Abstract
We found that Lactobacillus reuteri CRL1098, a lactic acid bacterium isolated from sourdough, is able to produce cobalamin. The sugar-glycerol cofermentation in vitamin B12-free medium showed that this strain was able to reduce glycerol through a well-known cobalamin-dependent reaction with the formation of 1,3-propanediol as a final product. The cell extract of L. reuteri corrected the coenzyme B12 requirement of Lactobacillus delbrueckii subsp. lactis ATCC 7830 and allowed the growth of Salmonella enterica serovar Typhimurium (metE cbiB) and Escherichia coli (metE) in minimal medium. Preliminary genetic studies of cobalamin biosynthesis genes from L. reuteri allowed the identification of cob genes which encode the CobA, CbiJ, and CbiK enzymes involved in the cobalamin pathway. The cobamide produced by L. reuteri, isolated in its cyanide form by using reverse-phase high-pressure liquid chromatography, showed a UV-visible spectrum identical to that of standard cyanocobalamin (vitamin B12).
Lactobacillus reuteri is a member of the gastrointestinal ecosystems of humans, poultry, swine, and other animals (5). This bacterium possesses certain probiotic properties, such as the hypocholesterolemic effect (21, 22), and has been shown to possess a unique property among bacteria in its ability to excrete reuterin (a mixture of monomeric, hydrated monomeric, and cyclic dimeric forms of 3-hydroxypropionaldehyde [3-HPA]), a broad-spectrum antimicrobial agent produced during anaerobic sugar-glycerol cofermentation (19, 20). The pathway for conversion of glycerol to 3-HPA is mediated through a cobalamin-dependent glycerol dehydratase (7).
Cobalamins, both as deoxyadenosylcobalamin and methylcobalamin, are involved as cofactors in a variety of enzymatic reactions and are synthesized by some bacteria and archaea (14, 17).
In cattle, sheep, and other ruminants, microorganisms present in the rumen can synthesize cobalamin, but humans do not have such microflora in their small intestines and must absorb the coenzyme from food (17). Albert et al. (1) reported that some apparently healthy southern Indian subjects harbored in their small intestines microorganisms from the genera Pseudomonas and Klebsiella that were able to synthesize cobalamin. These authors also found that among 12 lactobacilli tested, none produced detectable levels of coenzyme B12. In this respect, the information concerning the ability of lactic acid bacteria (LAB) to produce vitamins is very scarce, and most species are auxotrophic for these compounds (8, 15). Most studies on vitamin B12 in the genus Lactobacillus have dealt with the auxotrophic requirement of Lactobacillus delbrueckii subsp. lactis (Lactobacillus leichmannii), particularly strain ATCC 7830, a microorganism used in microbiological assays of the B12 content of food products (12, 18). In this work, we demonstrate that L. reuteri CRL1098 produces cobalamin. Unlike Pseudomonas and Klebsiella organisms, LAB possess GRAS (generally regarded as safe) status; therefore, the finding of a LAB strain able to produce cobalamin would be of remarkable importance for the food industry and in medical and veterinary fields. To our knowledge this is the first report of cobalamin biosynthesis by LAB.
Sugar-glycerol cofermentation.
L. reuteri is able to use glycerol as an external hydrogen acceptor during cofermentation with glucose (7). In this microorganism, glycerol is converted by a coenzyme B12-dependent glycerol dehydratase to 3-HPA. Subsequently, 3-HPA is reduced by NADH to 1,3-propanediol (1,3-PDL) mediated by the 1,3-PDL:NAD oxidoreductase (7). Since the glycerol dehydratase requires 5′desoxyadenosylcobalamin as a cofactor, the B12 has to be obtained from the medium or biosynthesized by the microorganism in order to be able to utilize glycerol. Therefore, we evaluated the ability of L. reuteri CRL1098 to coferment glycerol in a vitamin B12-free medium. The results of the high-performance liquid chromatography (HPLC) analysis, which are summarized in Table 1, clearly show that L. reuteri CRL1098 can utilize glycerol during glucose fermentation without exogenous cobalamin and with a concomitant production of 1,3-PDL. Besides, no significant stimulation was observed when exogenous cyanocobalamin (CN-Cbl) was added to the B12-free medium, which would indicate that endogenous cobalamin biosynthesis is repressed under this condition, as has been observed in other microorganisms (16).
TABLE 1.
Glycerol consumption and 1,3-PDL formation by L. reuteri CRL1098 during glucose-glycerol cofermentation in vitamin B12-free medium
| Time (h) | Sample no.a | Glycerol concn (mM) | 1,3-PDL concn (mM) | Growth (OD)b |
|---|---|---|---|---|
| 0 | 1 | 0.00 | 0.41 | 0.1 |
| 2 | 37.78 | 0.96 | 0.09 | |
| 3 | 36.7 | 0.82 | 0.09 | |
| 24 | 1 | 0.00 | 0.55 | 3.85 |
| 2 | 19.11 | 15.32 | 2.79 | |
| 3 | 19.54 | 16.68 | 2.98 |
Sample numbers correspond to growth and fermentation of samples in B12-free medium. Sample 1, without glycerol; sample 2, with glycerol; sample 3, with glycerol and 20 nM cyanocobalamin.
OD, optical density at 560 nm.
Preparation of cell extracts.
In order to analyze the production of cobalamin by L. reuteri, a culture of strain CRL1098 was inoculated in vitamin B12-free assay medium (Merck, Buenos Aires, Argentina) and transferred three times in the same medium. Cells grown in 50 ml of the defined medium at 37°C for 16 h were harvested and washed twice with 0.1 M phosphate buffer, pH 7.0. Washed cells were resuspended in 1 ml of extraction buffer (0.1 M Na2HPO4 [pH 4.5] [citric acid], 0.005% KCN) and disrupted with 1 g of glass beads (0.1-mm diameter) in a FastPrep FP120 (Qbiogene, Carlsbad, Calif.) sample preparation system. Extraction buffer was then added up to a final volume of 20 ml and autoclaved (at 120°C for 15 min). After centrifugation (at 8,000 × g for 10 min), the supernatant was passed over an Isolute solid-phase extraction (SPE) column (500-mg C18 end-capped column with a 3-ml reservoir volume) previously activated with 2 ml of acetonitrile. The column was washed twice with 2 volumes of distilled water to remove salts and other hydrophilic contaminants. After the cartridge was washed with water, the cobalamin was eluted with 1 volume of 50% acetonitrile and concentrated to dryness in vacuo at 30°C. The residue was dissolved in 1 ml of sterile distilled water and stored in the dark at −20°C until used.
Cobalamin detection and quantitative bioassay.
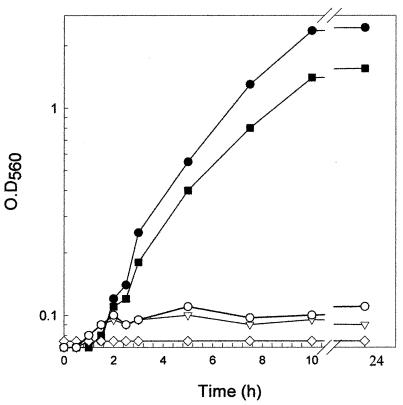
To demonstrate the production of cobalamin by L. reuteri CRL1098, three different bioassays were performed. First, L. delbrueckii subsp. lactis ATCC 7830, a strain that requires B12, was used to evaluate the cobalamin content in L. reuteri cell extract (CE) (12). The results shown in Fig. 1 demonstrate that L. delbrueckii ATCC 7830 grew in vitamin B12-free assay broth only when the CE from L. reuteri or standard CN-Cbl was added to the medium. On the other hand, no growth was detected when the CE from a control strain (Lactobacillus plantarum ATCC 8014 or L plantarum WCFS1) was used (data not shown). Quantification analyses using a CN-Cbl standard curve indicated that L. reuteri produced approximately 0.5 mg of cobalamin · liter−1. This value is based on the intracellular content since no cobalamin was found in the culture supernatant.
FIG. 1.
Growth of L. delbrueckii subsp. lactis ATCC 7830 in vitamin B12-free medium supplemented with CE of L. reuteri CRL1098 (▪), CN-Cbl (•), CE of L. plantarum ATCC 8014 (○), and saline solution (▿). Results for a CE sterility control, ◊, are also shown. O.D560, optical density at 560 nm. L. delbrueckii ATCC 7830 was preserved in MRS broth (9) plus glycerol at −70°C.
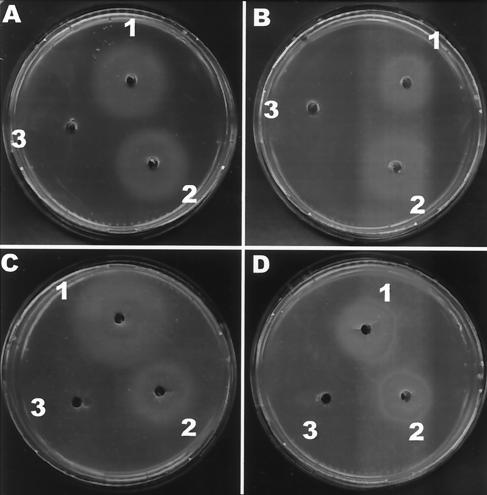
Although the L. delbrueckii B12 assay is a reference method according to the Official Methods of Analysis of AOAC International (11a), it presents a drawback, since this microorganism may also utilize the deoxyribonucleotide pool present in the sample. For this reason, two other bioassays using Salmonella enterica serovar Typhimurium AR2680 (metE cbiB) (16) and Escherichia coli RK4379 [(ΔargF-lac)U169 araD139 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR22 non-9 gyrA219 metE] (4) as indicator strains were performed. Salmonella serovar Typhimurium AR2680 requires cobalamin to grow in minimal medium due to the cobalamin-independent methionine synthase (MetE) mutation. Therefore, the only way to synthesize methionine is through the cobalamin-dependent methionine synthase (MetH). Since this strain also possesses a mutation in the cbiB gene (coding for cobinamide synthase), exogenous cobalamin (or a late precursor) has to be added to the medium. The CE from L. reuteri CRL1098 was examined for its ability to correct the cobalamin requirement of Salmonella serovar Typhimurium AR2680 in minimal medium. For this purpose, AR2680 cells grown for 16 to 18 h in TY medium (3) were collected by centrifugation, washed twice with 0.1 M phosphate buffer (pH 7.0), and resuspended to the original volume. This culture was seeded onto minimal agar A medium (4), and wells were made in each agar plate. Twenty microliters of CEs from L. reuteri CRL1098 and L. plantarum ATCC 8014 and standard CN-Cbl solution (50 nM) were loaded in each well and incubated overnight under anaerobic conditions at 37°C. The diameters of the growth area of different dilutions of the CE from L. reuteri were compared with those produced by standard dilutions of CN-Cbl. Figure 2A shows the minimal agar plates seeded with serovar Typhimurium AR2680 and inoculated with the CEs from L. reuteri and L. plantarum ATCC 8014. These results demonstrated that only the CE from L. reuteri and CN-Cbl were able to correct the cobalamin requirement of strain AR2680. A parallel bioassay using E. coli RK4379 (metE) confirmed the results obtained with AR2680 (Fig. 2C). Additionally, incubation of the minimal medium plates from both microoganisms in air showed decreases in the diameters of the growth halos (Fig. 2B and D). This variation between the levels of anaerobic and aerobic growth might be due to the cobalamin-dependent methionine synthase (MetH) activities of both strains, since the catalytic turnover of MetH is lower in the aerobic environment because the oxidation of cob(I)alamin to cob(II)alamin (inactive) is higher under this condition and the regeneration of the coenzyme methylcobalamin is reduced (10), which leads to smaller growth halos. Furthermore, control plates with methionine in the wells did not show significant differences in the growth halos (data not shown). In order to confirm that cobalamin was indeed the molecule responsible for the Salmonella and E. coli metE complementation, we performed another control assay using the strain E. coli NC17 [RK4379 ΔbtuF(11-239)::Km], a mutant defective in a periplasmic cobalamin-binding protein involved in cobalamin uptake (4). This strain was assayed with the methionine and ethanolamine assay described by Cadieux et al. (4) to test the ability of this mutant to use cobalamin for methionine synthesis and ethanolamine as a nitrogen source. As expected, due to the transport deficiency, neither L. reuteri CE nor CN-Cbl (up to 5 μM) complemented the growth of E. coli NC17 in either bioassay (data not shown). In consequence, these last experiments strongly suggest that cobalamin or other cobamides present in L. reuteri CE are responsible for the growth of E. coli and Salmonella in the minimal medium.
FIG. 2.
Bioassay of the CE from L. reuteri CRL1098 using S. enterica serovar Typhimurium AR2680 (A and B) and E. coli RK4379 (C and D) as test organisms in minimal glucose agar medium. Wells were inoculated with CE from L. reuteri (well 1), standard CN-Cbl (well 2), and CE from L. plantarum ATCC 8014 (well 3). Plates were incubated overnight under anaerobic (A and C) or aerobic (B and D) conditions.
A quantitative analysis using the Salmonella bioassay showed that the L. reuteri CE contained 50 μg of cobalamin · liter−1, a value 10-fold lower than that obtained with the L. delbrueckii bioassay. This difference may be explained by the fact that L. delbrueckii reacts not only to cobalamin but also to other cobamides, which might not be as active in the Salmonella bioassay. Deoxyribonucleotides present in the CE from strain CRL1098 might also enhance the growth of L. delbrueckii ATCC 783 when a small concentration of cobalamin is present.
Identification of cob genes in L. reuteri CRL1098.
To verify the presence of cob genes in L. reuteri, degenerate primers CP10f (5′-CTNYTNGGNGCNGGNCC-3′) and CP1r (5′-CCNACNACDATNARNGCNGG-3′) were designed based on the sequence homologies of uroporphyrinogen-III methyltransferases (CobA [not to be confused with the adenosyl transferase CobA from Salmonella] or CysGB) available from different microorganisms (17). These primers were used to amplify by PCR a 650-bp fragment by using genomic DNA from L. reuteri, which encoded a partial open reading frame (ORF) homologous to the cobA gene from Propionibacterium shermanii that is responsible for uroporphyrinogen-III methyltransferase activity. To amplify the 3′ and 5′ regions adjacent to this sequence, new primers were designed and used in a modified PCR technique, called Uneven PCR (6). Once the complete sequence was assembled, a new PCR was performed in order to amplify a 2,016-bp fragment from genomic DNA of L. reuteri CRL1098. The sequence (accession number AF067123) revealed one complete ORF spanning 1,392 bp, coding for a 464-amino-acid protein homologous to CobA/HemD from Selenomonas ruminantium (31.8%) (2), Listeria innocua (36.2%), and Listeria monocytogenes (36.9%) (11); CobA/HemD is a bifunctional protein with S-adenosyl-l-methionine-uroporphyrinogen-III methyltransferase and uroporphyrinogen-III synthase activities. Furthermore, two partial ORFs coding for precorrin-6-reductase (CbiJ) and anaerobic cobalt chelatase (CbiK), enzymes belonging to the cobalamin pathway, were also found (16). Likewise, for these partial ORFs, the higher homologies were found with the cobalamin biosynthesis proteins from Listeria monocytogenes (sequence identities of 45 and 42% for CbiK and CbiJ, respectively) and Listeria innocua (sequence identities of 44 and 42% for CbiK and CbiJ, respectively). No homology was found in a comparative analysis of the sequences of LAB genomes (data not shown) (draft sequences of LAB genomes are available at http://genome.ornl.gov/microbial/).
Purification of the cobalamin produced by L. reuteri.
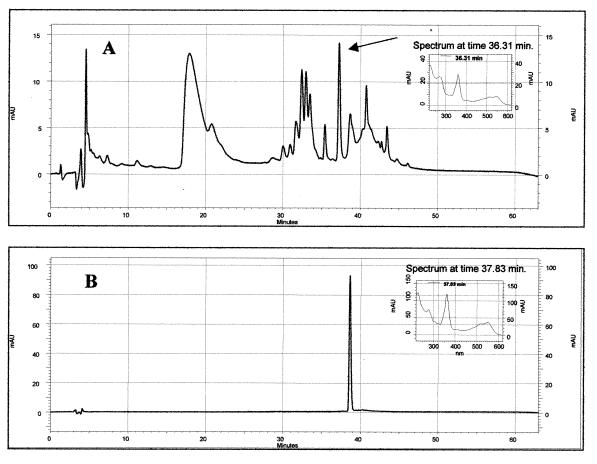
To isolate the cobamide produced by L. reuteri, the CE was passed through a C18 solid-phase extraction column and purified by reverse-phase (RP)-HPLC with a Waters (Milford, Mass.) 600E system automated gradient controller with a Waters 717 autosampler, a 250- by 3-mm Betasil phenyl column (Thermo Hypersil-Keystone, Waltham, Mass.), and an SPD-M10A VP diode array detector (Shimadzu Corporation, Kyoto, Japan). Figure 3 shows the HPLC chromatograms of the CE (Fig. 3A) and the standard CN-Cbl (Fig. 3B). The CE showed a peak with a retention time (RT) of 36.31 min that exhibited a UV-visible (UV-Vis) spectrum (Fig. 3A, inset) identical to the spectrum from CN-Cbl (Fig. 3B, inset), even though the vitamin B12 eluted from the column at 37.83 min (Fig. 3B). In addition, when the peak sample (RT, 36.31 min) was collected and analyzed by the Salmonella bioassay, it showed the same B12 complementation property as the crude CE (data not shown). Recently, Maggio-Hall and Escalante-Semerena (13), during an in vitro synthesis of the cobalamin nucleotide loop, isolated a cobamide with a UV-Vis spectrum identical to that of CN-Cbl but with a different RT in RP-HPLC (a difference of 3 min). Using mass spectrometry, they identified the compound as a phosphorylated form of CN-Cbl (CN-Cbl-5′-P). In our chromatograms the difference in RT between L. reuteri cobalamin and CN-Cbl was about 1 min, and although we cannot distinguish between the phosphorylated and desphosphorylated forms of the cobalamin, we can confirm the presence of cobalamin in the CE of L. reuteri CRL1098 based on the similarity of the UV-Vis spectra. On the other hand, quantification data from the HPLC analyses indicated similar cobalamin levels obtained with the E. coli and Salmonella bioassays (∼50 μg/liter).
FIG. 3.
(A) RP-HPLC chromatogram of the CE from L. reuteri CRL1098 and the UV-Vis spectrum of the peak (inset). (B) For reference, the chromatogram obtained with the standard CN-Cbl is shown as well as its corresponding UV-Vis spectrum (inset). mAU, milli-absorbance units.
The synthesis of this kind of compound by a LAB strain is a surprising finding based on the auxotrophic characteristics of many LAB to several vitamins and amino acids and the magnitude and complexity of the cobalamin biosynthetic pathways. From a biotechnological point of view, this microorganism might be a good candidate to increase the cobalamin content in fermented food. One important limitation to this projection is the fact that cobalamin is not excreted and remains in the cytoplasm. One alternative to circumvent this problem would be the isolation of autolytic mutants (e.g., during transit in the stomach) that would release the cobalamin in the gastrointestinal tract. Moreover, overexpression of key genes of the cobalamin biosynthetic pathway of L. reuteri CRL1098 might be an interesting alternative to consider in order to increase the coenzyme B12 production of this strain.
Acknowledgments
We are grateful to Martin Warren and Robert Kadner for sending us S. enterica and E. coli strains.
This work was supported in part by grants from CONICET and Agencia Nacional de Promoción Científica y Tecnológica. J. L. Vera is the recipient of a fellowship from CONICET (Buenos Aires, Argentina).
M. P. Taranto and J. L. Vera have contributed equally to this work.
REFERENCES
- 1.Albert, M. J., V. I. Mathan, and S. J. Baker. 1980. Vitamin B12 synthesis by human small intestinal bacteria. Nature 283:781-782. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, P. J., B. Entsch, and D. B. McKay. 2001. A gene, cobA+hemD, from Selenomonas ruminantium encodes a bifunctional enzyme involved in the synthesis of vitamin B12. Gene 281:63-70. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingstom, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1999. Short protocols in molecular biology, 4th ed. John Wiley and Sons, New York, N.Y.
- 4.Cadieux, N., C. Bradbeer, E. Reeger-Schneider, W. Köster, A. K. Mohanty, M. C. Wiener, and R. J. Kadner. 2002. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J. Bacteriol. 184:706-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casas, I. A., F. W. Edens, and W. J. Dobrogosz. 1998. Lactobacillus reuteri: an effective probiotic for poultry and other animals, p. 475-518. In S. Salminen and A. von Wright (ed.), Lactic acid bacteria: microbiology and functional aspects. Marcel Dekker, New York, N.Y.
- 6.Chen, X., and R. Wu. 1997. Direct amplification of unknown genes and fragments by Uneven polymerase chain reaction. Gene 185:195-199. [DOI] [PubMed] [Google Scholar]
- 7.Daniel, R., T. A. Bobik, and G. Gottschalk. 1998. Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes. FEMS Microbiol. Rev. 22:553-566. [DOI] [PubMed] [Google Scholar]
- 8.Deguchi, Y., and T. Morishita. 1992. Nutritional requirements in multiple auxotrophic lactic acid bacteria: genetic lesions affecting amino acid biosynthetic pathways in Lactococcus lactis, Enterococcus faecium, and Pediococcus acidilactici. Biosci. Biotechnol. Biochem. 56:913-918. [DOI] [PubMed]
- 9.de Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 10.Frasca, V., R. V. Banerjee, W. R. Dunham, R. H. Sands, and R. G. Matthews. 1988. Cobalamin-dependent methionine synthase from Escherichia coli B: electron paramagnetic resonance spectra of the inactive form and the active methylated form of the enzyme. Biochemistry 27:8458-8465. [DOI] [PubMed] [Google Scholar]
- 11.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 11a.Horwitz, W. (ed.). 2000. Official methods of analysis of AOAC International, 17th ed. AOAC International, Gaithersburg, Md.
- 12.Kelleher, B. P., and S. D. Broin. 1991. Microbiological assay for vitamin B12 performed in 96-well microtitre plates. J. Clin. Pathol. 44:592-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maggio-Hall, L. A., and J. C. Escalante-Semerena. 1999. In vitro synthesis of the nucleotide loop of cobalamin by Salmonella typhimurium enzymes. Proc. Natl. Acad. Sci. USA 96:11798-11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martens, J. H., H. Barg, M. J. Warren, and D. Jahn. 2002. Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 58:275-285. [DOI] [PubMed] [Google Scholar]
- 15.Morishita, T., Y. Deguchi, M. Yajima, T. Sakurai, and T. Yura. 1981. Multiple nutritional requirements of lactobacilli: genetic lesions affecting amino acid biosynthetic pathways. J. Bacteriol. 148:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raux, E., A. Lanois, F. Levillayer, M. J. Warren, E. Brody, A. Rambach, and C. Thermes. 1996. Salmonella typhimurium cobalamin (vitamin B12) biosynthetic genes: functional studies in S. typhimurium and Escherichia coli. J. Bacteriol. 178:753-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth, J. R., J. G. Lawrence, and T. A. Bobik. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50:137-181. [DOI] [PubMed] [Google Scholar]
- 18.Skeggs, H. R. 1963. Lactobacillus leichmanii assay for vitamin B12, p. 551-561. In F. Kavanagh (ed.), Analytical microbiology. Academic Press, New York, N.Y.
- 19.Talarico, T. L., I. A. Casas, T. C. Chung, and W. J. Dobrogosz. 1988. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 32:1854-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talarico, T. L., and W. J. Dobrogosz. 1989. Chemical characterization of an antimicrobial substance produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 33:674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taranto, M. P., M. Medici, G. Perdigón, A. P. Ruiz Holgado, and G. F. Valdez. 2000. Effect of Lactobacillus reuteri on the prevention of hypercholesterolemia in mice. J. Dairy Sci. 83:401-403. [DOI] [PubMed] [Google Scholar]
- 22.Taranto, M. P., M. Medici, G. Perdigón, A. P. Ruiz Holgado, and G. F. Valdez. 1998. Evidence for hypocholesterolemic effect of Lactobacillus reuteri in hypercholesterolemic mice. J. Dairy Sci. 81:2336-2340. [DOI] [PubMed] [Google Scholar]