Abstract
Vesicle encapsulation offers a biologically relevant environment for many soluble proteins and nucleic acids and an optimal immobilization medium for single-molecule fluorescence assays. Furthermore, the confinement of biomolecules within small volumes opens up new avenues to unique experimental configurations. Nevertheless, the vesicles' impermeability, even toward ions and other small molecules such as ATP, hinders more general applications. We therefore developed methods to induce pores into vesicles. Porous vesicles were then used to modulate the interaction between Escherichia coli RecA proteins and ssDNA by changing the extravesicular nucleotides. Repetitive binding and dissociation of the same RecA filament on the DNA was observed with a rebinding rate two orders of magnitude greater than in the absence of confinement, suggesting a previously unreported nucleation pathway for RecA filament. This method provides a biofriendly and simple alternative to surface tethering that is ideal for the study of transient and weakly interacting biological complexes.
Keywords: RecA, single molecule, vesicle encapsulation
The study of biochemical reactions at single-molecule resolution requires prolonged observation periods that normally necessitate their tethering to an artificial surface while minimizing alterations in the biological activities. For example, these measurements are frequently made on biological macromolecules with a specific single-point attachment on glass (1) or polymer-coated surfaces (2). Besides direct surface tethering methods, other approaches have been pursued by using molecular confinement by entrapping proteins inside silicate glass (3), polyacrylamide (4), agarose gels (5), elastic polymer chambers (6), or zero-mode metallic waveguides (7). However, although these innovative approaches have their own merits, potential interactions with the artificial surfaces could not be completely ruled out. Indeed, there have been reports of variability of surface environment and suspicion that the observed heterogeneity in dynamic properties of single molecules might be an artifact induced by the surroundings (8, 9).
Another promising approach for observing molecules for extended periods is to restrict molecules within small unilamellar vesicles (diameter ≈50–200 nm) that are anchored on the surface (10). Using this scheme, Rhoades et al. (11) were able to measure the equilibrium folding–unfolding fluctuations of single-protein molecules in real time. In contrast to the other immobilization practices, vesicles provide a more native environment for the biological entities, so long as the interaction with the membrane is minimal. In earlier studies, we adopted this vesicle encapsulation technique to test whether the markedly heterogeneous folding and unfolding dynamics of the hairpin ribozyme is intrinsic (12) and to ascertain that the extreme conformational diversity of human telomeric DNA is not caused by surface tethering (13). Although recognized as an “elegant” method, vesicle encapsulation comes with its own caveat (14); the lipid bilayer membrane of the vesicles acts as a barrier for the solutes, making the buffer exchange practically impossible.
Here, we overcome the limitation by making the vesicle porous and explore new avenues that this technique avails for single molecule fluorescence assays.
Vesicles as Biomimetic Nanocontainers
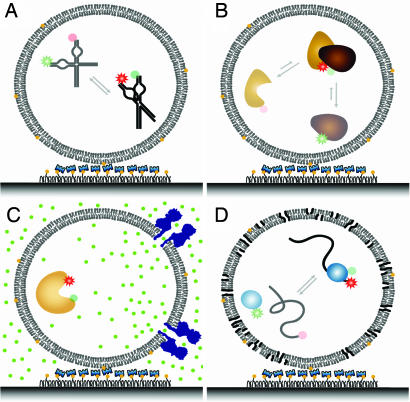
Inside the vesicles, molecules are essentially free of potential perturbations that might be induced upon direct surface tethering (Scheme 1A). In addition, vesicle encapsulation offers several appealing experimental possibilities.
Scheme 1.
Possible FRET assays with vesicle encapsulation. (A) Nucleic acids dynamics. Here the surface effects are minimized, and the molecular construct is simplified because the DNA/RNA needs no special modification for surface tethering. (B) Study of weak or transient interactions. Here a high local concentration of fluorescent molecules can be used without increasing noise in fluorescence detection. (C) Vesicles can be made porous with bacterial toxin. Here specific pores can be designed for selective permeation through the vesicle. (D) Vesicle readily porous at the lipid transition temperature. Here experiments such as a protein translocation, which would otherwise be a one-shot reaction, can be observed multiple times and under various chemical conditions.
First, because the volume enclosed in a vesicle is typically on the order of attoliters (10−18 liters), the effective concentrations of encapsulated molecules can be very high.¶ This feature of the encapsulation makes the fluorescence studies of the weakly interacting molecules, requiring high concentrations, readily realizable (Scheme 1B). The excess molecules that would normally lead to high background levels can be removed after vesicle immobilization, resulting in single-molecule detection without any compromise in signal-to-noise ratio. Also, the frequency of collision between a single enzyme and its substrate could be significantly enhanced within such small vesicles. For instance, molecules that would otherwise diffuse apart upon completion of the reaction will now remain in close proximity ready to react again. These frequent encounters among molecules inside the vesicle may enable repeated observations of the same biological events between the same set of molecules (Scheme 1D).
Porous Vesicles
The impermeability of vesicles made the encapsulation an unlikely replacement for the surface tethering methods in conventional studies and created a roadblock for further applications. If the vesicles could be made porous, the reactions inside would then be triggered on demand, and titrations of various small molecular weight reagents could be carried out for the encapsulated molecules. Likewise, the same interacting pair would be observed under different conditions by changing the external buffer solution without washing out the macromolecules (Scheme 1D). This unique feature might help address some of the elusive issues in the single molecule enzymology, such as molecular heterogeneities and memory effects (5, 15).
We propose two methods to tackle the permeability issue. The first approach is the use of a bacterial toxin, α-hemolysin (aHL), to introduce ≈2-nm diameter pores within the vesicle membranes (Scheme 1C). We were indeed able to change the solution conditions inside the vesicles containing aHL pores by flowing in different solutions to the surface-tethered vesicles as reported by the encapsulated RNA molecules by single-molecule fluorescence measurements (B.O., unpublished work). The second and simpler approach comes from a characteristic property of lipid membranes near the melting temperature (Tm) of the phospholipids that gives rise to defects in the lipid packing. Previous ensemble studies used this principle to form porous vesicles for engineering nanoscale bioreactors (16). At room temperature, for dimyristoyl phospatidylcholine [(DMPC) a phospholipid with two acyl chains of 14 carbons] vesicles, the pores were large enough to release ADP through the membrane with a half time of 2 h (16, 17). Although such pores allow the ATP exchange, previous observations suggested that even the smallest DNA oligomer, which is essentially of about twice the size of an ATP molecule, remains encapsulated inside a DMPC vesicle without any substantial leakage over long periods. We have previously reported that the surface-attached DMPC vesicles allowed the exchange of ions at room temperature and used them for single-molecule detection without hindrance to the encapsulated nucleic acid (13).
We would note here that, although the latter approach is much simpler, the aHL method may still prove superior for certain applications: for instance, the transient defects that form within the DMPC membrane are not controllable and depend on the temperature of the assay, whereas in contrast, aHL pores are extremely robust and stable over a wide range of conditions such as pH, temperature, and ionic strength.
Herein, we demonstrate that repeated interactions between the same set of proteins on DNA can be observed inside porous vesicles at the single-molecule level, with the added capability of changing the chemical condition inside the vesicles while keeping the macromolecules within. These measurements provide not only a proof of concept for the resolution of some of the previously discussed issues but also unique mechanistic insights on the RecA filament formation.
Results and Discussion
Escherichia coli RecA proteins bind to a ssDNA to form a filament, which is the active form of RecA in homologous recombination and SOS response that help preserve the genome (18, 19). To investigate RecA–DNA interaction within a confined volume, we encapsulated 400 nM ssDNA [(dT)20 labeled with Cy3 and Cy5 fluorescent dyes at the two ends] and 3-μM RecA inside lipid vesicles of 200-nm diameter, prepared by mixing biotinyl cap phosphoethanolamine with DMPC. The concentrations were chosen to obtain on average one ssDNA (12) and approximately seven RecA monomers per vesicle. Because the transition of DMPC (Tm = 23°C) between fluid and gel phases occurs around room temperature, heterogeneities are expected to arise on different regions of a vesicle, resulting in its porosity (20). Although the exact sizes of such pores on DMPC membrane are still unknown (16), our results below suggest the pores are small enough to retain a small protein such as RecA (38 kDa) and a short ssDNA inside the vesicles, yet large enough to allow the exchange of small molecules such as ATP (0.5 kDa).
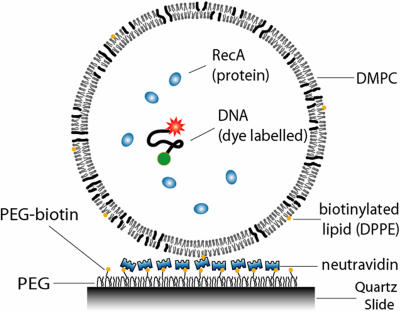
The vesicles were diluted before being immobilized to the PEG-coated surface (2, 19) through a biotin-neutravidin-biotin linkage (Scheme 2). The polymer coating also serves as a cushion and helps keep the vesicles intact upon surface fixation.
Scheme 2.
DNA and RecA encapsulated inside porous vesicle (not to scale).
The assembly and disassembly of RecA filament on ssDNA were detected using FRET (19). Here, we approximate the FRET efficiency E by using the ratio between the acceptor intensity and the total intensity. In the naturally coiled state of ssDNA, the proximity of the fluorescent dyes results in a strong dipole interaction (high FRET efficiency). Upon assembly, RecA filament stretches the ssDNA, pushing the dyes further apart, resulting in weaker dipole interactions between the dyes (low FRET efficiency). RecA monomers dissociate from the ends of the filament upon ATP hydrolysis (19, 21), making the filament a highly dynamic structure that can be precisely probed via single-molecule FRET.
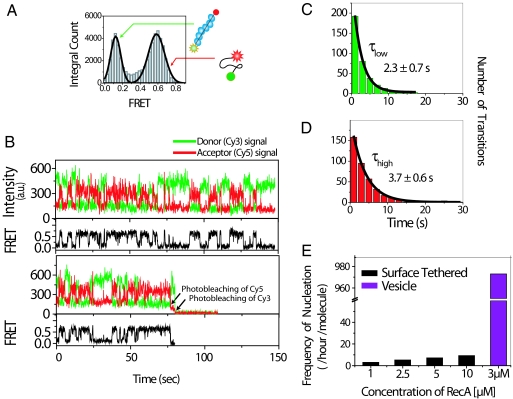
Upon introduction of our standard reaction buffer‖ and 1 mM ATP to the sample chamber containing encapsulated RecA and DNA, ssDNA molecules displayed fluctuations between high and low FRET states (Fig. 1 A and B). The low FRET state (E≈0.1) is clearly distinguishable from acceptor blinking or photobleaching, which would show E = 0. The high FRET value (E≈0.6) is identical to the FRET values we observed from the DNA-only sample, where the ssDNA was encapsulated under the identical conditions but in the absence of RecA (data not shown). Therefore, the fluctuations are attributed to the assembly (low FRET) and disassembly (high FRET) of RecA filament (19). Because three nucleotides are needed for the binding of a single RecA monomer (22), it is expected that the filament formed on the ssDNA used would consist of at most six monomers. Within the temporal resolution of 100 ms, there was no evidence of a stepwise transition through intermediate FRET states, suggesting that the RecA filament fully dissociates shortly after one RecA monomer dissociates from the ssDNA.** This observation is consistent with previous studies suggesting that the minimum number of RecA monomers required for stable filament formation is about five (19, 23).
Fig. 1.
Behavior of encapsulated molecules in presence of 1 mM ATP. (A) FRET histogram of 103 molecules showing two distinct states. (B) Time traces of single ssDNA molecules showing fluctuations between the two FRET states. (C) Dwell time plot of the low FRET state (filament assembly) of the 103 ssDNA with the exponential decay fit. (D) Dwell time plot of the high FRET state (filament unbound) obeying single exponential decay. (E) Comparison of transition frequencies between encapsulated molecules and surface tethered partial-duplex with 3′-(dT)19 with 1-, 2.5-, 5-, and 10-μM RecA.
From dwell-time analysis of low FRET states from >100 ssDNA molecules, the average lifetime of RecA filament is found to be 2.3 (±0.7) sec (Fig. 1C). This is in reasonable agreement with the dwell times obtained by using partial-duplex DNA with a 19-nucleotide single-strand overhang, (dT)19, when tethered to a PEG surface [6.0 (±2.0) sec] (19) or when encapsulated [3.7 (±0.8) seconds; see supporting information (SI) Fig. 3]. Because all of the encapsulated proteins and ssDNA must remain within 200-nm diameter, and the average stoichiometry between RecA and ssDNA is 7.5 to 1, it is highly probable that the same RecA molecules rebind the DNA in the reassembly of the RecA filament, a feature that will be impossible to achieve without an encapsulation or confinement of some type (6, 24).
Surprisingly, compared with what is observed with moleculestethered to a surface (19), filament reassembly is much more frequent [on average 973 transitions per DNA per hour, obtained from single exponential fitting of the dwell time histogram of the high FRET state (Fig. 1D)] for the encapsulated ssDNA. This reassembly frequency is two orders of magnitude greater than what could possibly be achieved by merely increasing the protein concentration in surface tethering experiments (average of 10 transitions per DNA per hour for 10 μM RecA) (Fig. 1E). This observation suggests that such drastic improvement in reaction rate is not a result of enhanced local RecA concentration but rather a unique consequence of the confinement aspect of this method. We suspect that this confinement-assisted rate enhancement is related to expected rate enhancement in molecular crowding (in vivo confinement because of the close vicinity of several dissimilar macromolecules) (25–27).
In our case, this higher rebinding rate would be understandable if, upon dissociation, RecA filament remains a polymer for at least several seconds to readily rebind to ssDNA. The dissociated polymer, which would in most cases diffuse away without confinement, inside the vesicle rebinds to the same DNA before falling apart into monomers.
It has long been known that RecA filament forms through a slow nucleation step followed by rapid extension (28–32), and recently the extension of the filament was seen to occur through successive addition of RecA monomers to the DNA-bound filament (19). However, the molecular mechanism of the nucleation of RecA filament was not deducible in previous assays. Because filament reformation is two orders of magnitude faster inside the vesicle, our observation supports a nucleation pathway where the preassembled filament binds ssDNA much more readily than de novo filament nucleation.
It must be emphasized, however, that our study does not address the mechanism of de novo filament formation, for example, whether it requires simultaneous binding of multiple monomers to the DNA or proceeds through binding of preformed oligomers. Rather, our work suggests that once formed, a nucleation cluster can stay assembled even after it dissociates from the DNA. A possible scenario inside the vesicle is that a nucleation cluster forms (N monomers) and stays bound to the DNA until a RecA monomer hydrolyzes an ATP and dissociates from the filament end, and the remaining filament (N−1 monomers) immediately dissociates from the DNA. Because DNA binding is necessary to stimulate RecA's ATPase activity, N−1-sized filament would stay intact in solution and would frequently collide with the DNA until another RecA monomer is added to stabilize the filament again (N monomers). In such a model, the exact number of excess RecA monomers over N would determine how frequently rebinding of the filament occurs, and indeed we have observed heterogeneous rebinding rates among vesicles (SI Fig. 4), possibly because of the probabilistic variation in the number of monomers in each vesicle.
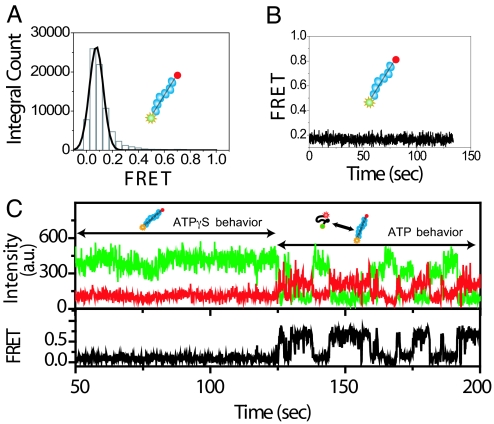
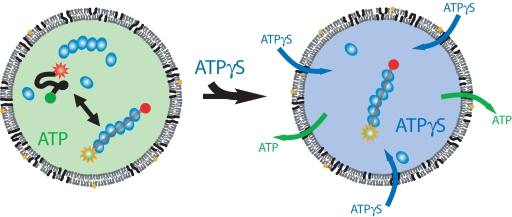
Next, when the solution inside the flow chamber was replaced with a solution containing 1 mM adenosine 5′-O-(3-thiotriphosphate) (ATPγS), a nonhydrolyzable analogue of ATP, the encapsulated molecules showed strictly one low FRET state (Fig. 2 A and B), likely because of increased stability of the filament in the absence of hydrolysis (Scheme 3).
Fig. 2.
Behavior at 1 mM ATPγS. (A) FRET histogram of 154 molecules. (B) Time trace of one ssDNA showing strictly one FRET state. (C) Time trace of an encapsulated ssDNA transitioning after the flow of 1 mM ATP to replace the solution of 1 mM ATPγS originally present inside the vesicle. The flow of ATP solution occurred before time = 0 sec.
Scheme 3.
A flow of ATPγS outside of the vesicles replaces the ATP inside the vesicle and induces a change in the interaction between the protein filament and the DNA.
Finally, after a solution containing ATP was reintroduced to replace the ATPγS inside the chamber, the ssDNA transitioned from a stable low FRET state back to the behavior of fluctuation between high and low FRET states (Fig. 2C), showing that ATPγS in the vesicle is replaced by ATP while keeping the DNA and proteins encapsulated. This response of molecules inside the vesicle to a change in the chemical environment outside the vesicle directly confirms the presence of pores on the vesicle membrane.
Conclusion
In summary, the dynamics of the same set of RecA proteins on a single strand of DNA is studied by encapsulating the molecules inside a porous vesicle. Our result supports a requirement on the minimum number of RecA for nucleation and further provides insight on the nucleation mechanism. The observation of frequent two-state fluctuations from which spun the nucleation model of RecA as a polymer is unique to the confinement aspect of this method. The unexpected enhancement of the filament formation rate might be reminiscent of a general mechanism used in nature to increase the efficiency of vital biological reactions by “molecular crowding” or “compartmentalization” (33).
The vesicle encapsulation method was previously used in single-molecule measurements to study protein folding (11) or complicated dynamics of nucleic acids alone (12, 13), but to our knowledge the detection of multicomponent macromolecular interactions inside vesicles was previously uncharacterized at the single-molecule level. This adaptation of porous vesicles to single-molecule techniques opens room for assays, otherwise unattainable, where a specific number of molecules must be constrained within a small volume, but the chemical condition needs to be changed in a controlled manner. Such assays allow a study of the behavior of the same set of molecules under various solution conditions free of surface tethering. Furthermore, because the volume of the vesicle and the number of encapsulated proteins remain unchanged upon buffer exchange, the local concentration of proteins also remains constant. Because this method recycles the proteins, it eliminates the need for replenishing the molecules each time the chemical condition is changed. The ability of reconditioning the chemical environment while maintaining a high effective protein concentration makes this method an ideal solution for the fluorescence study of weak and transient biological interactions (34). With the control and versatility provided by this porous vesicle encapsulation method, we anticipate immediate applications in single-molecule enzymology, bioengineering, pharmaceutical research, and biological chemistry. We expect this method to be well suited not only for DNA–protein interaction, but also RNA–protein, protein–protein, and other complex enzyme–substrate interactions.
Materials and Methods
Lipid films were prepared by mixing biotinyl cap phycoerythrin with DMPC dissolved in chloroform (1:100 molar ratio) then vacuumed for 2 h. A solution of 1 mM 2-mercaptoethanol/10 mM Mg(CH3COO)2/100 mM Na(CH3COO)/25 mM Tris·CH3COOH, pH 7.5/1 mM ATP was made (Buffer A). A concentration of 400 nM of the ssDNA [ssDNA constructs of 20 nucleotides, (dT)20, doubly labeled with Cy3 and Cy5 fluorescent dyes at the two ends were used] and 3 μM E. coli RecA proteins were incubated in buffer A for 30 min. To encapsulate the molecules in self-assembling multilamellar vesicles, the ssDNA and RecA solution was used to hydrate the lipid film. Freeze and thaw cycles (in liquid nitrogen then water) were conducted seven times, followed by size extrusion to obtain unilamellar vesicles 200 nm in diameter. The 200-nm diameter was carefully chosen such that 1 RecA per 3-nt ratio is to be kept, whereas the local concentration of RecA is in the micromolar for reasonable binding. Data obtained with 100-nm-diameter vesicles with 3 μM RecA concentration is presented in SI Fig. 5.
For imaging, total internal reflection fluorescence microscopy was used (35). The sample chamber consists of a flow channel assembled from quartz slides and glass coverslips glued by two-sided adhesive tape. To eliminate nonspecific adhesion of vesicles (or nonencapsulated molecules) to the chamber and to facilitate the specific binding of biotinylated vesicles, the surfaces of the chamber were coated with a mixture of PEG and biotinylated PEG†† (2, 19). The surface-immobilized vesicles remained stable for several hours even after multiple washing cycles. To increase the photostability of the fluorescent dyes, imaging Buffer A was made to contain 1 mg/ml glucose oxidase, 0.04 mg/ml catalase, 1% (vol/vol) 2-mercaptoethanol, and 0.4% glucose and injected to the flow chamber.
The imaging was conducted at room temperature (23 ± 1°C), and the observation period varied between 2 and 5 min until most molecules photobleached. Vesicles with no ssDNA have no fluorescence and are therefore not detected. Among the vesicles showing both donor and acceptor signals, 57% showed multistep photobleaching likely because of several encapsulated ssDNA, 20% showed constantly high FRET states with no stable low FRET states and likely because of vesicles containing less than the minimum number of RecA monomers required for nucleation, and ≈23% showed multiple transitions between high- and low-FRET states as well as evidence for single ssDNA in the form of single-step photobleaching. Only this last class of vesicles with traces consistent with one labeled ssDNA and with enough RecA for filament formation is chosen for analysis.
Supplementary Material
Acknowledgments
We thank Tim Lohman (Washington University, St. Louis, MO) for the labeled ssDNA, Sua Myong for insightful discussion, and Salman Syed for timely provision of logistics that made this work possible. We also acknowledge the early participation of Stephanus Fengler in the aHL project. This work was funded by National Institutes of Health Grant GM074526 (to T.H.) and National Science Foundation Grant PHY0646550. T.H. is an Investigator with the Howard Hughes Medical Institute.
Abbreviations
- DMPC
dimyristoyl phospatidylcholine
- aHL
α-hemolysin
- ATPγS
adenosine 5′-O-(3-thiotriphosphate).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610673104/DC1.
The interior volume of a 100-nm diameter vesicle is ≈0.5 attoliters, and the effective concentration of one molecule within that volume is ≈3 μM.
Buffer A is composed of 1 mM 2-mercaptoethanol, 10 mM Mg(CH3COO)2, 100 mM Na(CH3COO), 25 mM Tris·CH3COOH (pH 7.5), and 1 mM ATP. To increase the photostability of the fluorescent molecules, we include 1 mg/ml glucose oxidase/0.04 mg/ml catalase/1% (v/v) 2-mercaptoethanol/0.4% glucose.
Monomeric dissociation, should it occur, would be detected in the 100-ms resolution.
When the PEG surface was not covered with neutravidin, essentially no binding was observed. In sharp contrast, even a lower dilution of the sample yielded a good coverage on the surface after neutravidin treatment. The binding was therefore specific to only the biotinylated molecules, which would be the vesicles.
References
- 1.Wennmalm S, Edman L, Rigler R. Proc Natl Acad Sci USA. 1997;94:10641–10646. doi: 10.1073/pnas.94.20.10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 3.Ellerby LM, Nishida CR, Nishida F, Yamanaka SA, Dunn B, Valentine JS, Zink JI. Science. 1992;255:1113–1115. doi: 10.1126/science.1312257. [DOI] [PubMed] [Google Scholar]
- 4.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 5.Lu HP, Xun L, Xie XS. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 6.Rondelez Y, Tresset G, Tabata KV, Arata H, Fujita H, Takeuchi S, Noji H. Nat Biotechnol. 2005;23:361–365. doi: 10.1038/nbt1072. [DOI] [PubMed] [Google Scholar]
- 7.Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 8.Friedel M, Baumketner A, Shea JE. Proc Natl Acad Sci USA. 2006;103:8396–8401. doi: 10.1073/pnas.0601210103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talaga DS, Lau WL, Roder H, Tang J, Jia Y, DeGrado WF, Hochstrasser RM. Proc Natl Acad Sci USA. 2000;97:13021–13026. doi: 10.1073/pnas.97.24.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boukobza ES A, Haran G. J Phys Chem B. 2001;105:12165–12170. [Google Scholar]
- 11.Rhoades E, Gussakovsky E, Haran G. Proc Natl Acad Sci USA. 2003;100:3197–3202. doi: 10.1073/pnas.2628068100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okumus B, Wilson TJ, Lilley DM, Ha T. Biophys J. 2004;87:2798–2806. doi: 10.1529/biophysj.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JY, Okumus B, Kim DS, Ha T. Proc Natl Acad Sci USA. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amirgoulova EV, Groll J, Heyes CD, Ameringer T, Rocker C, Moller M, Nienhaus GU. ChemPhysChem. 2004;5:552–555. doi: 10.1002/cphc.200400024. [DOI] [PubMed] [Google Scholar]
- 15.English BP, Min W, van Oijen AM, Lee KT, Luo G, Sun H, Cherayil BJ, Kou SC, Xie XS. Nat Chem Biol. 2006;2:87–94. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- 16.Monnard PA. J Membr Biol. 2003;191:87–97. doi: 10.1007/s00232-002-1046-0. [DOI] [PubMed] [Google Scholar]
- 17.Chakrabarti AC, Deamer DW. Biochim Biophys Acta. 1992;1111:171–177. doi: 10.1016/0005-2736(92)90308-9. [DOI] [PubMed] [Google Scholar]
- 18.Kowalczykowski SC. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 19.Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 20.Bolinger PY, Stamou D, Vogel H. J Am Chem Soc. 2004;126:8594–8595. doi: 10.1021/ja049023u. [DOI] [PubMed] [Google Scholar]
- 21.Cox MM. Prog Nucleic Acid Res Mol Biol. 1999;63:311–366. doi: 10.1016/s0079-6603(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 22.Egelman EH, Yu X. Science. 1989;245:404–407. doi: 10.1126/science.2667137. [DOI] [PubMed] [Google Scholar]
- 23.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 24.Cai L, Friedman N, Xie XS. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 25.Cheung MS, Klimov D, Thirumalai D. Proc Natl Acad Sci USA. 2005;102:4753–4758. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pielak GJ. Proc Natl Acad Sci USA. 2005;102:5901–5902. doi: 10.1073/pnas.0501812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornberg A. Trends Biochem Sci. 2003;28:515–517. doi: 10.1016/j.tibs.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Pugh BF, Cox MM. J Biol Chem. 1987;262:1337–1343. [PubMed] [Google Scholar]
- 29.Pugh BF, Cox MM. J Biol Chem. 1988;263:76–83. [PubMed] [Google Scholar]
- 30.Pugh BF, Cox MM. J Mol Biol. 1988;203:479–493. doi: 10.1016/0022-2836(88)90014-9. [DOI] [PubMed] [Google Scholar]
- 31.Sattin BD, Goh MC. Biophys J. 2004;87:3430–3436. doi: 10.1529/biophysj.104.045567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bar-Ziv R, Tlusty T, Libchaber A. Proc Natl Acad Sci USA. 2002;99:11589–11592. doi: 10.1073/pnas.162369099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szostak JW, Bartel DP, Luisi PL. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 34.Laurence TA, Weiss S. Science. 2003;299:667–668. doi: 10.1126/science.1081025. [DOI] [PubMed] [Google Scholar]
- 35.Axelrod D. Methods Cell Biol. 1989;30:245–270. doi: 10.1016/s0091-679x(08)60982-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.