Abstract
Optical imaging and analysis of single molecules continue to unfold as powerful ways to study the individual behavior of biological systems, unobscured by ensemble averaging. Current expansion of interest in this field is great, as evidenced by new meetings, journal special issues, and the large number of new investigators. Selected recent advances in biomolecular analysis are described, and two new research directions are summarized: superresolution imaging using single-molecule fluorescence and trapping of single molecules in solution by direct suppression of Brownian motion.
Keywords: biophysics, microscopy, superresolution, trapping
Optical fluorescence imaging and analysis of single molecules continues to unfold as a powerful way to study the individual behavior of biological systems, unobscured by ensemble averaging. It has become abundantly clear that the ability to measure the distribution of behaviors, as opposed to only the average behavior, provides insight in cases where static or dynamic heterogeneity is present, such as in a complex system. For biomolecules in particular, a variety of cellular operations occur as individual enzymes work, one by one, on various tasks, and understanding how these individual nanomachines operate is an appealing and exciting challenge.
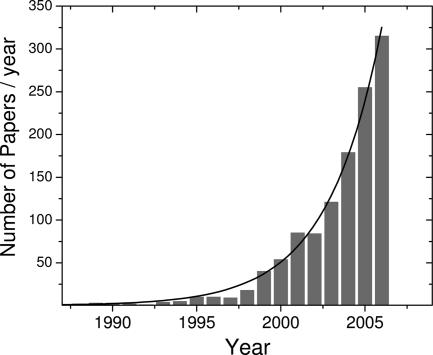
The early single-molecule optical studies in solids at low temperatures (1, 2), followed by extensions to room temperature in the mid-1990s (3–6), laid the groundwork for the broad range of biological studies occurring since the year 2000 (7–9). [Optical trapping methods, in which a dielectric bead is trapped in a laser tweezers device, yield exquisite precision in position and force measurements when a single biomolecule can be tethered to a large bead; work in these areas has been separately reviewed (10–12).] Because the basic methods of single-molecule fluorescence microscopy and spectroscopy are now well known (13–15) and relatively easy to apply with modern microscopes, lasers, and detectors (16), a wealth of new investigators have recently entered the field. Applications to the study of single proteins, DNA, RNA, and enzymes, both inside and outside of cells are numerous and increasing, as illustrated by the wide selection of papers in this special issue. Collections of review articles have also appeared in Annual Review of Physical Chemistry in 2004 (124–127) and in the journals Accounts of Chemical Research (128) and ChemPhysChem in 2005 (129), spanning both biological and nonbiological studies of polymers and materials. One way to illustrate the rapid growth can be generated by a search of the PubMed database for all papers with the words “single molecule” in the title. Although this is only an approximate and very simple measure, the number of such papers per year, plotted in Fig. 1, shows that the field appears to be in an exponential growth phase with a doubling time of 2.2 years.
Fig. 1.
Numbers of papers indexed in the PubMed database with “single molecule” in the title (image courtesy of Taekjip Ha); exponential growth with doubling time of 2.2 years.
Further attesting to the interest and progress in single-molecule studies, in the past few years, several comprehensive symposia at major international meetings have occurred. For example, the Telluride workshop “Single-Molecule Measurements: Theory and Experiment” began in 2005, a critical development, because further progress in extracting the maximum amount of information from single-molecule data depends not only on experimental techniques but also on theoretical models and statistical methods for extracting unbiased information (17). From another point of view, the wealth of new information available as one molecule is observed to fluctuate on a complex energy landscape has in fact stimulated new theoretical approaches too numerous to review here. More broadly, a new Gordon Research Conference on Single Molecule Approaches to Biology was established at Colby–Sawyer College (New London, NH) in the summer of 2006, with biannual meetings planned into the future. Focused symposia at annual meetings of the American Chemical Society (fall 2006 and 2007); biannual single-molecule biophysics meetings in Aspen, CO; and annual meetings in Berlin (Germany) on single-molecule detection have all occurred. These meetings span the areas of enzymatic fluctuations (18), diffusion analysis (19), protein/RNA folding (20, 21), DNA processing (22), DNA sequencing (23), cellular entry (24), and many other biological problems where single-molecule measurements can provide new information.
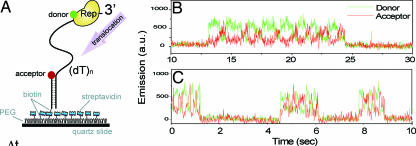
A more specific example of one type of measurement of current interest may be drawn from current work by Taekjip Ha and coworkers on DNA processing enzymes (25, 26). By placing both donor and acceptor fluorophores at precise locations on the molecule of interest, information on distance (and angle) changes between the two fluorophores can be extracted as a function of time through Förster resonant energy transfer (FRET), by now a well known method (14). In Fig. 2A, the basic setup is illustrated, where a single-stranded DNA segment is anchored to a surface, with an acceptor label (red) at the end. The Escherichia coli Rep enzyme with the donor label (green) binds to the distal end of the DNA with a low FRET signal and then slides along the DNA segment until it reaches the acceptor. Interestingly, each time this happens, the Rep enzyme quickly slides back to the end of the DNA, and the process repeats, as shown in the time traces of Fig. 2 B (22°C) and C (37°C). The back-and-forth repetitive shuttling process may be involved in the restarting of a stalled replication fork.
Fig. 2.
Explorations of the sliding behavior of Rep on a single-stranded DNA segment attached to a surface. (A) Schematic of the labeling arrangement for FRET measurements. Traces (B, 22°C; C, 37°C) of donor (green) and acceptor (red) fluorescence signals for a single Rep molecule are shown. [Reproduced with permission from ref. 25 (Copyright 2005, MacMillian Publishers, Ltd.).]
Although the FRET method continues to provide useful insight about spontaneous and active conformational changes, advanced versions of FRET have also been developed (i) where alternating excitation of the donor and the acceptor separately allows cases of absent acceptors to be removed from the analysis (27, 28); or (ii) where, by careful statistical analysis, precise measurement errors can be extracted (29). The single-molecule FRET method continues to be a powerful tool used by many investigators (30–33).
Another area of current interest involves further extensions of single-molecule studies to the interior of living cells. Although tracking of moving single molecules on the plasma membrane or moving in the cytoplasm began some years ago (34–36), many new investigators are stepping up to the additional challenge of imaging in the higher background of the cell (37–40). Naturally, autofluorescent proteins (41–43) are a powerful way to achieve genetically directed labeling and thus to follow intracellular events at the single-molecule level (44–49). A continuing challenge is to identify additional classes of small-molecule labels for improved brightness and readout capability at the single-copy level (50–52). In recent work, the detection of emission from freshly translated GFP or from the activity of an enzyme acting on fluorogenic substrates was used to probe gene expression events (53, 54).
In the remainder of this paper, recent work in two selected areas will be described in slightly more detail. First, methods to simultaneously localize the positions of multiple single fluorophores by precisely determining their individual positions are now yielding impressive gains in resolution for optical microscopy, far beyond the diffraction limit. Second, new techniques to suppress Brownian motion have been devised that allow extended study of single molecules in solution, without attachment to surfaces or entrapment in vesicles or in the pores of gels.
Superresolution Imaging
One continuing driving force in single-molecule fluorescence studies is the study of biomolecules, in vitro and in vivo (55). As is well known, biological fluorescence microscopy depends upon a variety of labeling techniques to light up different structures in cells, but the price often paid for using visible light is the relatively poor spatial resolution compared with x-ray or electron microscopy. The basic problem is that in conventional microscopes, fundamental diffraction effects limit the resolution to a dimension of roughly the optical wavelength λ divided by two times the N.A. of the imaging system, λ/(2×N.A.). Because the largest values of N.A. for state-of-the-art highly corrected microscope objectives are in the range of ≈1.3–1.6, the spatial resolution of optical imaging has been limited to ≈180 nm for visible light of 500-nm wavelength.
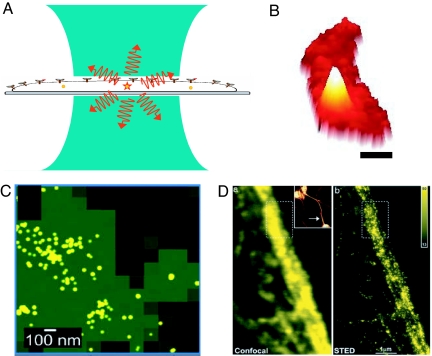
In fact, the light from single fluorescent molecular labels ≈1–2 nm in size provides a way around this problem, that is, a way to provide “superresolution,” or resolution far better than the diffraction limit. How can single molecules help? The sketch in Fig. 3A illustrates the typical imaging problem at room temperature: the single molecule is far smaller than the focused laser spot, yet if only one molecule is pumped, information related to one individual molecule and its local “nanoenvironment” can be extracted by detecting the photons from that molecule alone (56). In terms of spatial resolution, however, when the laser beam is scanned, the observed “peak” from the single-nanoscale source of light maps out the point-spread function (PSF) of the microscope, because the molecule is a nanoscale light absorber, far smaller than the size of the PSF. More specifically, the molecule absorbs light with a probability proportional to the square of the dot product between the local optical electric field and the molecule's transition dipole moment. This point was realized at the very beginning of work in this field, where the fluorescence excitation signal from one molecule was used to map out the size of the focused pumping laser beam (57). Fig. 3B shows this PSF for emission from a single molecule of the bacterial actin protein MreB [Fig. 3B (the white mountain, labeled by fusion to a yellow fluorescent protein) in a bacterial cell (the red crescent shape)] (48). It is this mountain-like image from a single molecular emitter that forms the basis for current superresolution efforts based on single-molecule microscopy. To keep the PSFs from different molecules from overlapping, very low concentrations of the emitting molecule are usually required, although the early low-temperature work in the field used spectral selection to identify the different individual molecules in the same laser focal volume (1, 2).
Fig. 3.
Overview of superresolution imaging. (A) Schematic of a tightly focused laser beam (blue) of diffraction-limited diameter of ≈200 nm irradiating a cell. One molecule is in the focal volume, which emits fluorescence (red). (B) Wide-field fluorescence image of a bacterial cell (red) containing a single protein fusion between the bacterial actin MreB and EYFP (mountain). Acquisition time, 100 ms. (Scale bar, 0.5 μm.) (C) Fluorescence PALM image of PA-GFP molecules on a glass substrate, with green regions showing the approximate blur region of diffraction-limited imaging and yellow dots showing the actual detected positions of the single molecules. [Reproduced with permission from ref. 78 (Copyright 2006, Biophysical Society).] (D) Confocal (Left) and STED (Right) images of a neurofilament in a human neuroblastoma cell labeled by immunofluorescence. [Reproduced with permission from ref. 88 (Copyright 2006, National Academy of Sciences).]
Recently, several researchers have begun to take advantage of the nanoscale size of single-molecule emitters more directly. Simply by measuring the shape of the PSF, the position of its center can be determined much more accurately than its width. This idea, digitizing the PSF, a form of simple deconvolution, has been known for many years, but deconvolution without knowledge of the PSF can generate spurious features in the presence of noise. The knowledge that a single small object is emitting means that a very good estimate of the PSF can be extracted almost trivially just by recording the shape of the detected images. Put another way, the knowledge that only one tiny nanoscale emitter is present allows the experimenter to interpret the center of the PSF as a measurement of the location of the emitter. This idea was applied early on to single-nanoscale fluorescent beads with many emitters (58) and then to low-temperature single-molecule images (59, 60), where both spatial information and the secondary variable, laser wavelength, were used to separate molecules.
Importantly, the accuracy with which a single molecule can be located by digitizing the PSF depends fundamentally upon the Poisson process of photon detection, so the most important variable is the total number of photons detected above background, with a weaker dependence on the size of the detector pixels (61, 62). Clearly, then, emitters with the largest numbers of emitted photons before photobleaching are preferable. A detailed statistical analysis of the measurement process based on Fisher information has been completed (63, 64). Digitization of the PSF for single Cy3 fluorescently labeled myosin molecules was used to extract position information down to a few nanometers by Yildiz et al. (65), and a new acronym was proposed [fluorescence imaging with 1-nm accuracy (FIONA)]. Digitization of the PSF for individual semiconductor quantum dot emitters has allowed observation of time-dependent nanometer-sized steps produced by motors in cells by Nan et al. (66). In fact, the single MreB protein shown in Fig. 3B is part of an unlabeled filament of MreB molecules and, over time, the molecule moves linearly through the filament by a treadmilling process (48). By fitting the sequence of PSFs to Gaussian profiles, an image of the shape of the filament can be obtained with 30-nm resolution.
A variation on the digitization of the PSF for one single molecule occurs when a variable, such as excitation color or wavelength, allows different molecules in the same volume to be separately localized. If this can be done, there is no need to reduce the concentration of single molecules to levels so low that only one molecule is present in the pumped laser volume. This idea was central to the early low-temperature fluorescence excitation work of the early 1990s, where hundreds of molecules in the same volume were separated by excitation wavelength, and was generalized to other variables at room temperature in 1995 (67). By separately imaging two fluorophores (Cy3 and Cy5) attached to two different calmodulin molecules that bind to the “legs” of the same single molecule of myosin V, distance measurements accurate to ≈10 nm were achieved, and another acronym was generated (68, 69) [single-molecule high-resolution colocalization (SHREC) of fluorescent probes].
However, there is still a problem for imaging of complex structures in cells: it is not easily possible to label with more than just a few colors at room temperature, so to use the techniques described thus far, one must keep the concentration of the labeled biomolecules at a very low value, so that the PSF functions from individual emitters do not overlap in the images. One way to do this is to use the naturally occurring photobleaching; eventually all molecules will bleach except one. Adding further to the exploding menagerie of acronyms, this basic idea was demonstrated by Gordon et al. (70) for Cy3 labels on DNA [single-molecule high-resolution imaging with photobleaching (SHRImP)] and by Qu et al. (71) using Cy3-labeled PNA probes on DNA [nanometer localized multiple single (NALMS) molecule fluorescence microscopy]. Lidke et al. (72) showed that superresolution beyond the diffraction limit can also be achieved with the blinking of fluorescent semiconductor quantum dots.
To obtain more control over the process of digitization of the PSF for densely spaced single-molecule fluorophores in cells, researchers have begun to use photoinduced spectral changes of fusions to autofluorescent proteins such as GFP and its relatives. Indeed, reversible photoswitching of the emission of certain GFP mutants was reported in the first single-molecule observations of this amazingly useful cellular label in 1997 (73). In the years since, much progress has been made in the development of improved photoactivatable (turn-on of emission by excitation at a control wavelength) and photoswitchable fluorescent proteins with colorful names such as Kaede (74), PA-GFP (75), and DRONPA (76). Several of these cellular labels are used in the method of Betzig et al. (77) termed photoactivated localization microscopy (PALM), where light-induced photoactivation of GFP mutant fusions is used to randomly turn on only a few single molecules at a time in fixed cell sections or fixed cells. In this tour de force experiment, individual PSFs were recorded in detail to find their positions to ≈20 nm, then were photobleached so that others could be turned on, and so on until many thousands of PSF positions were determined. After 2–12 h of imaging, a high-resolution image was extracted that correlated well with a transmission electron microscopy image. Essentially simultaneously, Hess et al. (78) published a nearly identical approach with a very similar acronym, termed fluorescence PALM (F-PALM), which also utilizes a photoactivatable GFP with PSF localization to obtain superresolution. Fig. 3C shows an example image of PA-GFP molecules on a glass coverslip, where the green region represents the blurred-out image from conventional evanescent-wave fluorescence imaging, and the yellow dots represent the determination of the positions of individual single-molecule emitters. It is this impressive improvement of resolution that is causing obvious excitement in superresolution at the present time.
In another approach, Rust et al. (79) have used controlled photoswitching of a single photoswitchable fluorophore for superresolution demonstrations [stochastic optical reconstruction microscopy (STORM)]. This method uses a Cy3-Cy5 emitter pair in close proximity that show a novel property: restoration of Cy5's photobleached emission can be achieved by brief pumping of the Cy3 molecule. In this way, the emission from a single Cy5 on DNA or an antibody is turned on and off, again and again, to measure its position multiple times. After many such determinations, localization accuracy can approach ≈20-nm resolution, and labeled antibodies (labeled with >1 Cy3, ≪1 Cy5) were used to localize RecA proteins bound to DNA. The controllability of a reversible photoswitch based on these good single-molecule emitters is appealing, but because these molecules cannot be genetically encoded in cells like autofluorescent proteins, other methods of implanting properly formed Cy3-Cy5 pairs in cells need to be developed.
Recently, an alternative approach has been reported by Sharonov et al. (80) based on accumulated binding of diffusible probes, which are quenched in solution yet dequench in close proximity of the surface of the object to be imaged [points accumulation for imaging in nanoscale topography (PAINT)]. The method relies on the photophysical behavior of molecules with a twisted intermolecular charge transfer state such as Nile red (81). PAINT has the advantages that the object to be imaged need not be labeled, and many individual fluorophores are used for the imaging, thus relaxing the requirement on the total number of photons detected from each single molecule. The feasibility of this approach in the restricted cytoplasm needs to be explored.
In contrast to the previous approaches, it is important to note that there are also promising superresolution imaging methods (82) that do not specifically require single emitters, random photobleaching/blinking events, or photoswitching to be sure that only one emitter is present in a diffraction-limited volume. The basic idea proposed by Hell et al. (83) involves using optical saturation of the emission to provide a nonlinear response, which directly alters the shape of the PSF itself. This approach has been termed RESOLFT (reversible saturable optical fluorescence transitions) (84), because reversible photoswitching into dark states may be regarded as a type of optical saturation that produces a nonlinear dependence upon the local pumping intensity, at much lower power levels. The key point here is that these methods make the PSF itself much smaller, a step that can improve in principle any superresolution method based on confocal scanning [such as the 4Pi method (85) or even single-molecule imaging] at the cost of somewhat higher complexity.
A powerful but sophisticated implementation of these ideas developed in the laboratory of S. Hell (86) has been termed stimulated emission depletion (STED) microscopy (86). In STED, pulsed excitation of the absorbing molecules with a focused diffraction-limited excitation laser pulse places them in the electronically excited manifold as usual, and the molecules quickly relax to the lowest excited singlet state. However, before the molecules can emit, a second depleting laser pulse at a longer wavelength with a ring- or donut-shaped focal spot profile excites the sample. This pulse causes depletion of the excited state by stimulated emission, that is, the molecules not in the very center of the originally pumped region are forced down to the ground-state manifold. The stimulated photons from the molecule are at the same wavelength as the depleting beam and are attenuated by filtering. Thus, the only region of the sample that is allowed to emit in the range not blocked by filters is the much smaller central portion that was not irradiated by the depleting pulse. This method has recently yielded resolution far below the diffraction limit for both GFP-based labeling (87) and other forms of fluorescent labeling; Fig. 3D shows confocal and STED images of a neurofilament in a human neuroblastoma labeled by immunofluorescence (88). In principle, STED can operate with any nonlinear response of the sample.
In a different approach that also effectively suppresses the size of the PSF, Gustafsson (89) has proposed and demonstrated a wide-field superresolution technique that also relies upon nonlinear response but features structured illumination (90) with a large number of differently oriented standing-wave intensity patterns from two interfering laser beams. The nonlinearity changes the physical response to the sinusoidal illumination into a function with many higher spatial frequencies that ultimately alias back into the acceptance angle of the collection lens. With a complex computerized deconvolution, a superresolution image is obtained.
All of these tantalizing new approaches to superresolution in biological imaging have advantages and disadvantages, and which method will achieve widespread use and useful time resolution for observing nanoscale cellular dynamics still has yet to be determined. Better single-molecule emitters with photoactivation and/or photoswitching for cellular labeling would certainly help, because most superresolution methods have accuracy limitations arising from the finite number of photons available from a single molecule before photobleaching (with the exception of PAINT). Some of the key questions to be addressed are: How much dynamical information can be obtained in the superresolution limit? How can one deal with the randomness of molecules that blink on and off? and so on. For the former question, the highest-resolution method (PALM) has required long acquisition times on fixed cells, so extension to the observation of cellular dynamics will likely require tradeoffs. For the latter question, it may be that the STED method has a controllability advantage, because it uses high laser intensities to achieve stimulated emission. Considering the STORM method, where the on/off switching is somewhat more controlled than other photoswitchable molecules (yet still a Poisson process), one can regard each emission time of the molecule as a new position measurement, so that over time a large number of determinations are made of the position of the molecule. Each of these determinations is performed with fewer photons than if switching did not occur, thus each has larger mean-squared error. However, taken as a whole, the many position determinations of the same molecule under photoswitching conditions should be equivalent to one long acquisition of all of the photons, as long as the molecule does not move, and central-limit statistics applies. The situation is roughly analogous to trying to measure a photon emission stream in time, where one can use small time constants and many determinations or one long time constant. As is well known, if there is any chance of dynamical changes (either unwanted motion or drifts), one would prefer the short time acquisitions, because then dynamical effects (such as motion of the emitters) can be extracted, either directly or by various forms of correlation analysis. Such considerations will be an important topic of future work, along with many new applications of superresolution imaging using the methods already demonstrated.
Trapping of Single Molecules in Solution
In single-molecule studies, one would like to observe each molecule for as long a time as possible to extract maximal information from the molecule, a requirement that is relatively easy to achieve for solid samples (91), but that is quite challenging for small biomolecules in their native aqueous environment. Brownian motion is severe in solution at room temperature: a single 10-nm object in water diffuses through a diffraction-limited laser spot ≈0.3 μm in diameter in ≈1 ms. To address this, investigators have previously used a variety of strategies, such as immobilization on a transparent glass surface (92, 93) or encapsulation within the water-filled pores of a gel (5, 94, 95). Although these approaches can be useful, interactions with the glass surface or the gel-forming material can perturb the behavior of the biomolecules of interest. Another approach has been to try to enclose the biomolecules of interest in a vesicle (96), a promising approach especially for membrane-bound molecules, but bringing ligands or nucleotides in and out of the vesicle can be challenging in some cases. In the fluorescence correlation spectroscopy method (97), diffusion of a single biomolecule in solution brings it into the tiny 100-fl volume of a focused laser beam, but only for a time on the order of 1 ms. Analysis of the correlation function resulting from many single-molecule passages has been successfully used to tease out internal dynamics of fluorescent biomolecules (98–100).
One may ask, why not use an optical trap, such as the highly successful laser-tweezer trap, first proposed in the mid-1980s by Ashkin et al. (101)? Indeed, in the ensuing decades, this approach has led to major strides in biophysical understanding resulting from extremely precise biophysical force and subnanometer position measurements (10, 11, 102). However, it is worth remembering that the object actually trapped in a laser-tweezer system is a large dielectric bead, such as a poly(styrene) or glass sphere ≈1 μm in diameter. The biomolecule of interest is always attached to the sphere by various types of tethering/attachment chemistry. In fact, laser tweezers cannot trap a single small biomolecule directly, for the following reason. The gradient optical forces in a tweezers system are proportional to the polarizability α of the trapped object, which scales as the volume d3 (with d the diameter). Thus, if 10 mW of focused laser power is used trapping a 1-μm-size object, trapping a 100-nm object of a similar material would require 10 W of laser power, and trapping a 10-nm object would require 10 kW of laser power. The polarizability of a single small protein molecule is simply too small.
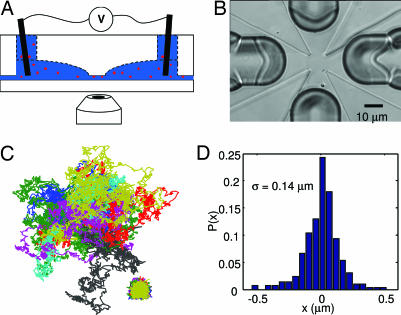
Feedback control, widely used to reduce stochastic fluctuations, provides a potential solution to these difficulties, which profits from the high processing speed of modern computers and image processing software (103). Recent theoretical proposals have discussed using feedback to track the Brownian motion of individual fluorescent molecules in solution (104–106). For nanoscale objects, electrokinetic (electrophoretic and electroosmotic) forces are far stronger than magnetic (107), dielectrophoretic (108), or optical forces and are thus most suitable. Recently, a new trap was proposed and demonstrated by A. E. Cohen and W.E.M., the anti-Brownian electrokinetic trap (ABEL trap), which scales quite favorably for trapping very small objects in solution (109). In the ABEL trap, a decrease in the diameter of a particle by a factor of k requires an increase in the speed of the ABEL trap by only a factor of k to maintain the same trapping strength, so extremely small objects can be trapped. The ABEL trap uses low-frequency electric fields and real-time feedback control to manipulate and trap individual nanoscale objects in solution at ambient temperature. Referring to Fig. 4(A, side-view section showing only two of four electrodes; B, top view of the microfluidic cell), the ABEL trap works by (i) monitoring the Brownian motion of the particle by directly measuring the particle's position with standard single-molecule fluorescence imaging microscopy and then (ii) applying a feedback voltage to the solution, so that the resulting electrokinetic drift (which may be either electrophoretic or electroosmotic in character) cancels the Brownian motion within the bandwidth of the feedback system. The particle is confined in the z direction by a thin channel on the order of 500 nm in thickness, and the microfluidic cell may be fabricated out of poly(dimethyl siloxane) (110), glass (111), or quartz (112) (shown in Fig. 4B). The control fields are applied in the x-y plane by four macroscopic electrodes placed in deep channels extending away from the trapping region in all four x-y directions.
Fig. 4.
Trapping single molecules in solution with the ABEL trap. (A) Schematic side view of the ABEL trap showing that the microfluidic cell sits above the oil-immersion objective of an inverted fluorescence microscope. Confinement in the z direction along the axis of the microscope is produced by the thin gap between the upper transparent structure and a flat coverslip. Four electrodes are placed in the solution far away from the central trapping region. (B) Top view of the microfluidic cell, showing the trapping region ≈10 × 10 μm in size in the center. Four deep milled channels extend out in the +/− x and +/− y directions. The four sharply pointed raised regions serve to define the thickness of the trap in the z direction normal to the page. (C) Measured (lower right) and pseudofree (center) trajectories of 13 trapped particles of TMV. [Reproduced with permission from ref. 113 (Copyright 2006, National Academy of Sciences).] (D) Position probability distribution of a single fluorescently labeled molecule of the chaperonin, GroEL, trapped in buffer. The standard deviation is shown.
The initial implementations of the ABEL trap used software-based feedback and electron-multiplying CCD imaging technology, in which the update time could be made as small as 4.5 ms (113). In this configuration, the ABEL trap was used to trap a variety of small objects in solution, ranging from 20-nm fluorescent spheres to single fluorescently labeled tobacco mosaic virus (TMV) particles (113). To trap smaller objects such as single copies of B-phycoerythrin, fluorescently labeled GroEL protein molecules, or fluorescent semiconductor nanocrystals, sucrose or glycerol was added to the solution to reduce the diffusion coefficient of the particle (113). Fig. 4C lower right shows the x-y trajectory of single TMV particles, and one can see that a small residual motion of the particle naturally occurs. This may be understood by thinking about the trapping algorithm in a little more detail: an image of the trapped object taken at a particular moment is used to calculate the required electrokinetic drift force and direction that would move the center of mass back to the center of the trap. It takes a certain amount of time (≈3.3 ms) to acquire the image and more time to perform the feedback calculation. The applied force certainly moves the object in the required direction, but the random Brownian forces acting continuously on the particle will always prevent it from being exactly at the center of the trap when the next image acquisition is complete. Rather than being a nuisance, interestingly, this jiggling of the particle in the trap contains useful information (114). A record of the actual positions of the particle and the applied electric fields at each update time can be used to remove the effects of the trapping and calculate a pseudofree trajectory, which is statistically similar to the trajectory the particle would have followed had it not been trapped (shown for the TMV particles in the main part of Fig. 4C) (113). Under various assumptions, the pseudofree trajectory can be used in principle to extract information about both the mobility and the diffusion coefficient of the trapped particle, and development of a fully general decomposition algorithm is a topic of current research (112).
To go faster, and therefore to trap single molecules without the need to artificially increase the solution viscosity, a hardware version of the ABEL trap has been constructed by using rotation of the laser focus (104). This method allows update times as small as ≈25 μs, and Fig. 4D shows the displacement histogram of a single fluorescently labeled molecule of GroEL trapped in buffer (112).
Tracking and trapping nanoscale objects is a fascinating new direction showing potential for improvements along many lines. Single molecules of DNA can be trapped without the need for attachment to a bead, and their shape fluctuations may be analyzed in detail to extract information about DNA dynamics as shown on p. 12622 of this issue of PNAS (130), as well as information about DNA mechanical properties (131). The ABEL trap principle can be extended to three dimensions by adding more electrodes and by sensing z displacements by a defocusing algorithm similar to that used in CD players. Tracking can also be accomplished by moving the stage itself in three dimensions, but this approach can be limited in its speed by the requirement to move a massive stage to follow the nanoscale object. As mentioned above, rotation of the laser focus is a method for producing position information at a very high speed (104), and this scheme has been demonstrated in a 3D geometry by using two-photon excitation of fluorescence (115) and for small nanoparticles in two dimensions showing shot-noise-limited localization (116). Going beyond fluorescence, 3D tracking has recently been achieved by detecting scattered light from a 250-nm gold colloid by using a 3D defocus detection scheme and stage motion (117, 118).
Outlook
The recent progress in single-molecule imaging and microscopy has been impressive from both the methodological and the applications perspectives. At the same time, there has been a welcome increase in contributions from the theoretical community to the analysis and interpretation of single-molecule data (17, 119–123). With many new investigators continuing to enter the field, the applications of single-molecule fluorescence imaging methods should continue to expand. It is an exciting time for study of single biomolecules willing to tell us how each individual behaves, as a window into the true nature of complex nanoscale systems.
Acknowledgments
I thank Julie Biteen, Adam E. Cohen, Marcelle Koenig, and Samuel J. Lord for a careful reading of portions of the manuscript; Taekjip Ha (University of Illinois, Urbana–Champaign) for Fig. 1; So Yeon Kim, (Stanford University, Stanford, CA) and Anika Kinkhabwala (Stanford University) for the data in Fig. 3B; and Adam E. Cohen (Stanford University) for the data in Fig. 4. This work was supported in part by National Science Foundation Grant CHE-0554681 and by National Institutes of Health Grants 1P20-HG003638, 1R21-GM075166, and 1R21-RR023149.
Abbreviations
- FRET
Förster resonant energy transfer
- PSF
point-spread function
- PALM
photoactivated localization microscopy
- ABEL
anti-Brownian electrokinetic
- TMV
tobacco mosaic virus
- STED
stimulated emission depletion.
Footnotes
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Moerner WE, Kador L. Phys Rev Lett. 1989;62:2535–2538. doi: 10.1103/PhysRevLett.62.2535. [DOI] [PubMed] [Google Scholar]
- 2.Orrit M, Bernard J. Phys Rev Lett. 1990;65:2716–2719. doi: 10.1103/PhysRevLett.65.2716. [DOI] [PubMed] [Google Scholar]
- 3.Betzig E, Chichester RJ. Science. 1993;262:1422–1428. doi: 10.1126/science.262.5138.1422. [DOI] [PubMed] [Google Scholar]
- 4.Nie S, Chiu DT, Zare RN. Science. 1994;266:1018–1021. doi: 10.1126/science.7973650. [DOI] [PubMed] [Google Scholar]
- 5.Xie XS. Acc Chem Res. 1996;29:598–606. [Google Scholar]
- 6.Ambrose WP, Goodwin PM, Jett JH, VanOrden A, Werner JH, Keller RA. Chem Rev. 1999;99:2929–2956. doi: 10.1021/cr980132z. [DOI] [PubMed] [Google Scholar]
- 7.Rigler R, Orrit M, Basché T, editors. Single Molecule Spectroscopy: Nobel Conference Lectures, Springer Series in Chemical Physics. Vol 67. Berlin: Spirnger; 2001. [Google Scholar]
- 8.Moerner WE. J Phys Chem B. 2002;106:910–927. [Google Scholar]
- 9.Tinnefeld P, Sauer M. Angew Chem Int Ed. 2005;44:2642–2671. doi: 10.1002/anie.200300647. [DOI] [PubMed] [Google Scholar]
- 10.Bustamante C, Bryant Z, Smith SB. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 11.Neuman KC, Block SM. Rev Sci Instr. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hummer G, Szabo A. Acc Chem Res. 2005;38:504–513. doi: 10.1021/ar040148d. [DOI] [PubMed] [Google Scholar]
- 13.Ha T, Laurence TA, Chemla DS, Weiss S. J Phys Chem B. 1999;103:6839–6850. [Google Scholar]
- 14.Ha T. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 15.Kapanidis AN, Weiss S. J Chem Phys. 2002;117:10953–10964. [Google Scholar]
- 16.Moerner WE, Fromm DP. Rev Sci Instr. 2003;74:3597–3619. [Google Scholar]
- 17.Barkai E, Jung Y, Silbey RJ. Annu Rev Phys Chem. 2004;55:457–507. doi: 10.1146/annurev.physchem.55.111803.143246. [DOI] [PubMed] [Google Scholar]
- 18.Min W, English BP, Luo G, Cherayil BJ, Kou SC, Xie XS. Acc Chem Res. 2005;38:923–931. doi: 10.1021/ar040133f. [DOI] [PubMed] [Google Scholar]
- 19.Vrljic M, Nishimura SY, Moerner WE, McConnell HM. Biophys J. 2005;88:334–347. doi: 10.1529/biophysj.104.045989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuler B, Lipman EA, Eaton WA. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 21.Zhuang X, Rief M. Curr Opin Struct Biol. 2003;13:88–97. doi: 10.1016/s0959-440x(03)00011-3. [DOI] [PubMed] [Google Scholar]
- 22.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 23.Braslavsky I, Hebert B, Kartalov E, Quake SR. Proc Natl Acad Sci USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Proc Natl Acad Sci USA. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 26.Joo C, McKinney SA, Nakamura M, Rasnik I, Ha T. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 27.Kapanidis AN, Laurence TA, Lee NK, Margeat E, Kong X, Weiss S. Acc Chem Res. 2005;38:523. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- 28.Lee NK, Kapanidis AN, Koh HR, Korlann Y, Ho SO, Kim Y, Gassman N, Kim SK, Weiss S. Biophys J. 2007;92:303–312. doi: 10.1529/biophysj.106.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nir E, Michalet X, Hamadani KM, Laurence TA, Neuhauser D, Kovchegov Y, Weiss S. J Phys Chem B. 2006;110:22103–22124. doi: 10.1021/jp063483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slaughter BD, Allen MW, Unruh JR, Urbauer RJB, Johnson CJ. J Phys Chem B. 2004;108:10388–10397. [Google Scholar]
- 31.Diez M, Zimmermann B, Börsch M, König M, Schweinberger E, Steigmiller S, Reuter R, Felekyan S, Kudryavtsev V, Seidel CAM, et al. Nat Struct Mol Biol. 2004;11:135. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- 32.Kozuka J, Yokota H, Arai Y, Ishii Y, Yanagida T. Nat Chem Biol. 2006;2:83–86. doi: 10.1038/nchembio763. [DOI] [PubMed] [Google Scholar]
- 33.Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sako Y, Minoghchi S, Yanagida T. Nat Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 35.Kues T, Peters R, Kubitscheck U. Biophys J. 2001;80:2954–2967. doi: 10.1016/S0006-3495(01)76261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harms GS, Cognet L, Lommerse PHM, Blab GA, Kahr H, Gamsjaeger R, Spaink HP, Soldatov NM, Romanin C, Schmidt T. Biophys J. 2001;81:2639–2646. doi: 10.1016/S0006-3495(01)75907-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moerner WE. Trends Anal Chem. 2003;22:544–548. [Google Scholar]
- 38.Konopka MC, Weisshaar JC. J Phys Chem A. 2004;108:9814–9826. [Google Scholar]
- 39.Ichinose J, Sako S. Trends Anal Chem. 2004;23:587–594. [Google Scholar]
- 40.Xie XS, Yu J, Yang WY. Science. 2006;312:228–230. doi: 10.1126/science.1127566. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Campbell RE, Ting AY, Tsien RY. Nat Rev. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 42.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 43.Remington SJ. Curr Opin Struct Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Femino AM, Fay FS, Fogarty K, Singer RH. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 45.Harms GS, Cognet L, Lommerse PHM, Blab GA, Schmidt T. Biophys J. 2001;80:2396–2408. doi: 10.1016/S0006-3495(01)76209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moerner WE. J Chem Phys. 2002;117:10925–10937. [Google Scholar]
- 47.Deich J, Judd EM, McAdams HH, Moerner WE. Proc Natl Acad Sci USA. 2004;101:15921–15926. doi: 10.1073/pnas.0404200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Proc Natl Acad Sci USA. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Courty S, Luccardini C, Bellaiche Y, Cappello G, Dahan M. Nano Lett. 2006;6:1491–1495. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- 50.Chen I, Ting A. Curr Opin Biotechnol. 2005;16:35. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 51.Willets KA, Nishimura SY, Schuck PJ, Twieg RJ, Moerner WE. Acc Chem Res. 2005;38:549–556. doi: 10.1021/ar0401294. [DOI] [PubMed] [Google Scholar]
- 52.Nishimura SY, Lord SJ, Klein LO, Willets KA, He M, Lu ZK, Twieg RJ, Moerner WE. J Phys Chem B. 2006;110:8151–8157. doi: 10.1021/jp0574145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai L, Friedman N, Xie XS. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 54.Yu J, Xiao J, Ren X, Lao K, Xie XS. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 55.Weiss S. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 56.Moerner WE. Science. 1994;265:46–53. doi: 10.1126/science.265.5168.46. [DOI] [PubMed] [Google Scholar]
- 57.Ambrose WP, Basché T, Moerner WE. J Chem Phys. 1991;95:7150. [Google Scholar]
- 58.Gelles J, Schnapp BJ, Sheetz MP. Nature. 1988;4:450–453. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- 59.Güttler F, Irngartinger T, Plakhotnik T, Renn A, Wild UP. Chem Phys Lett. 1994;217:393. [Google Scholar]
- 60.van Oijen AM, Köhler J, Schmidt J, Müller M, Brakenhoff GJ. Chem Phys Lett. 1998;292:183–187. [Google Scholar]
- 61.Thompson RE, Larson DR, Webb WW. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michalet X, Weiss S. Proc Natl Acad Sci USA. 2006;103:4797–4798. doi: 10.1073/pnas.0600808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ober RJ, Ram S, Ward ES. Biophys J. 2004;86:1185–1200. doi: 10.1016/S0006-3495(04)74193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ram S, Ward ES, Ober RJ. Proc Natl Acad Sci USA. 2006;103:4457–4462. doi: 10.1073/pnas.0508047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yildiz A, Forkey JN, McKinner SA, Ha T, Goldman YE, Selvin PR. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 66.Nan X, Sims PA, Chen P, Xie XS. J Phys Chem B. 2005;109:24220–24224. doi: 10.1021/jp056360w. [DOI] [PubMed] [Google Scholar]
- 67.Betzig E. Opt Lett. 1995;20:237–239. doi: 10.1364/ol.20.000237. [DOI] [PubMed] [Google Scholar]
- 68.Churchman LS, Oekten Z, Rock RS, Dawson JF, Spudich JA. Proc Natl Acad Sci USA. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Churchman LS, Flyvbjerg H, Spudich JA. Biophys J. 2006;90:668–671. doi: 10.1529/biophysj.105.065599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gordon MP, Ha T, Selvin PR. Proc Natl Acad Sci USA. 2004;101:6462–6465. doi: 10.1073/pnas.0401638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qu X, Wu D, Mets L, Scherer NF. Proc Natl Acad Sci USA. 2004;101:11298–11303. doi: 10.1073/pnas.0402155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lidke KA, Rieger B, Jovin TM, Heintzmann R. Opt Exp. 2005;13:7052–7062. doi: 10.1364/opex.13.007052. [DOI] [PubMed] [Google Scholar]
- 73.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 74.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. Proc Natl Acad Sci USA. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patterson GH, Lippincott-Schwartz J. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 76.Ando R, Mizuno H, Miyawaki A. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 77.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 78.Hess ST, Girirajan TPK, Mason MD. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rust MJ, Bates M, Zhuang X. Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharonov A, Hochstrasser RM. Proc Natl Acad Sci USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mei E, Gao F, Hochstrasser RM. Phys Chem Chem Phys. 2006;8:2077–2082. doi: 10.1039/b601670g. [DOI] [PubMed] [Google Scholar]
- 82.Hell SW. Nat Biotechnol. 2003;21:1347–1355. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- 83.Hell SW, Jakobs S, Kastrup L. Appl Phys A. 2003;77:859–860. [Google Scholar]
- 84.Hofmann M, Eggeling C, Jakobs S, Hell SW. Proc Natl Acad Sci USA. 2005;102:17565–17569. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schrader M, Hell SW, van der Voort HTM. J Appl Phys. 1998;84:4033–4042. [Google Scholar]
- 86.Hell SW, Wichmann J. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 87.Willig KI, Kellner RR, Medda R, Hein B, Jakobs S, Hell SW. Nat Methods. 2006;3:721–723. doi: 10.1038/nmeth922. [DOI] [PubMed] [Google Scholar]
- 88.Donnert G, Keller J, Medda R, Andrei MA, Rizzoli SO, Luehrmann R, Jahn R, Eggeling C, Hell SW. Proc Natl Acad Sci USA. 2006;103:11440–11445. doi: 10.1073/pnas.0604965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gustafsson MGL. Proc Natl Acad Sci USA. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gustafsson MGL, Agard DA, Sedat JW. J Microsc. 1999;195:10–16. doi: 10.1046/j.1365-2818.1999.00576.x. [DOI] [PubMed] [Google Scholar]
- 91.Werley CA, Moerner WE. J Phys Chem B. 2006;110:18939–18944. doi: 10.1021/jp057570b. [DOI] [PubMed] [Google Scholar]
- 92.Yang H, Luo G, Karnchanaphanurach P, Louie T, Rech I, Cova S, Xun L, Xie XS. Science. 2003;302:262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 93.Rasnik I, McKinney SA, Ha T. Acc Chem Res. 2005;38:542–548. doi: 10.1021/ar040138c. [DOI] [PubMed] [Google Scholar]
- 94.Dickson RM, Norris DJ, Tzeng Y, Moerner WE. Science. 1996;274:966–969. doi: 10.1126/science.274.5289.966. [DOI] [PubMed] [Google Scholar]
- 95.Peterman EJG, Brasselet S, Moerner WE. J Phys Chem A. 1999;103:10553–10560. [Google Scholar]
- 96.Boukobza E, Sonnenfeld A, Haran G. J Phys Chem B. 2001;105:12165–12170. [Google Scholar]
- 97.Magde DL, Elson EL, Webb WW. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 98.Eigen M, Rigler R. Proc Natl Acad Sci USA. 1994;91:5740–5747. doi: 10.1073/pnas.91.13.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gell C, Brockwell DJ, Beddard GS, Radford SE, Kalverda AP, Smith DA. Single Mol. 2001;2:177–181. [Google Scholar]
- 100.Hess ST, Huang S, Heikal AA, Webb WW. Biochemistry (NY) 2002;41:697–705. doi: 10.1021/bi0118512. [DOI] [PubMed] [Google Scholar]
- 101.Ashkin A, Dziedzic JM, Bjorkholm JE, Chu S. Opt Lett. 1986;11:288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 102.Grier DG. Nature. 2003;424:810–816. doi: 10.1038/nature01935. [DOI] [PubMed] [Google Scholar]
- 103.Armani M, Chaudhary S, Probst R, Shapiro B. Proc IEEE Int Conf MicroElectroMech Syst (MEMS) 2005:855–858. [Google Scholar]
- 104.Enderlein J. Appl Phys B. 2000;71:773–777. [Google Scholar]
- 105.Berglund AJ, Mabuchi H. Appl Phys B. 2004;78:653–659. [Google Scholar]
- 106.Andersson SB. Appl Phys B. 2005;80:809–816. [Google Scholar]
- 107.Gosse C, Croquette V. Biophys J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Voldman J, Braff RA, Toner M, Gray ML, Schmidt MA. Biophys J. 2001;80:531–541. doi: 10.1016/S0006-3495(01)76035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cohen AE, Moerner WE. Appl Phys Lett. 2005;86:093109. [Google Scholar]
- 110.Cohen AE, Moerner WE. ProcSPIE. 2005;5699:296–305. [Google Scholar]
- 111.Cohen AE, Moerner WE. ProcSPIE. 2005;5930:191–198. [Google Scholar]
- 112.Cohen AE. Stanford, CA: Stanford University; 2006. PhD thesis. [Google Scholar]
- 113.Cohen AE, Moerner WE. Proc Natl Acad Sci USA. 2006;103:4362–4365. doi: 10.1073/pnas.0509976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cohen AE. Phys Rev Lett. 2005;94:118102. doi: 10.1103/PhysRevLett.94.118102. [DOI] [PubMed] [Google Scholar]
- 115.Levi V, Ruan Q, Gratton E. Biophys J. 2005;88:2919–2928. doi: 10.1529/biophysj.104.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Berglund AJ, McHale K, Mabuchi H. Opt Lett. 2007;32:145–147. doi: 10.1364/ol.32.000145. [DOI] [PubMed] [Google Scholar]
- 117.Cang H, Wong CM, Xu CS, Rizvi AH, Yang H. Appl Phys Lett. 2006;88:223901. [Google Scholar]
- 118.Xu CS, Cang H, Montiel D, Yang H. J Phys Chem C. 2007;111:32–35. [Google Scholar]
- 119.Andrec M, Levy RM, Talaga DS. J Phys Chem A. 2003;107:7454–7464. doi: 10.1021/jp035514+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Watkins LP, Yang H. Biophys J. 2004;86:4015–4029. doi: 10.1529/biophysj.103.037739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Watkins LP, Yang H. J Phys Chem B. 2005;109:617–628. doi: 10.1021/jp0467548. [DOI] [PubMed] [Google Scholar]
- 122.Witkoskie JB, Cao J. J Chem Phys. 2004;121:6373–6379. doi: 10.1063/1.1785784. [DOI] [PubMed] [Google Scholar]
- 123.Gopich I, Szabo A. J Phys Chem B. 2005;109:6845–6848. doi: 10.1021/jp045398q. [DOI] [PubMed] [Google Scholar]
- 124.Peterman EJ, Sosa H, Moerner WE. Annu Rev Phys Chem. 2004;55:79–96. doi: 10.1146/annurev.physchem.55.091602.094340. [DOI] [PubMed] [Google Scholar]
- 125.Yeung ES. Annu Rev Phys Chem. 2004;55:97–126. doi: 10.1146/annurev.physchem.54.011002.103820. [DOI] [PubMed] [Google Scholar]
- 126.Barkai E, Jung YJ, Silbey R. Annu Rev Phys Chem. 2004;55:457–507. doi: 10.1146/annurev.physchem.55.111803.143246. [DOI] [PubMed] [Google Scholar]
- 127.Kilzer F, Orrit M. Annu Rev Phys Chem. 2004;55:585–611. doi: 10.1146/annurev.physchem.54.011002.103816. [DOI] [PubMed] [Google Scholar]
- 128.Barbara P, editor. Acc Chem Res. Vol. 38. 2005. pp. 503–610. [DOI] [PubMed] [Google Scholar]
- 129.Haran G, Vöhringer P, editors. ChemPhysChem. Vol. 6. 2005. pp. 753–998. [DOI] [PubMed] [Google Scholar]
- 130.Cohen AE, Moerner WE. Proc Natl Acad Sci USA. 2007;104:12622–12627. doi: 10.1073/pnas.0610396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cohen AE, Moerner WE. Phys Rev Lett. 2007;98:116001. doi: 10.1103/PhysRevLett.98.116001. [DOI] [PMC free article] [PubMed] [Google Scholar]