archaeal thermophile Methanococcus igneus (Table 2), which grows at 85°C. These observations support the overall thesis that posttranscriptional modification in tRNA serves in part to provide crucial elements of control in fine-tuning of tertiary structure for optimal function (16, 43), which is of paramount importance in thermophiles (27, 50) but less so at significantly lower temperatures (10), as in the notable case of M. burtonii.
It is important to note that the overall extent of modification is also subject to the influence of phylogeny, the effects of which cannot always be clearly distinguished from those associated with temperature. An unusual but notable example is the similarity of modification levels between M. maripaludis and M. igneus (Table 2), which grow optimally at very different temperatures. However, if the levels of specific modifications closely tied to thermostability are considered, a clearer picture of temperature effects emerges: e.g., the trimethylated guanosine 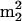 Gm, believed to be a stabilizing entity at tRNA stem junctions, is present in M. igneus (85°C) but absent in M. maripaludis (37°C) (32). (Further discussion of
Gm, believed to be a stabilizing entity at tRNA stem junctions, is present in M. igneus (85°C) but absent in M. maripaludis (37°C) (32). (Further discussion of 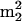 Gm is given below in the section on S. hydrogenophila.) For comparison of organisms not closely related the phylogeny, temperature distinction is also confounded by the present lack of sufficient data to gain a refined picture, particularly with regard to bacterial thermophiles, for comparison with a large body of data on archaeal thermophiles (17, 27, 33).
Gm is given below in the section on S. hydrogenophila.) For comparison of organisms not closely related the phylogeny, temperature distinction is also confounded by the present lack of sufficient data to gain a refined picture, particularly with regard to bacterial thermophiles, for comparison with a large body of data on archaeal thermophiles (17, 27, 33).
The differences between M. burtonii and S. hydrogenophila in overall modification, when considered in terms of the numbers of structural motifs employed at the nucleoside level (Table 2), are similarly great. The finding of 31 different modified nucleosides in S. hydrogenophila tRNAs is the greatest number of which we are aware, based on earlier reports of tRNA modification in various archaeal hyperthermophiles (17, 32, 33) and eukarya, including unfractionated calf liver tRNA (21). The occurrence of nine different methylated guanosines is particularly striking. The structural diversity of nucleosides in M. burtonii, 17 modified nucleosides, is greater than that in the bacterium ANT-300 but still significantly below that of other representative organisms (Table 2). The strong apparent correlation between temperature of growth and number of modification motifs recruited by the RNA-processing enzymes seems to be more pronounced at higher temperatures, at least within the archaea. This probably reflects the essential need for mechanisms of tRNA stabilization at growth temperatures generally above the melting points of unmodified tRNA base-paired stems (a summary of Tm values in relation to G+C content and the role of modification can be found in reference 27). Additional comparisons of modification patterns, between closely related archaeal mesophiles and thermophiles from selected methanococci, have been reported (32).
M. burtonii is shown (Fig. 3) to contain significant amounts of D, a nucleoside associated with maintenance of polynucleotide flexibility in RNA (12). This modification is absent in S. hydrogenophila and in most other archaeal hyperthermophiles and mesophiles examined (17), unlike its wide occurrence in eukarya and bacteria, including the bacterial thermophiles (45). Within archaea, the finding of D in abundance only in M. burtonii supports and extends the interpretation of its functional importance in psychrophilic bacteria (10). The occurrence of more than one D residue per tRNA molecule in M. burtonii is about the same as that in E. coli (1.79 mol%) (11) but is greater than that in other archaea studied to date. Thus, posttranscriptional enzymatic processing to form D appears to be a common mechanism for adaptation to the cold in both archaea and bacteria.
Quantitative data for D incorporation is not available for other cold-adapted archaea. Recently, however, putative D synthase genes were identified in the partial genome sequences of M. burtonii and the psychrophilic methanogen Methanogenium frigidum (44). This indicates that D incorporation may be a general characteristic of cold-adapted archaea. No convincing evidence was obtained that levels of D are responsive to a decrease in growth temperature (Fig. 3), although it will be interesting to examine regulation of expression of the D synthase gene.
We know of no previous effort to document the presence versus the absence of D or the level of D as a function of temperature in tRNA from any given organism. Thus, there is no evidence for a response opposite to that found for bacterial (2, 51) and archaeal (27) thermophiles (increase in abundance of selected stabilizing modifications with culture temperature). However, it was shown that in small D-containing oligonucleotides the fractional population of the inherently more flexible C-2′-endo ribose conformer of D increases at lower temperatures (12). Therefore, in contrast to a presence-versus-absence mechanism for preservation of conformational flexibility at lower temperatures, the influence of D in those organisms may reside to some extent in a thermodynamic preference for favored ribose conformers in existing D residues present in the D and T loops.
Both M. burtonii and S. hydrogenophila contain tRNA modifications that have been formed potentially under the direction of small RNAs known under the generic name of guide RNAs (15, 38). These modifications are pseudouridines (in both organisms) and ribose-methylated nucleosides (in S. hydrogenophila). Modification targeting by sRNAs has been studied mostly with regard to rRNA modification in eukarya but has been reported to operate also in some archaeal tRNAs (15, 38, 46). The extent to which these mechanisms of modification operate in the present case is unknown, and progress is confounded by the absence of knowledge regarding isoacceptor tRNA sequences, which remains a worthwhile area for future investigation.
Modifications in M. burtonii.
The strikingly low levels of tRNA modification that occur in M. burtonii include nine nucleosides that are common to RNA in all three phylogenetic domains (34) (Archaea, Bacteria, and Eukarya) and thus constitute a minimalist core of modifications that probably evolved prior to the separation of these domains. These modified species are D, Ψ, and the simple monomethylated nucleosides Cm, Gm, m1G, m7G, m1A, and m6A. Two of the modifications, m1Ψ and G+, are unique hallmarks of archaeal modification that occur widely but are each highly conserved at single sequence locations, tRNA positions 54 and 15, respectively (45). The remaining nucleosides are evenly divided from the perspective of phylogenetic commonality: s4U, hn6A, and ms2hn6A are shared with bacteria, while the guanosine derivatives m2G, 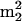 G, and imG* are otherwise characteristically eukaryotic (34). Notably absent in tRNA from M. burtonii is the complex anticodon nucleoside mnm5s2U, recently detected in four mesophilic and thermophilic methanococci (32). However, mnm5s2U is usually considered a bacterial modification (in tRNA-Glu and -Lys [45]) which is otherwise unknown in archaeal tRNA and may be restricted only to the methanococci. Overall, these findings substantially add to our limited understanding of the mechanisms of adaptation in psychrotolerant archaea (6).
G, and imG* are otherwise characteristically eukaryotic (34). Notably absent in tRNA from M. burtonii is the complex anticodon nucleoside mnm5s2U, recently detected in four mesophilic and thermophilic methanococci (32). However, mnm5s2U is usually considered a bacterial modification (in tRNA-Glu and -Lys [45]) which is otherwise unknown in archaeal tRNA and may be restricted only to the methanococci. Overall, these findings substantially add to our limited understanding of the mechanisms of adaptation in psychrotolerant archaea (6).
Modifications in S. hydrogenophila.
A significant fraction of the unusual number and diversity of modifications in S. hydrogenophila, 9 of 31 nucleosides, are modified by methylation at O-2′ of ribose and are therefore assumed to exert influences through points of regional stabilization of the folded tRNA molecule. This effect is attributed to thermodynamic stabilization of the C-3′-endo sugar conformation, thus avoiding base-O-2′ steric interactions in the alternate C-2′-endo conformation (14), resulting in higher Tms (9) and reduced dynamic motion at elevated temperatures. Previous studies have demonstrated increased levels of ribose methylation as a function of culture temperature, in both tRNA (2, 27, 28) and rRNA (37). Two nucleosides found in S. hydrogenophila, ac4Cm and 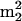 Gm (and the related modification m2Gm), were reported to be temperature responsive in tRNA from the archaeal hyperthermophile Pyrococcus furiosus (27). The family of structurally related nucleosides, m2G,
Gm (and the related modification m2Gm), were reported to be temperature responsive in tRNA from the archaeal hyperthermophile Pyrococcus furiosus (27). The family of structurally related nucleosides, m2G, 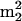 G, m2Gm, and
G, m2Gm, and 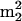 Gm, are from known archaeal tRNA sequences, conserved at only two locations, positions 10 and 26 (45). Nucleotides at these locations are at the junctions of the acceptor and D-loop stems and of the D-loop and anticodon stems, respectively, where they play crucial roles in the control and stabilization of the tertiary L fold of the tRNA (42). The unusual abundance of
Gm, are from known archaeal tRNA sequences, conserved at only two locations, positions 10 and 26 (45). Nucleotides at these locations are at the junctions of the acceptor and D-loop stems and of the D-loop and anticodon stems, respectively, where they play crucial roles in the control and stabilization of the tertiary L fold of the tRNA (42). The unusual abundance of 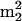 Gm in Fig. 2, taking UV molar absorptivities into account, corresponds to an average of approximately 0.6
Gm in Fig. 2, taking UV molar absorptivities into account, corresponds to an average of approximately 0.6 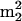 Gm residues per tRNA molecule, while the estimated sum of the four modifications is approximately two residues per molecule. This implies that essentially all tRNAs from S. hydrogenophila are modified at both positions, in contrast to M. burtonii (Fig. 1, components 10 and 12), in which an average of only ∼0.2 residues per molecule are modified in this fashion. This result further extends the implication of these residues in tRNA stabilization in the archaeal thermophiles (27, 32) and their greatly reduced role in tRNA from M. burtonii.
Gm residues per tRNA molecule, while the estimated sum of the four modifications is approximately two residues per molecule. This implies that essentially all tRNAs from S. hydrogenophila are modified at both positions, in contrast to M. burtonii (Fig. 1, components 10 and 12), in which an average of only ∼0.2 residues per molecule are modified in this fashion. This result further extends the implication of these residues in tRNA stabilization in the archaeal thermophiles (27, 32) and their greatly reduced role in tRNA from M. burtonii.
Acknowledgments
We are grateful to J. Barciszewski for discussion of guanosine oxidation in RNA, to T. Cole for information on use of C30 reversed-phase HPLC columns for nucleoside analysis, to S. C. Pomerantz for preparation of figures, and to H. Ertan and T. Thomas for comments during preparation of the manuscript.
This study was supported by grants from the National Institute of General Medical Sciences (J.A.M.), by the Fonds der Chemischen Industrie (M.T.), and by grants from the Australian Research Council (R.C.).
REFERENCES
- 1.Agris, P. F. 1996. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol. 53:79-129. [DOI] [PubMed] [Google Scholar]
- 2.Agris, P. F., P. Koh, and D. Söll. 1973. The effect of growth temperature on the in vivo ribose methylation of Bacillus stearothermophilus. Arch. Biochem. Biophys. 154:277-282. [DOI] [PubMed] [Google Scholar]
- 3.Andachi, Y., F. Yamao, A. Muto, and S. Osawa. 1989. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. J. Mol. Biol. 209:37-54. [DOI] [PubMed] [Google Scholar]
- 4.Auffinger, P., and E. Westhof. 1998. Location and distribution of modified nucleotides in tRNA, p. 569-576. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 5.Buck, M., M. Connick, and B. N. Ames. 1983. Complete analysis of tRNA-modified nucleosides by high performance liquid chromatography: the 29 modified nucleosides of Salmonella typhimurium and Escherichia coli tRNA. Anal. Biochem. 129:1-13. [DOI] [PubMed] [Google Scholar]
- 6.Cavicchioli, R., T. Thomas, and P. M. G. Curmi. 2000. Cold stress response in archaea. Extremophiles 4:321-331. [DOI] [PubMed] [Google Scholar]
- 7.Crain, P. F. 1998. Detection and structure analysis of modified nucleosides in RNA by mass spectrometry, p. 47-57. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 8.Crain, P. F. 1990. Preparation and enzymatic hydrolysis of RNA and DNA for mass spectrometry. Methods Enzymol. 193:782-790. [DOI] [PubMed] [Google Scholar]
- 9.Cummins, L. L., S. R. Owens, L. M. Risen, E. A. Lesnik, S. M. Freier, D. McGee, C. J. Guinosso, and P. D. Cook. 1995. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 23:2019-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalluge, J. J., T. Hamamoto, K. Horikoshi, R. Y. Morita, K. O. Stetter, and J. A. McCloskey. 1997. Posttranscriptional modification of transfer RNA in psychrophilic bacteria. J. Bacteriol. 179:1918-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalluge, J. J., T. Hashizume, and J. A. McCloskey. 1996. Quantitative measurement of dihyrouridine in RNA using isotope dilution chromatography-mass spectrometry (LC/MS). Nucleic Acids Res. 24:3242-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalluge, J. J., T. Hashizume, A. E. Sopchik, J. A. McCloskey, and D. R. Davis. 1996. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res. 24:1073-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davanloo, P., M. Sprinzl, K. Watanabe, M. Albani, and H. Kersten. 1979. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res. 6:1571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, D. R. 1998. Biophysical and conformational properties of modified nucleosides in RNA (nuclear magnetic resonance studies), p. 85-102. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 15.Dennis, P. P., A. Omer, and T. Lowe. 2001. A guided tour: small RNA function in Archaea. Mol. Microbiol. 40:509-519. [DOI] [PubMed] [Google Scholar]
- 16.Derrick, W. B., and J. Horowitz. 1993. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA: evidence for stable base modification-dependent conformers. Nucleic Acids Res. 21:4948-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edmonds, C. G., P. F. Crain, R. Gupta, T. Hashizume, C. H. Hocart, J. A. Kowalak, S. C. Pomerantz, K. O. Stetter, and J. A. McCloskey. 1991. Posttranscriptional modification of transfer RNA in thermophilic archaea. J. Bacteriol. 173:3138-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edmonds, C. G., S. C. Pomerantz, F. F. Hsu, and J. A. McCloskey. 1988. Thermospray liquid chromatography/mass spectrometry in deuterium oxide. Anal. Chem. 60:2314-2317. [DOI] [PubMed] [Google Scholar]
- 19.Felden, B., K. Hanawa, J. F. Atkins, H. Himeno, A. Muto, R. F. Gesteland, J. A. McCloskey, and P. F. Crain. 1998. Presence and location of modified nucleotides in E. coli tmRNA: structural mimicry with tRNA acceptor branches. EMBO J. 17:3188-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franzmann, P. D., N. Springer, W. Ludwig, E. Conway de Macario, and M. Rohde. 1992. A methanogenic archaeon from Ace Lake, Antarctica: Methanococcoides burtonii sp. nov. Syst. Appl. Microbiol. 15:573-581. [Google Scholar]
- 21.Gehrke, C. W., and K. C. Kuo. 1990. Ribonucleoside analysis by reversed-phase high performance liquid chromatography, p. A3-A64. In C. W. Gehrke and K. C. Kuo (ed.), Chromatography and identification of nucleosides, part A. Journal of chromatography library, vol. 45A. Elsevier, New York, N.Y. [DOI] [PubMed]
- 22.Hall, R. 1971. The modified nucleosides in nucleic acids, p. 17-207. Columbia University Press, New York, N.Y.
- 23.Hayakawa, H., M. Kuwano, and M. Sekiguchi. 2001. Specific binding of 8-oxoguanine-containing RNA to polynucleotide phosphorylase protein. Biochemistry 40:9977-9982. [DOI] [PubMed] [Google Scholar]
- 24.Horie, N., M. Hara-Yokoyama, S. Yokoyama, K. Watanabe, Y. Kuchino, S. Nishimura, and T. Miyazawa. 1985. Two tRNA Ile species from an extreme thermophile, Thermus thermophilus HB8: effect of 2-thiolation of ribothymidine on the thermostability of tRNA. Biochemistry 24:5711-5715. [DOI] [PubMed] [Google Scholar]
- 25.Jochimsen, B., S. Peinemann-Simon, H. Völker, D. Stüben, R. Botz, P. Stoffers, P. R. Dando, and M. Thomm. 1997. Stetteria hydrogenophila, gen. nov. and sp. nov., a novel mixotrophic sulfur-dependent crenarchaeote isolated from Milos, Greece. Extremophiles 1:67-73. [DOI] [PubMed] [Google Scholar]
- 26.Kasai, H., M. Goto, K. Ikeda, M. Zama, Y. Mizuno, S. Takemura, S. Matsuura, T. Sugimoto, and T. Goto. 1976. Structure of wye (Yt base) and wyosine (Yt) from Torulopsis utilis phenylalanine transfer ribonucleic acid. Biochemistry 15:898-904. [DOI] [PubMed] [Google Scholar]
- 27.Kowalak, J. A., J. J. Dalluge, J. A. McCloskey, and K. O. Stetter. 1994. Role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33:7869-7876. [DOI] [PubMed] [Google Scholar]
- 28.Kumagi, I., K. Watanabe, and T. Oshima. 1980. Thermally induced biosynthesis of 2′-O-methylguanosine in tRNA from an extreme thermophile, Thermus thermophilus HB27. Proc. Natl. Acad. Sci. USA 77:1922-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Limbach, P. A., P. F. Crain, and J. A. McCloskey. 1994. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 22:2183-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCloskey, J. A. 2001. Appendix: modified nucleosides from RNA, p. 309-316. In D. Söll, S. Nishimura, and P. B. Moore (ed.), RNA. Elsevier, Amsterdam, The Netherlands.
- 31.McCloskey, J. A., P. F. Crain, C. G. Edmonds, R. Gupta, T. Hashizume, D. W. Phillipson, and K. O. Stetter. 1987. Structure determination of a new fluorescent tricyclic nucleoside from archaebacterial tRNA. Nucleic Acids Res. 15:683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCloskey, J. A., D. E. Graham, S. Zhou, P. F. Crain, M. Ibba, J. Konisky, D. Söll, and G. J. Olsen. 2001. Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 29:4699-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCloskey, J. A., X.-H. Liu, P. F. Crain, E. Bruenger, R. Guymon, T. Hashizume, and K. O. Stetter. 2000. Posttranscriptional modification of transfer RNA in the submarine hyperthermophile Pyrolobus fumarii. Nucleic Acids Symp. Ser. 44:267-268. [DOI] [PubMed] [Google Scholar]
- 34.Motorin, Y., and H. Grosjean. 1998. Appendix 1: chemical structures and classification of posttranscriptionally modified nucleosides in RNA, p. 543-549. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 35.Moyer, C. L., and R. Y. Morita. 1989. Effect of growth rate and starvation-survival on the viability and stability of a psychrophilic marine bacterium. Appl. Environ. Microbiol. 55:1122-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moyer, C. L., J. M. Tiedje, F. C. Dobbs, and D. M. Karl. 1996. A computer-simulated restriction fragment length polymorphism analysis of bacteria small-subunit rRNA genes: efficiency of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl. Environ. Microbiol. 62:2501-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noon, K. R., E. Bruenger, and J. A. McCloskey. 1998. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J. Bacteriol. 180:2883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omer, A. D., S. Ziesche, W. A. Decatur, M. J. Fournier, and P. P. Dennis. 2003. RNA-modifying machines in archaea. Mol. Microbiol. 48:617-629. [DOI] [PubMed] [Google Scholar]
- 39.Pomerantz, S. C., and J. A. McCloskey. 1990. Analysis of RNA hydrolyzates by LC/MS. Methods Enzymol. 193:796-824. [DOI] [PubMed] [Google Scholar]
- 40.Reddy, D. M., P. F. Crain, C. G. Edmonds, R. Gupta, T. Hashizume, K. O. Stetter, F. Widdel, and J. A. McCloskey. 1992. Structure determination of two new amino acid-containing derivatives of adenosine from tRNA of thermophilic bacteria and archaea. Nucleic Acids Res. 20:5607-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozenski, J., P. F. Crain, and J. A. McCloskey. 1999. The RNA modification database—1999 update. Nucleic Acids Res. 27:196-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saenger, W. 1984. Principles of nucleic acid structure, p. 334-337. Springer-Verlag, New York, N.Y.
- 43.Sampson, J. R., and O. C. Uhlenbeck. 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. USA 85:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saunders, N. F. W., T. Thomas, P. M. G. Curmi, J. S. Mattick, E. Kuczek, R. Slade, J. Davis, P. D. Franzmann, D. Boone, K. Rusterholtz, R. Feldman, C. Gates, S. Bench, K. Sowers, K. Kadner, A. Aerts, P. Dehal, C. Detter, T. Glavina, S. Lucas, P. Richardson, F. Larimer, L. Hauser, M. Land, and R. Cavicchioli. 2003. Mechanisms of thermal adapatation revealed from the genomes of the Antarctic archaea Methanogenium frigidum and Methanococcoides burtonii. Genome Res. 13:1580-1588. [DOI] [PMC free article] [PubMed]
- 45.Sprinzl, M., C. Horn, M. Brown, A. Ioudovitch, and S. Steinberg. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang, T.-H., J.-P. Bachellerie, T. Rozhdestvensky, M.-L. Bortolin, H. Huber, M. Drungowski, T. Elge, J. Brosius, and A. Hüttenhofer. 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA 99:7536-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas, T., N. Kumar, and R. Cavicchioli. 2001. Effects of ribosomes and intracellular solutes on activities and stabilities of elongation factor 2 proteins from psychrotolerant and thermophilic methanogens. J. Bacteriol. 183:1974-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Knippenberg, P. H. 1985. Structural and functional aspects of the N6,N6-dimethyladenosine in 16S ribosomal RNA, p. 412-424. In B. Hardesty and G. Kramer (ed.), Structure, function, and genetics of ribosomes. Springer-Verlag, New York, N.Y.
- 49.van Knippenberg, P. H., J. M. Van Kimmenade, and H. A. Heus. 1984. Phylogeny of the conserved 3′ terminal structure of the RNA of small ribosomal subunits. Nucleic Acids Res. 12:2595-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe, K., T. Oshima, K. Iijima, Z. Yamaizumi, and S. Nishimura. 1980. Purification and thermal stability of several amino acid-specific tRNAs from an extreme thermophile, Thermus thermophilus HB8. J. Biochem. (Tokyo) 87:1-13. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe, K., M. Shinma, and T. Oshima. 1976. Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem. Biophys. Res. Commun. 72:1137-1144. [DOI] [PubMed] [Google Scholar]
- 52.Yanagawa, H., Y. Ogawa, and M. Ueno. 1992. Redox ribonucleosides. Isolation and characterization of 5-hydroxyuridine, 8-hydroxyguanosine and 8-hydroxyadenosine from Torula yeast RNA. J. Biol. Chem. 267:13320-13326. [PubMed] [Google Scholar]