Abstract
We report fluorescence assays for a functionally important conformational change in bacteriophage T7 DNA polymerase (T7 pol) that use the environmental sensitivity of a Cy3 dye attached to a DNA substrate. An increase in fluorescence intensity of Cy3 is observed at the single-molecule level, reflecting a conformational change within the T7 pol ternary complex upon binding of a dNTP substrate. This fluorescence change is believed to reflect the closing of the T7 pol fingers domain, which is crucial for polymerase function. The rate of the conformational change induced by a complementary dNTP substrate was determined by both conventional stopped-flow and high-time-resolution continuous-flow fluorescence measurements at the ensemble-averaged level. The rate of this conformational change is much faster than that of DNA synthesis but is significantly reduced for noncomplementary dNTPs, as revealed by single-molecule measurements. The high level of selectivity of incoming dNTPs pertinent to this conformational change is a major contributor to replicative fidelity.
DNA polymerases (DNA pols) are essential components of replicases that ensure faithful replication of cellular DNA (1). In addition to their role in replication, some DNA pols specialize in DNA repair (2). DNA pols also have important applications in molecular biology, such as in DNA sequencing (3) and in the PCR (4).
DNA pols catalyze the formation of a phosphodiester bond between the 3′-OH of a DNA primer strand and the α-phosphorous atom of an incoming dNTP. Continuous rounds of catalysis lead to a DNA duplex through extension of a nascent DNA primer having a sequence complementary to the template. The error rate during DNA replication can be as low as 10−9 (5). Although Watson–Crick base pairs are more stable than other base pair combinations, the thermodynamic contributions corresponding to an error rate of 10−2 are not sufficient to account for the extraordinarily high base selectivity of DNA pols. The precise mechanism responsible for base selectivity is an important and as yet unsolved problem, despite many attempts to fashion an all-encompassing general solution (2, 5–9).
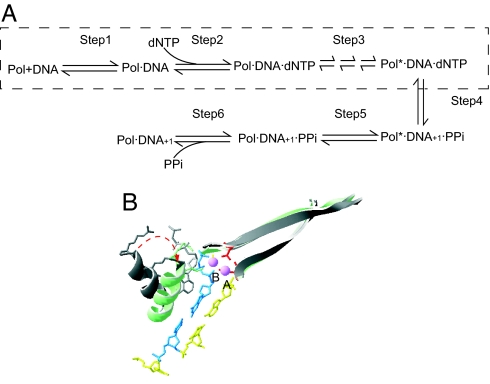
Kinetic studies have shown that all DNA pols share a common multiple-step scheme for the synthesis of DNA (7), as depicted in Fig. 1A. With a few exceptions, selection of the complementary dNTP occurs at the initial dNTP binding (step 2). This step contributes ≈102- to 103-fold to base selectivity for high-fidelity DNA pols (5). For many polymerases, such as T7 pol (10) and the Klenow fragment of Escherichia coli polymerase I (KF) (11), the conformational change (step 3) has been proposed to be rate-limiting for complementary dNTPs. This supposition has been supported by evidence from the elemental effect of αS-dNTP utilization, pulse–chase, and pulse–quench experiments (10, 12). Step 3 is important for base selection by most polymerases because, in addition to the initial dNTP binding step, it serves as a kinetic checkpoint to select against noncomplementary dNTPs (7). The selectivity arising from the difference in catalytic rates between complementary and noncomplementary substrates contributes a factor of 103 to 104 to replication fidelity (5).
Fig. 1.
Kinetic scheme and conformational change of DNA pols. (A) Minimum kinetic scheme shared by different DNA pols. To extend a primer, the following steps are required: Step 1, binding of DNA to form a binary complex; Step 2, binding of a dNTP substrate to form a ternary complex; Step 3, a series of conformational changes in the ternary complex that ensure proper alignment of the reactants; Step 4, phosphoryl transfer; Step 5, another conformational change that relaxes the complex; Step 6, pyrophosphate release and translocation of the DNA, bringing the P/T into a position poised for the next synthetic step. This scheme was adapted from ref. 7. The states accessible in our fluorescence assay are shown within the dashed rectangle. (B) Conformational change of T7 pol from the open form (gray; PDB entry 1SL0; ref. 7) to the closed form (in color; PDB entry 1T7P; ref. 7). Two structures are superimposed by aligning the polymerase palm domains. The conformational change occurs through coordination of Mg2+ (pink) by both the triphosphate moiety of the dNTP (blue) and two conserved residues with carboxylic side chains (red).
It is of great interest to understand the molecular basis for the high fidelity exhibited by replicative DNA pols. Structural studies on T7 pol (13), HIV reverse transcriptase (14), human pol β (polβ) (15), Bacillus stearothermophilus pol I (16), and Klentaq polymerase (17) have shown that binding of the complementary dNTP results in formation of a ternary complex in which the fingers subdomain of DNA pols has closed so that the incoming dNTP is encapsulated, with all possible hydrogen bonds and electrostatic interactions satisfied. As shown in Fig. 1B, T7 pol provides a good example of these features. Stopped-flow fluorescence measurements with 2-aminopurine (2AP), a probe for base stacking, have been used to detect conformation changes in polβ (18), T4 pol (19), and KF (20). The results suggested that the fingers domain may be in rapid equilibrium between the open and closed states before the putative rate-limiting step. Consistent with this observation, a recent stopped-flow study with fluorescence resonance energy transfer (FRET) on Klentaq indicated that the motion of the fingers subdomain is also a fast step for this enzyme (21). In contrast, a kinetic study of a fluorescently labeled T7 pol showed that a conformational change, which is partially rate-limiting, could be responsible for base selectivity (22).
In light of the apparent discrepancies among different experimental approaches, additional assays probing the conformational change triggered by dNTP binding are necessary to fully understand the relationship between conformational dynamics and function of DNA pols. Here we report an assay to measure the T7 pol conformational change, based on the fluorescence intensity variation of a Cy3-labeled primer/template (P/T)·T7 pol complex due to environmental changes surrounding the dye (23). Upon the binding of a complementary dNTP, a fast conformational change is observed that is not rate-limiting for DNA synthesis.
This change can be observed at both the single-molecule and ensemble-average levels. In general, real-time single-molecule studies (24–26) have the potential to identify transient intermediates without the need for synchronization, thus helping to elucidate enzymatic reaction mechanisms (27–29). Although spontaneous conformational fluctuations have been directly measured by single-molecule measurements (30, 31), less information is available about functionally important conformational changes at the single-molecule level. Here we use single-molecule techniques to directly observe stochastic events of switching between two conformational states of T7 pol. One particular advantage offered by single-molecule measurements is that rate constants of conformational changes for both the forward and reverse directions can be determined independently from a single time trace.
In this particular system, the fast forward conformational change cannot be resolved with single-molecule observations because of limited time resolution. To measure the fast kinetics, we have used a conventional stopped-flow apparatus and continuous flow in a microfluidic device with high time resolution. Michaelis–Menten kinetics for the conformational change upon binding of a complementary dNTP was observed and found not to be rate-limiting. We have assigned this conformational change to closing of the fingers domain in T7 pol.
We found that the rate constant of this conformational change is highly dependent on the complementarity of the dNTP substrate. Slow conformational changes induced by noncomplementary dNTP binding are difficult to observe at the ensemble level because they are rare events. However, infrequent conformational changes can easily be identified in a single-molecule time trace. Our results indicate that the rate constant of the conformational change is markedly reduced with noncomplementary bases. Therefore, this conformational change, presumably reflecting fingers domain closing, plays a crucial role in determining base selectivity and, thus, DNA replication fidelity.
Results and Analysis
Single-Molecule Binding and Dissociations of T7 Pol·Cy3-P/T Complexes.
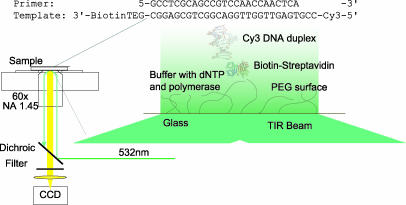
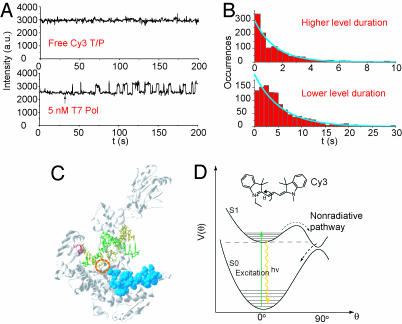
The single-molecule fluorescence experimental setup is shown in Fig. 2[also see supporting information (SI) Appendix, Section 2 for details]. We first performed single-molecule experiments to observe the binding between T7 pol and immobilized Cy3-P/T. The fluorescence intensity of a single Cy3-P/T was stable. However, upon introduction of T7 pol in phosphate buffer, the fluorescence of Cy3-P/T fluctuated between two levels: a baseline level and a level with a 40% higher intensity (Fig. 3A). We attribute the increase in fluorescence intensity to the binding of T7 pol to the Cy3-P/T, which changes with the local environment of Cy3 (Fig. 3D, and see Discussion). This change is due to the reduced relaxation rate of Cy3 from its excited state, consistent with the higher fluorescence quantum yield of Cy3 in more viscous solvents.
Fig. 2.
Experimental setup for our single-molecule fluorescence observations. Cy3-P/T was immobilized on the surface of a glass coverslip through a biotin–streptavidin linkage. DNA pol and dNTP were supplied in the solution above the coverslip. TIR, total internal reflection.
Fig. 3.
Detection of T7 pol binding by Cy3 fluorescence. (A) Fluorescence intensity trajectory of a surface-immobilized Cy3-P/T duplex. The arrow marks the time when the polymerase solution was flowed in. Fluorescence intensity trajectories shown here were all filtered with a nonlinear filter (32). (B) Histograms of the durations of the polymerase on and off states, with exponential fits (blue curves). (C) Estimation of the Cy3 position in the T7 pol binary complex. The base (red) corresponds to the second to last base from the 5′ end of the template (yellow) where Cy3 is attached. The residues deleted from the wt T7 pol in the Sequenase are shown in blue. In the binary complex, Cy3 should be located between the base (red) and the residues (blue). The circle shows the approximate position of Cy3. (D) Cy3 structure (Inset) and the potential energy surface of Cy3, adapted from ref. 23. The excited-state process following the dashed arrows is a nonradiative relaxation pathway sensitive to local environment.
When the concentration of T7 pol was reduced, the durations of the lower fluorescence intensity level were lengthened, but the duration of the higher intensity level remained constant. The distribution of the duration of lower and higher fluorescence intensity levels is related to polymerase binding and dissociation kinetics, and both fit single exponential functions (Fig. 3B). The association and dissociation rate constants (3.6 × 107 M−1·s−1 and 0.67 s−1, respectively) are comparable to previously reported ensemble measurements (10).
Interestingly, when Sequenase (USB, Cleveland, OH), an exonuclease-deficient T7 pol with a 28-residue deletion (33), binds to the Cy3-P/T, the fluorescence intensity of the Cy3-P/T does not change significantly. We have tested the reactivity of Sequenase by using fluorescently labeled dCTP and Cy3-P/T and have confirmed that Sequenase still binds the DNA substrate. On the basis of these observations, the region of the T7 pol required for Cy3 fluorescence enhancement can be tentatively identified and is highlighted in Fig. 3C, with the estimated position of the Cy3 probe circled.
Single-Molecule Observations of T7 Pol Conformational Changes Upon Complementary dNTP Binding.
To evaluate the effect of the complementary incoming dNTP on Cy3 fluorescence, we introduced Mg2+, which is essential for both synthesis and exonuclease activity. To focus on probing the conformational change in the absence of DNA synthesis and nucleotide excision, we used a nonextendable dideoxy-terminated primer (ddP) and a phosphorothioate linkage between the penultimate and 3′ terminal nucleotide residue, which prevents exonuclease digestion (34) (see Materials and Methods). This primer was annealed to the same Cy3-labeled template.
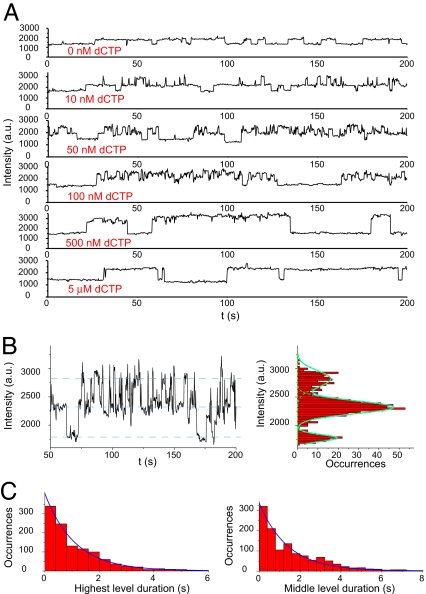
Fig. 4A shows the intensity trajectories of the Cy3-ddP/T in the presence of T7 pol and a complementary dCTP in which the dCTP concentration was varied from 10 nM to 5 μM. After addition of dCTP, the intensity fluctuation pattern of Cy3 fluorescence in the complex changed significantly. Three different intensity levels of Cy3 fluorescence are seen in the intensity histogram (Fig. 4B). As mentioned above, the lowest level represents the free Cy3-ddP/T, the middle level corresponds to the T7 pol binary complex, and the highest level corresponds to the environment of Cy3 associated with the T7 pol ternary complex with dCTP. We also observed a similar fluorescence increase in Cy3-ddP/T·T7 pol complex upon ddCTP binding (see SI Appendix, Section 15).
Fig. 4.
Conformational change of T7 pol upon dCTP binding detected by Cy3 fluorescence change. (A) Fluorescence intensity trajectories of the Cy3-ddP/T duplex in a mixture containing 10 nM T7 pol, 20 μM ddATP, and 0–5 μM dCTP in 50 mM Tris buffer, pH 7.5, with 10 mM MgCl2. (B) Part of the fluorescence intensity trajectory of the duplex Cy3-ddP/T with an expanded y axis, obtained with 10 nM T7 pol and 50 nM dCTP. The fluorescence intensity histogram, along with a fit of Gaussian peaks on the right, clearly shows three intensity levels. (C) Statistics of the lifetimes of the T7 pol binary and ternary states with 50 nM of dCTP, with exponential fits (blue curves).
Because the incoming dCTP is bound at the O helix (Fig. 1B) (13), some distance from the Cy3 probe, there is no direct interaction between Cy3 and the dCTP. We attribute the highest intensity level to a conformational state induced by dCTP binding. The reverse conformational change can be observed because the subsequent chemical step is blocked.
Individual binding and dissociation events can be clearly resolved in the single-molecule time traces at low concentrations of dCTP (≈10–100 nM). The durations of T7 pol binding on DNA (the middle and the highest levels) are longer than those in the absence of dCTP, indicating that the binding of the complementary dCTP significantly stabilized the complex between T7 pol and DNA (see SI Appendix, Section 8 and Fig. S5).
The rate constants of the forward and backward conformational changes are easily extracted from the single-molecule time traces (Fig. 4C). The distribution of time intervals of the highest level is exponential, with a decay constant of ≈0.9 s−1, reflecting the rate constant of the reverse conformational change. This rate constant is independent of dCTP concentration. The distribution of middle intensity level durations is also exponential, with a decay rate constant proportional to dCTP concentration. At 50 nM dCTP, this pseudo-first-order rate constant is ≈0.7 s−1. At higher concentrations of dCTP (500 nM to 5 μM), the middle level cannot be observed because of the limited time resolution.
We distinguish two different situations, as follows:
and
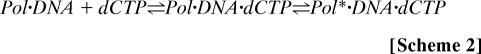 |
The intermediate intensity level represents the binary complex on the left sides of the equations, but it is not clear whether the highest intensity level represents a nascent ternary (Pol·DNA·dCTP) complex (Scheme 1) or an ensuing conformer of the ternary complex (Pol*·DNA·dCTP in Scheme 2). The rates of Cy3 fluorescence increases have different dCTP concentration dependencies for Scheme 1 and Scheme 2. The dependence is linear at low dCTP concentrations for both schemes; however, Scheme 2 would be expected to exhibit saturation behavior at high dCTP concentrations. Unfortunately, our single-molecule measurements do not provide sufficiently high time resolution at high dCTP concentrations to enable us to distinguish between the two schemes.
Ensemble-Averaged Stopped-Flow and Continuous-Flow Experiments to Determine the Rate of the Conformational Change.
To achieve higher time resolution and to distinguish the two schemes above, we performed ensemble-averaged measurements to monitor rates of change in Cy3 fluorescence upon dCTP binding.
The traces in Fig. 5A show the temporal profiles of Cy3 fluorescence after the T7 pol·Cy3-ddP/T binary complex was mixed with increasing dCTP concentration by using a stopped-flow instrument. The fluorescence scans fit best to double exponentials. The fast phase, which is proportional to dCTP concentrations in the range of ≈1–20 μM, accounts for the majority (≈90% amplitude) of the intensity increase. This phase likely corresponds to the conformational change induced by dCTP binding as seen in the single-molecule fluorescence trajectories (Fig. 4 A and B). The bimolecular rate constant obtained from stopped-flow experiments (2.2 × 107 M−1·s−1, obtained at higher dCTP concentrations) is consistent with the rate obtained from single-molecule observations (1.4 × 107 M−1·s−1, obtained with 50 nM dCTP). The minor increase in fluorescence intensity (≈10% amplitude) of the slow phase (≈1 s−1) is independent of dCTP concentration and may reflect a shift in equilibrium, converting the remaining free Cy3-ddP/T to bind T7 pol because of the depletion of the T7 pol·Cy3-ddP/T binary complex as a consequence of dCTP binding. This minor phase is not considered in evaluating the rate constant.
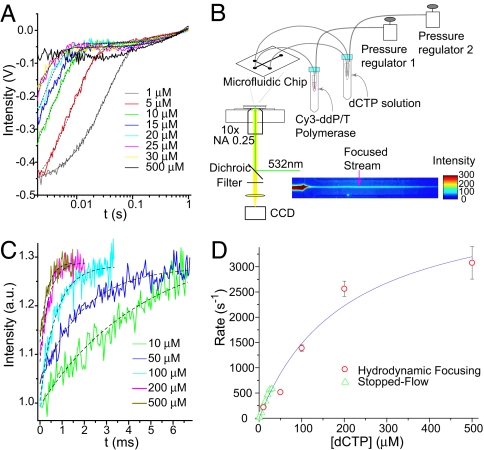
Fig. 5.
Ensemble-averaged measurements of the conformational change of T7 pol complex upon dCTP binding. (A) Stopped-flow fluorescence intensity traces of the Cy3 probe after mixing a solution of Cy3-ddP/T·T7 pol complex with a solution of different concentrations of dCTP. The black dashed lines represent double exponential fits of the data. (B) Experimental setup for continuous-flow measurement based on hydrodynamic focusing. (C) Fluorescence intensity traces of the Cy3-ddP/T·T7 pol after mixing with dCTP, obtained with a continuous-flow device. The black dashed lines are single exponential fits of the data. (D) dCTP concentration dependence of the observed rates of conformational change. A Michaelis–Menten fit was used to extract the rate of conformational change and Kd value for this process.
At higher dCTP concentrations, the rate of the fluorescence change is so fast (>1,000 s−1) that the stopped-flow instrument cannot capture it. We therefore constructed a continuous-flow device, using hydrodynamic focusing to achieve fast mixing at the microsecond time scale (35) (Fig. 5B; also see SI Appendix, Section 14 and Figs. S9 and S10 for details). The measured temporal profiles of Cy3-ddP/T·T7 pol fluorescence increase after mixing with dCTP are shown in Fig. 5C. The rate constants can be obtained from single exponential fits of the traces.
The dependence of the rates of the major fluorescence enhancement on dCTP concentrations from both measurements follows the Michaelis–Menten equation kobs = kmax [dCTP]/([dCTP] + Kd) (Fig. 5D). The kmax and Kd for dCTP binding are 4,500 s−1 and 200 μM, respectively. Had the fluorescence increase arisen solely from dCTP binding, the concentration dependence would be linear. The fact that we observed Michaelis–Menten kinetics provides direct proof that the fluorescence increase is related to a conformational change triggered by substrate binding (Scheme 2). This result rules out Scheme 1. Moreover, we reach the important conclusion that the observed conformational change is not rate-limiting in DNA synthesis because kmax is much greater than the nucleotidyl transfer rate (≈300 s−1) (10). We note that other conformational changes could occur after this conformational change and might limit the rate for the nucleotidyl transfer reaction (22).
The Effect of Mg2+.
We found that Mg2+ is required for the observed conformational change. Removal of Mg2+ from the solution by EDTA completely abolished the fluorescence increase of Cy3 upon dCTP binding. Single-molecule measurements of dCTP binding were also performed in the presence of 20 and 100 μM of Mg2+ (see SI Appendix, Section 5 and Fig. S2). At lower Mg2+ concentrations, we found a significantly lower rate for the forward transition at equivalent dCTP concentrations. The rate of the reverse transition was independent of Mg2+ concentration. We characterized the effect of Mg2+ on the affinity of dCTP by titrating fluorescence intensity with dCTP at different Mg2+ concentrations (see SI Appendix, Sections 4, 10, and 11 and Fig. S7A) and then determined that the binding constant for Mg2+ was ≈5 mM (see SI Appendix and Fig. S7B).
Even at very low Mg2+ concentration (≈3 μM) (see SI Appendix, Section 6 and Fig. S3), dCTP binding still induced a conformational change in the complex between T7 pol and the extendable Cy3-P/T without resulting in dCMP incorporation (SI Appendix, Section 9 and Fig. S6). We used a FRET quencher, Dabcyl-labeled dNTP, to test whether base incorporation occurs at this low concentration of Mg2+ (SI Appendix, Section 9), but no incorporation was found in this time period. Therefore, under these conditions, a conformational change can occur when only one Mg2+ is present in the catalytic center (B site), but nucleotidyl transfer will not occur. This result indicates that the two essential Mg2+ ions play different roles in the catalytic center and is consistent with previous reports (36, 37).
Single-Molecule Measurements with Noncomplementary dNTP.
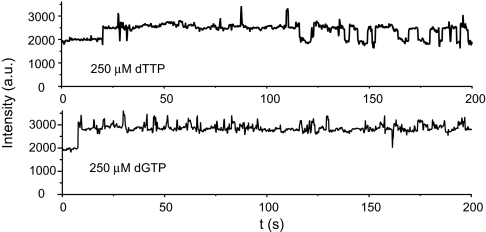
Can a noncomplementary dNTP induce a conformational change? The fluorescence increase upon noncomplementary dNTP binding is difficult to detect at the ensemble level because the equilibrium strongly favors reversal of the conformational change. When a high concentration (0.25 mM) of the noncomplementary dTTP or dGTP was added, together with the T7 pol, to the immobilized Cy3-P/T, we observed enhancement in the single-molecule fluorescence time traces, but the frequency was much lower than with the complementary dNTP (Fig. 6). The rate constants for the forward transitions were estimated to be 0.02 and 0.2 s−1 for dTTP and dGTP, respectively, ≈104 times lower than for dCTP. However, the reverse transition (2 s−1 for dTTP and 1.7 s−1 for dGTP) was only slightly faster than that observed with the complementary dCTP, indicating that the conformation of the transition state more closely resembles the final, rather than the initial, state (see Discussion).
Fig. 6.
Fluorescence intensity trajectories of the Cy3-ddP/T duplex showing a slow T7 pol conformational change induced in the presence of 0.25 mM noncomplementary substrates (dTTP or dGTP).
The observation of ≈104-fold discrimination against noncomplementary dNTPs in the forward conformational change upon dNTP binding indicates that such conformational change is crucial for base selectivity and is a major factor contributing to the high fidelity of DNA replication.
Discussion
Cy3 as a Sensitive Probe for the Functionally Important Conformational Change.
We selected Cy3 as a probe because of its high fluorescence quantum yield and its environment-dependent fluorescence. The photophysical properties of this dye have been studied extensively (38). The torsional motion of double bonds (Fig. 3D) in its excited state can bring Cy3 back to its electronic ground state without photon emission. Constraints of the torsional motion hinder the nonradiative decay pathway, giving a higher fluorescence quantum yield. In fact, the fluorescence quantum yield of Cy3 has strong solvent viscosity dependence. This property makes Cy3 a valuable probe for detecting subtle conformational alterations within an intact protein or protein complex.
The fluorescence enhancement of Cy3-P/T upon T7 pol binding is due to the steric hindrance of the excited-state torsional motion, as indicated in Fig. 3C. In the T7 pol complex, Cy3 may reside within a “cavity” formed by the DNA major groove and the polymerase thumb, finger, and exonuclease domains. Residues in this cavity can hinder the internal torsion of Cy3 and enhance its fluorescence. We noted that the fluorescence intensity of Cy3 barely increases upon Sequenase binding, possibly because of the 28-aa deletion and the subsequent disruption of this proposed cavity in Sequenase.
From analysis of available kinetic and structural information, we propose that the rise in Cy3 fluorescence reflects a conformational change in the polymerase ternary complex subsequent to dNTP binding. Additional evidence in support of this proposal is as follows. (i) With a noncomplementary dNTP, the events of fluorescence increase occurred much less frequently than with the complementary dNTP. (ii) In the absence of Mg2+, there was no fluorescence increase upon complementary dNTP binding. (iii) If substrate binding alone was able to induce a change in Cy3 fluorescence, then we would not have observed Michaelis–Menten behavior when the complementary dNTP was added to the T7 pol·DNA binary complex.
Fluorescence Change Reflects Fingers Domain Closing.
We have assigned the Cy3 fluorescence changes to conformational changes in the T7 pol complex. The only major conformational change in the T7 pol ternary complex, observed by x-ray crystallography after dNTP binding, is the open-to-closed transition in the fingers domain. The incoming dNTP initially binds to the fingers subdomain in the open state by means of interactions between the triphosphate tail and the conserved, positively charged side chains in the O helix (38).
A key relating the observed fluorescence change to the actual structural change in T7 pol is the Mg2+ dependence. It is known that Mg2+ is crucial for the open-to-closed transition because it stabilizes the closed complex (13), forming a coordination complex with the dNTP triphosphate tail as well as with the highly conserved Asp and Glu residues in the palm domain. Formation of the Mg2+ coordination complex in the active site also requires closing of the fingers to bring the dNTP triphosphate tail and the conserved Asp and Glu residues close enough for them to ligate the Mg2+. The dNTP-induced fluorescence increase in the T7 pol complex was observed only in the presence of Mg2+, indicating that the fluorescence change relies on the formation of the Mg2+ coordination complex. Because of the geometric constraints of the Mg2+ ligands, such a fluorescence change can only occur when the fingers domain of T7 pol is closed. This connection is further reinforced by the observation that the conformational change can also be induced by ddCTP, which produced an equivalent conformational state as shown by x-ray crystallography (13).
Closing of the fingers subdomain (Fig. 1B), by itself, does not appear to have a major effect on the environment of Cy3, according to the x-ray structure. Instead, the conformational change in the DNA template has a more significant effect on the environment of Cy3. In the open form of the DNA pol complex, the DNA templating base is flipped out of its helical conformation (38). Accompanying the closing of fingers, the templating base rotates into the active site and assumes a position as if it were actually part of a duplex. This movement of the templating base repositions the Cy3 probe so that it senses a different environment, giving rise to a the observed increase in fluorescence intensity. Additional evidence for this explanation also comes from previous reports. In particular, kinetic studies have shown that the incoming dNTP is likely to form H bonds with the templating base while T7 pol is still in the open state (38). Likewise, a recent crystal structure of the T7 RNA polymerase, which has close structural homology to T7 DNA pol, shows that such interaction occurs in the open form of the T7 RNA pol complex (39).
Closing of T7 Pol Fingers Domain Is a Fast Step Triggered by dNTP Binding.
It has not been resolved in the literature whether reversible open-to-closed conformational changes occur before binding of dNTP substrates (21). Our single-molecule experiment can address this question. Because no increase of fluorescence intensity of the T7 pol·Cy3-P/T complex was found in the single-molecule fluorescence time traces in the absence of dNTP, there is no evidence for the existence of dynamic equilibrium between open and closed states before dNTP binding. Therefore, binding of an incoming dNTP is required to induce fingers closing.
Our observations show that the fingers closing is a rapid step in the T7 pol nucleotidyl transfer reaction and is not rate-limiting, which is in agreement with the kinetic results reported for polβ (20), KF (20), and Klentaq1 (21). We note that fast conformational change upon binding of the complementary base is necessary for high replication efficiency. If the conformational change upon binding of complementary dNTP were to be rate-limiting, the overall DNA replication rate would be significantly reduced because of competitive inhibition by noncomplementary dNTPs and NTPs.
We note that other conformational changes could occur after the one that we observed. These ensuing conformational changes might be rate-limiting for the nucleotidyl transfer reaction. In fact, Tsai and Johnson (22) observed a much slower conformational change by a fluorescence label at a different location. A possible change could consist of a small adjustment of amino acid side chains that ligate Mg2+ in the A metal site, as suggested in ref. 40.
Conformational Change Plays a Major Role in Determining DNA Replication Fidelity.
Because the nucleotidyl transfer step is precluded in our experiment, we have been able to determine the rate of the conformational change. We observed that binding of noncomplementary dNTPs can also give rise to an increase of Cy3 fluorescence, with amplitude similar to that for complementary dNTP. However, a much lower rate of fluorescence intensity transitions was obtained with noncomplementary vs. complementary dNTPs. For noncomplementary bases, the rate of the conformational change is comparable to the overall rate of incorporation and could be the rate-limiting step (41).
The catalytic efficiency kcat/Km for complementary vs. noncomplementary dNTP substrates determines the extent of base discrimination by DNA pols (5, 22). The rate of fingers closing for complementary dNTP substrates is ≈103 to 105 times higher than for noncomplementary dNTPs. Therefore, the conformational change we observed serves as a major kinetic checkpoint to discriminate against noncomplementary dNTP incorporation.
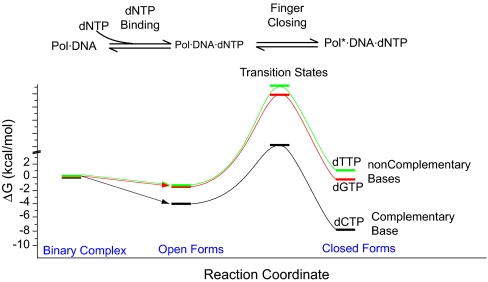
Fig. 7 shows an energy diagram for the dNTP-binding step and the open-to-closed transition of the T7 pol complex for complementary and noncomplementary dNTPs. The variation in stability of closed forms with different dNTP substrates affects the rate of fingers closing significantly but has a minimal effect on the rate of fingers opening. This implies that the transition state for the fingers closing has a structure that more closely resembles the closed, rather than open, form. Perturbations affecting the closed form should affect the transition state in a similar way. As a result, the more stable the T7 pol closed complex is, the lower the barrier (and higher the rate) for the open-to-closed transition. Therefore, it is the molecular interactions between the T7 pol·DNA binary complex and the incoming dNTP that give rise to the enhanced replication fidelity, in addition to the fidelity provided by the Watson–Crick base pairing (42–45).
Fig. 7.
Free-energy diagram for complementary and noncomplementary dNTP substrate binding (calculated from previous reports) and for the open-to-closed conformational change of the T7 pol ternary complex (calculated from our experimental data).
In summary, the photophysical properties of Cy3 have allowed us to observe the conformational change in the T7 pol ternary complex before phosphodiester bond formation. Structural information, together with Cy3 fluorescence enhancement mechanisms and Mg2+ dependence of conformational change, suggest that the transition we observed is assigned to closing of the fingers subdomain. Based on the ensemble-averaged observations, this conformational change is a very rapid, non-rate-limiting step. The conformational change makes a major contribution to fidelity because it allows efficient discrimination against the incorporation of noncomplementary bases.
Materials and Methods
Materials.
All reagents were the highest grade obtainable and were used without further purification. See SI Appendix, Section 1 for details.
P/T Preparation.
If not otherwise specified, the following oligonucleotides were used as the primer and template strands of the P/T: primer, 5′-GCCTCGCAGCCGTCCAACCAACTCA-3′ (P); template, 3′-BiotinTEG-CGGAGCGTCGGCAGGTTGGTTGAGTGCC-Cy3–5′ (Cy3-T). The duplex Cy3-P/T was prepared by mixing the Cy3-T and P oligos at a 1/1.5 ratio in 50 mM phosphate buffer, pH 7.5.
We made observations by using 40 mM Tris buffer, pH7.5, containing 10 mM MgCl2 in experiments in which the dNTP substrate was present. A thiophosphoryl primer with the same sequence as primer P, 5′-GCCTCGCAGCCGTCCAACCAACT∗CddA-3′ (ddP), where ∗ is the phosphorothioate linkage between the deoxynucleoside residues and ddA is the dideoxynucleoside residue (see SI Appendix, Section 1), was prepared and annealed to the Cy3-T. In experiments involving the ddP primer, 20 μM ddATP was added to the reaction mixture to ensure that the primer always had a ddA residue at its 3′ terminus, in case it is cut by the exonuclease activity of T7 pol.
Microscope Sample Preparation and Single-Molecule Imaging.
Biotin-functionalized glass coverslips and flow cells were prepared as described in ref. 46. See SI Appendix, Section 2 for details of sample preparation and single-molecule fluorescence imaging.
Ensemble-Averaged Fluorescence Measurements.
For stopped-flow fluorescence measurements, see SI Appendix, Section 13. For continuous-flow measurements using hydrodynamic focusing (35), see SI Appendix, Section 14.
Supplementary Material
Acknowledgments
We thank Drs. Antoine M. van Oijen, Paul Blainey, Hong Zhang, and Jeremy Agresti and Mr. Brian English for their suggestions and help. This work was supported by the Department of Energy, Office of Science, Office of Basic Energy Science, Chemical Sciences (X.S.X.) and by U.S. Public Health Service Grant GM063276-01 (to W.H.K.).
Abbreviations
- DNA pol
DNA polymerase
- KF
the Klenow fragment of Escherichia coli polymerase I
- polβ
human pol β
- P/T
primer/template
- T7 pol
T7 DNA pol.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700920104/DC1.
References
- 1.Kornberg A, Baker TA. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 2.Goodman MF. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 3.Tabor S, Richardson CC. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 5.Johnson KA. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 6.Showalter AK, Tsai MD. Biochemistry. 2002;41:10571–10576. doi: 10.1021/bi026021i. [DOI] [PubMed] [Google Scholar]
- 7.Joyce CM, Benkovic SJ. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 8.Kunkel TA, Bebenek R. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 9.Kunkel TA. J Biol Chem. 1992;267:18251–18254. [PubMed] [Google Scholar]
- 10.Patel SS, Wong I, Johnson KA. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 11.Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ. Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 12.Dahlberg ME, Benkovic SJ. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 13.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 14.Huang H, Chopra R, Verdine GL, Harrison SC. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 15.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 16.Johnson SJ, Taylor JS, Beese LS. Proc Natl Acad Sci USA. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Korolev S, Waksman G. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunlap CA, Tsai MD. Biochemistry. 2002;41:11226–11235. doi: 10.1021/bi025837g. [DOI] [PubMed] [Google Scholar]
- 19.Fidalgo da Silva E, Mandal SS, Reha-Krantz LJ. J Biol Chem. 2002;277:40640–40649. doi: 10.1074/jbc.M203315200. [DOI] [PubMed] [Google Scholar]
- 20.Purohit V, Grindley NDF, Joyce CM. Biochemistry. 2003;42:10200–10211. doi: 10.1021/bi0341206. [DOI] [PubMed] [Google Scholar]
- 21.Rothwell PJ, Mitaksov V, Waksman G. Mol Cell. 2005;19:345–355. doi: 10.1016/j.molcel.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 22.Tsai YC, Johnson KA. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aramendia PF, Negri RM, Roman ES. J Phys Chem. 1994;98:3165–3173. [Google Scholar]
- 24.Xie XS, Trautman JK. Annu Rev Phys Chem. 1998;49:441–480. doi: 10.1146/annurev.physchem.49.1.441. [DOI] [PubMed] [Google Scholar]
- 25.Weiss S. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 26.Moerner WE, Orrit M. Science. 1999;283:1670–1676. doi: 10.1126/science.283.5408.1670. [DOI] [PubMed] [Google Scholar]
- 27.Zhuang X, Kim H, Pereira MJB, Babcock HP, Walter NG, Chu S. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 28.Lu HP, Xun LY, Xie XS. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 29.English BP, Min W, van Oijen AM, Lee KT, Luo GB, Sun HY, Cherayil BJ, Kou SC, Xie XS. Nat Chem Biol. 2006;2:87–94. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- 30.Min W, Luo GB, Cherayil BJ, Kou SC, Xie XS. Phys Rev Lett. 2005;94:198302. doi: 10.1103/PhysRevLett.94.198302. [DOI] [PubMed] [Google Scholar]
- 31.Yang H, Luo GB, Karnchanaphanurach P, Louie TM, Rech I, Cova S, Xun L, Xie XS. Science. 2003;302:262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 32.Chung SH, Kennedy RA. J Neurosci Methods. 1991;40:71–86. doi: 10.1016/0165-0270(91)90118-j. [DOI] [PubMed] [Google Scholar]
- 33.Tabor S, Richardson CC. J Biol Chem. 1989;264:6447–6458. [PubMed] [Google Scholar]
- 34.Brautigam CA, Steitz TA. J Mol Biol. 1998;277:363–377. doi: 10.1006/jmbi.1997.1586. [DOI] [PubMed] [Google Scholar]
- 35.Knight JB, Vishwanath A, Brody JP, Austin RH. Phys Rev Lett. 1998;80:2737–2740. [Google Scholar]
- 36.Zhong X, Patel SS, Tsai MD. J Am Chem Soc. 1998;120:235–236. [Google Scholar]
- 37.Bakhtina M, Lee S, Wang Y, Dunlap C, Lamarche B, Tsai MD. Biochemistry. 2005;44:5177–5187. doi: 10.1021/bi047664w. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Dutta S, Doublié S, Bdour HM, Taylor JS, Ellenberger T. Nat Struct Mol Biol. 2004;11:784–790. doi: 10.1038/nsmb792. [DOI] [PubMed] [Google Scholar]
- 39.Temiakov D, Patlan V, Anikin M, McAllister WT, Yokoyama S, Vassylyev DG. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 40.Yang L, Beard WA, Wilson SH, Broyde S, Schlick T. J Mol Biol. 2002;317:651–671. doi: 10.1006/jmbi.2002.5450. [DOI] [PubMed] [Google Scholar]
- 41.Wong I, Patel SS, Johnson KA. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 42.Petruska J, Sowers LC, Goodman MF. Proc Natl Acad Sci USA. 1986;83:1559–1562. doi: 10.1073/pnas.83.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reineks EZ, Berdis AJ. Biochemistry. 2004;43:393–404. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- 44.Morales JC, Kool ET. Nat Struct Mol Biol. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 45.Kool ET. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 46.van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS. Science. 2003;301:1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.