Abstract
Studies of 25-hydroxyvitamin D3-24-hydroxylase (CYP24A1) have demonstrated that it is a bifunctional enzyme capable of the 24-hydroxylation of 1α,25-(OH)2D3, leading to the excretory form, calcitroic acid, and 23-hydroxylation, culminating in 1α,25-(OH)2D3-26,23-lactone. The degree to which CYP24A1 performs either 23- or 24-hydroxylation is species-dependent. In this paper, we show that the human enzyme that predominantly 24-hydroxylates its substrate differs from the opossum enzyme that 23-hydroxylates it at only a limited number of amino acid residues. Mutagenesis of the human form at a single substrate-binding residue (A326G) dramatically changes the regioselectivity of the enzyme from a 24-hydroxylase to a 23-hydroxylase, whereas other modifications have no effect. Ala-326 is located in the I-helix, close to the terminus of the docked 25-hydroxylated side chain in a CYP24A1 homology model, a result that we interpret indicates that substitution of a glycine at 326 provides extra space for the side chain of the substrate to move deeper into the pocket and place it in a optimal stereochemical position for 23-hydroxylation. We discuss the physiological ramifications of these results for species possessing the A326G substitution, as well as implications for optimal vitamin D analog design.
Keywords: 1α,25-(OH)2D3; cytochrome P450; dual metabolic pathways; substrate docking
The hormone 1α,25-dihydroxyvitamin D3 [1α,25-(OH)2D3] plays multiple roles in the animal body, regulating extracellular calcium and phosphate homeostasis as well as the expression of a number of genes involved in cell division, cell differentiation, and immunomodulation in various tissues (1, 2). The vitamin D signal transduction cascade involves cytochrome P450 (CYP)-based activating enzymes (CYP2R1, CYP27A1, and CYP27B1), a specific plasma transport protein (vitamin D-binding protein), a dedicated nuclear receptor [1α,25-dihydroxyvitamin D3 receptor (VDR)], and a catabolic enzyme (CYP24A1) (1–3). CYP24A1, originally referred to as the 25-hydroxyvitamin D3-24-hydroxylase and thought to catalyze a single hydroxylation step in 25-hydroxyvitamin-D3 (25-OH-D3) metabolism, is now known to carry out multiple steps of hydroxylation involving the side chains of both 25-OH-D3 and 1α,25-(OH)2D3 (4). CYP24A1 hydroxylates 1α,25-(OH)2D3, at either C24 or C23, to give the terminal products of calcitroic acid or 1α,25-(OH)2D3-26,23-lactone, respectively (5–7) (Fig. 1). Studies using CYP24A1-knockout (8) and VDR-knockout mice (8, 9) demonstrate the total absence of calcitroic acid and 1α,25-(OH)2D3-26,23-lactone formation when the vitamin D-inducible, VDR-mediated CYP24A1 is not expressed. The in vivo result of CYP24A1 ablation is decreased viability, with 50% of mice not surviving beyond weaning because of hypercalcemia and nephrocalcinosis (8, 10). Although it has been well established that calcitroic acid is the major biliary product of 1α,25-(OH)2D3 in the rat (11, 12), there have been claims that intermediates of the C24 oxidation pathway retain some biological activity (13) and that the terminal product of 23-hydroxylation, namely 1α,25-(OH)2D3-26,23-lactone, belongs to a family of potent VDR antagonists with potential for use in the treatment of Paget's disease (14, 15).
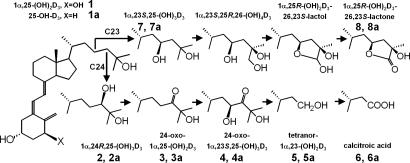
Fig. 1.
CYP24A1 catalyzes the catabolism of 1α,25-(OH)2D3 (1) and 25-OH-D3 (1a) via C24- or C23-hydroxylation pathways. Products of 1α,25-(OH)2D3 (1) are denoted by the numerals 2–8. Products of 25-OH-D3 (1a) are denoted as 2a–8a. The metabolite 23-oxo-25-OH-D3 (data not shown), denoted as 3b, is a product of the C23-hydroxylation pathway.
The degree to which CYP24A1 performs either of these pathways varies with the species. It had been shown in the mid-1980s by using isolated kidney mitochondria from a variety of species, that the 24-hydroxylase and 23-hydroxylase activities copurify (16). Although both pathways are used in humans, certain species, such as the guinea pig (17, 18), were shown to 23-hydroxylate, whereas other species, such as the rat, primarily 24-hydroxylate the substrate, 25-OH-D3 (16). These studies suggested that the two enzyme activities might have different kinetic parameters, but it was not clear whether the activities resided on a single or distinct polypeptide chains. With the cloning and expression of the recombinant protein from different species (19, 20), it became clear that a single polypeptide chain was capable of both C23- and C24-hydroxylation activities (5–7).
Recent studies of rat and human CYP24A1 have used homology modeling to reveal the general structure of the protein and highlight active-site residues for mutagenesis studies (21, 22). Masuda et al. (21) have shown the importance of residues in the B′-helix, B′/C loop, F-helix, and β-5 hairpin of the human enzyme. Recently, Hamamoto et al. (23) focused on amino acid residues 416 (β-3) and 500 (β-5) in the rat and human sequences as a potential determinant of species-based differences in the extent of C24- or C23-hydroxylation. On the basis of a modest increase in 23-hydroxylation activity demonstrated by the “humanized” T416M rat enzyme, these authors (23) concluded that a threonine possibly assisted by an active-site water must be largely responsible for the hydroxylation differences observed in regioselectivity. In our human (h)CYP24A1 homology model, Met-416 is distant from the A-ring of the docked substrate and thus is distant from the side chain, which prompted us to study other potentially key differences in the structure of CYP24A1 that could explain species-based differences in activity and regioselectivity.
In the current study, we set out to (i) compare available CYP24A1 sequences in the various databases using a hCYP24A1 model to establish their relative importance to enzyme function; (ii) select cell lines from species showing extremes of 24-hydroxylation or 23-hydroxylation, to establish the analytical methods needed to best show side-chain hydroxylation patterns; and (iii) perform mutagenesis studies on selected substrate-contact residues within hCYP24A1 and examine the resultant metabolic patterns obtained.
Our results show that residue Ala-326 plays a crucial role in side-chain hydroxylation and its modification to a glycine causes a dramatic change in the pattern of products formed, a finding consistent with 3D models of the substrate binding pocket of hCYP24A1.
Results
Based on a review of 25-OH-D3-24-hydroxylase and 23-hydroxylase enzyme activity data (16–18), the results from cultured cell lines (3, 7, 24), a survey of available cultured cell lines containing CYP24A1 and the published CYP24A1 sequences in available databases (Fig. 2; figure 2 in ref. 22; data not shown), we selected V79-CYP24 cells containing hCYP24A1 (7) and the opossum kidney (OK) (24) cell lines as representative models for 24- and 23-hydroxylation by CYP24A1, respectively. We then set out to optimize cell culture and incubation conditions, substrate concentrations, and chromatographic methods required to demonstrate the 24- and 23-hydroxylation pathway intermediates (Fig. 1).
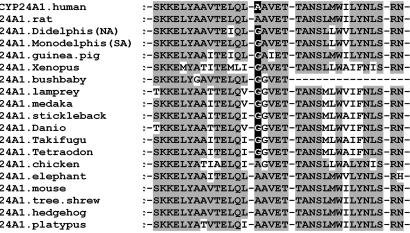
Fig. 2.
Alignment of the I-helix (residues 312–345) of CYP24A1 orthologs from selected species. The highlighted residue is Ala-326 in the human enzyme mutated in the studies reported here.
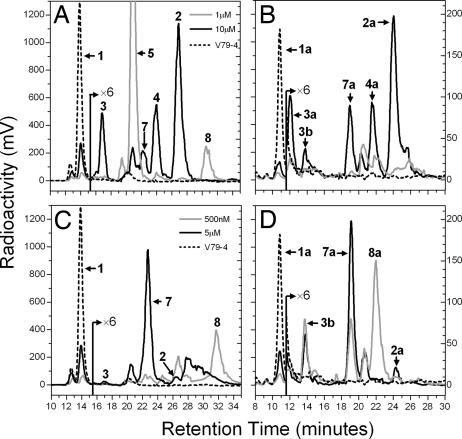
The radiochromatograms depicted in Fig. 3 illustrate typical metabolic patterns obtained from these two CYP24A1-expressing cell lines when incubated with either [1β-3H]1α,25-(OH)2D3 (Fig. 3 A and C) or [26,27-3H]25-OH-D3 (Fig. 3 B and D), where the V79-CYP24 cell line (Fig. 3 A and B) showed predominantly C24-hydroxylated products (7), whereas the OK cell line (Fig. 3 C and D) showed mainly C23-hydroxylated products (24). Nanomolar 1α,25-(OH)2D3 maximized the terminal pathway products: tetranor-1α,23-(OH)2D3 (peak 5) or 1α,25-(OH)2D3-26,23-lactone (peak 7), whereas micromolar substrate maximized initial pathway intermediates such as 1α,24,25-(OH)3D3 (peak 2) or 1α,23,25-(OH)3D3 (peak 7). The patterns using a higher 25-OH-D3 substrate concentration were essentially analogous with V79-CYP24 showing predominantly 24,25-(OH)2D3 (peak 2a), whereas OK cells showed mainly 23,25-(OH)2D3 (peak 7a). Studies of the metabolism of lower concentrations of [26,27-3H]25-OH-D3, where terminal pathway products were expected, were confounded in both cell lines but especially using V79-CYP24, by the loss of 26,27-3H label, its being distal to the side-chain C23-cleavage site rendering metabolites, such as peak 5a, devoid of radioactivity. Because peak 8a is made by OK cells from [26,27-3H]25-OH-D3 at low substrate concentrations, it retains significant 26,27-3H label, possesses the appropriate ions on liquid chromatography (LC)-MS, and has been identified by others in OK extracts (24), we conclude that it is 25-OH-D3-26,23-lactone.
Fig. 3.
Metabolism of 1α,25-(OH)2D3 and 25-OH-D3 by V79-CYP24 or OK cells. (A and B) Representative HPLC chromatograms are shown for incubations of 1 or 10 μM 1α,25-(OH)2D3 (A) and 1 or 10 μM 25-OH-D3 with V79-CYP24 cells (B). (C and D) In comparison, chromatograms are also shown for incubations of 500 nM or 5 μM 1α,25-(OH)2D3 (C) and 500 nM or 5 μM 25-OH-D3 with OK cells (D).
We generated sufficient quantities of most metabolites from either cell line to definitively identify the metabolic products of both 1α,25-(OH)2D3 and 25-OH-D3 by a combination of techniques including LC-MS, which gave several characteristic ions including molecular ions [M+H]+ and their dehydration products, as summarized in Table 1. The major OK cell metabolite, peak 7, was identified as 1α,23,25-(OH)3D3 by its molecular mass and chromatographic properties before and after sodium m-periodate treatment [Table 1; supporting information (SI) Fig. 6]. The lack of cleavage of peak 7 indicated that the side chain was unchanged by periodate and thus hydroxyls are not vicinal, suggesting that the metabolite must be 1α,23,25-(OH)3D3, analogous to the metabolite observed by Yamada's group (24).
Table 1.
HPLC and LC-MS-based characterization of the catabolic products of 1α,25-(OH)2D3 and 25-OH-D3
| Peak | RT, min | Molecular mass, Da | Characteristic MS1 ions, m/z | Putative ID of metabolite |
|---|---|---|---|---|
| 1 | 13.79 | 416 | 439, 417, 399, 381, 515 | 1α,25-(OH)2D3 (substrate) |
| 2 | 25.78* | 432 | 455, 433, 415, 397, 447, 547 | 1α,24,25-(OH)3D3 |
| 2* | 12.06* | 372 | 373, 355, 337, 387, 427, 519 | |
| 7 | 21.69* | 432 | 455, 433, 415, 397, 447, 547 | 1α,23,25-(OH)3D3 |
| 7* | 16.20* | 432 | 455, 433, 415, 397, 447, 547 | |
| 3 | 16.54 | 430 | 453, 413, 529, 531, 591 | 24-Oxo-1α,25-(OH)2D3 |
| 4 | 23.24 | 446 | 469, 429, 411, 529, 545, 577 | 24-Oxo-1α,23,25-(OH)3D3 |
| 5 | 20.01 | 360 | 383, 361, 343, 325, 451, 491 | Tetranor-1α,23-(OH)2D3 |
| 6 | 10.68† | 374 | 397, 375, 357, 339, 473, 505 | Calcitroic acid |
| 8 | 29.12 | 444‡ | 467, 445, 427, 409, 559, 591 | 1α,25-D3-26,23-lactone |
| 1a | 10.79 | 400 | 423, 401, 383, 505 | 25-OH-D3 (substrate) |
| 2a | 24.05 | 416 | 439, 417, 399, 471, 537, 581 | 24,25-(OH)2D3 |
| 7a | 19.00 | 416 | 439, 417, 399, 471, 537, 591 | 23,25-(OH)2D3 |
| 3a | 12.08 | 414 | 437, 415, 397, 469, 519, 577 | 24-Oxo-25-OH-D3 |
| 3b | 13.73 | 414 | 437, 415, 397, 469, 519, 577 | 23-Oxo-25-OH-D3 |
| 4a | 21.56 | 430 | 453, 431, 413, 454, 485, 537 | 24-Oxo-23,25-(OH)2D3 |
| 5a | —§ | 344 | 367, 345, 399 | Tetranor-23-OH-D3 |
| 6a | —§ | 358 | 381, 359, 341, 423, 463, 489 | 23-Acid |
| 8a | 21.71 | 428 | 451, 429, 411, 446 | 25-OH-D3-26,23-lactone |
Retention times (RTs) on Zorbax-SIL were as follows: 91:7:2 hexane/isopropanol/methanol, 1 ml/min for products of peak 1; 96:3:1 hexane/isopropanol/methanol, 1 ml/min for products of peak 1a. M+H+ ions are shown in bold.
*RTs of sodium m-periodate-treated peaks 2 or 7 on Zorbax-SIL where untreated peaks 2 and 7 ran at 20.00 and 16.19 min, respectively.
†RT on Zorbax-SB-C18; 69:30:1 water/acetonitrile/10% GAA in water-gradient system.
‡Synthetic 1α,25-(OH)2D3-26,23-lactone was used for LC-MS comparison.
§Peaks 5a and 6a were not detected because of loss of 26,27-3H label.
Our original comparison of the two CYP24A1 sequences from human and South American opossum (Monodelphis domesticus) revealed ≈80 aa differences, 30 of which were considered irrelevant because they are in the highly variable, N-terminal region containing the membrane-associated domain. After consideration of the homology model of hCYP24A1 (21) and the sequence alignments of a wider range of CYP24A1 species orthologs, our studies focused on three residues that differ, including Ala-326 (Fig. 2), Asn-448, and Gln-471 (SI Fig. 7), as potential factors in substrate contact that may play a role in regioselectivity. In this survey, it was also noted that amino acid differences between human and rat sequences at residues (human) Met-416 and Thr-500 (23) are not consistently different in nonhuman species including opossum, and it was thus unlikely that they were the primary determinants of the observed differences in regioselectivity.
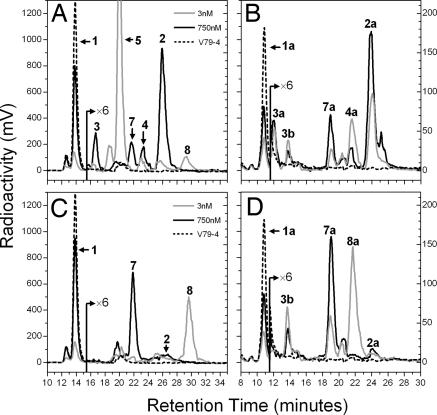
Mutagenesis studies were undertaken to change hCYP24A1 at key residues Ala-326, Asn-448, and Gln-471 into the corresponding opossum (Monodelphis) sequence. Wild-type human and mutant forms of CYP24A1 were transiently expressed in V79-4 cells and incubated with 1α,25-(OH)2D3 or 25-OH-D3 under similar conditions used for cell line studies. As expected, the wild-type human enzyme (Fig. 4 A and B) gave similar metabolic patterns with [1β-3H]1α,25-(OH)2D3 (Fig. 4A) or [26,27-3H]25-OH-D3 (Fig. 4B) to those found with the V79-CYP24 cell line, showing predominantly 24-hydroxylated products, confirming that metabolic patterns observed for different CYP24A1 orthologs in transfection and in situ cell systems are mainly determined by CYP24A1 sequence, not by the incubation conditions. The mutants N448H and Q471H gave the same patterns as the wild-type hCYP24A1 (data not shown). In contrast, the A326G mutant (Fig. 5 C and D) showed completely different patterns that represent a dramatic change from the 24-hydroxylation patterns seen for the wild-type hCYP24A1 in Fig. 4 A and B. Moreover, the metabolic patterns with A326G are almost identical to those previously observed in OK cells (Fig. 3), with 1α,25-(OH)2D3-26,23-lactone (peak 8) and putative 25-OH-D3-26,23-lactone (peak 8a) formed at low substrate concentrations; whereas 1α,23,25-(OH)3D3 (peak 7) and 23,25-(OH)2D3 (peak 7a) are the principal products at higher substrate concentrations.
Fig. 4.
Metabolism of 1α,25-(OH)2D3 and 25-OH-D3 by V79-4 cells transiently transfected with wild-type (A and B) or A326G mutated (C and D) hCYP24A1. Representative HPLC chromatograms are shown for incubations of 3 or 750 nM 1α,25-(OH)2D3 (A and C) or 3 or 750 nM 25-OH-D3 (B and D).
Fig. 5.
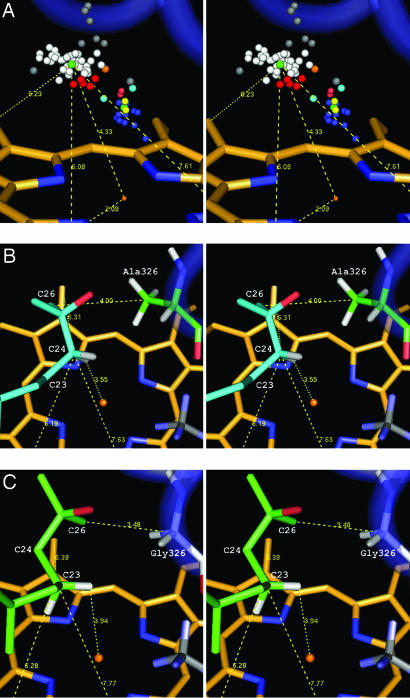
Substrate docking of 1α,25-(OH)2D3 in a homology model of hCYP24A1. (A) Stereoview of hydroxylation-susceptible carbon atoms and heme-ligating heteroatoms in 67 crystal structures triangulated off the heme methyl groups and transformed onto the heme of CYP24A1. White (P450cam), red (P450eryF), gray (CYP2C5, CYP2C9, and P450cam outliers), and green (average, triangulated) spheres correspond to carbon atoms in substrate-like ligands. The cluster of colored spheres axial to the heme iron correspond to atoms in inhibitor-type ligands. (B) Docked conformation for C24-hydroxylation (cyan) showing triangulation of C24 and the proximity of C26 to Ala-326 methyl in the wild-type enzyme. (C) Docked conformation for C23-hydroxylation (green) showing triangulation of C23 and the proximity of C26 to Gly-326 α-carbon in the A326G mutant enzyme.
To quantitatively assess the dramatic shift in regioselectivity, we calculated the relative 24-/23-hydroxylation ratio found with wild-type and A326G mutant by using two methods (Table 2). The weighted activity method gave ratio values heavily favoring 24-hydroxylation with the hCYP24A1 ranging from 6 to 24 when 1α,25-(OH)2D3 was used as substrate, or 3.83 to 8 when 25-OH-D3 metabolism was studied. In contrast, the A326G mutant, the ratios were 0.12–0.57 using 1α,25-(OH)2D3 as substrate or 0.05–0.70 using 25-OH-D3, confirming that 23-hydroxylation is favored and resembling values found for the opossum CYP24A1. The simple ratio of peak 2/peak 7 (Table 2) gave the same overall conclusion. The radical shift in 24-hydroxylation/23-hydroxylation ratio observed for the A326G hCYP24A1 mutant, suggests that this amino acid residue is largely responsible for the regioselectivity differences observed between the human and opossum CYP24A1 orthologs.
Table 2.
Regioselectivity in CYP24A1-expressing cells
| Cell line | Substrate | Substrate concentration, nM* | C24/C23 activity ratio† | Ratio of peaks 2/7 or 2a/7a |
|---|---|---|---|---|
| V79-CYP24 | 1α,25-(OH)2D3 | 10,003 | 24.06 | 6.66 |
| V79-CYP24 | 1α,25-(OH)2D3 | 1,003 | 6.07 | — |
| OK | 1α,25-(OH)2D3 | 5,003 | 0.04 | 0.04 |
| OK | 1α,25-(OH)2D3 | 503 | 0.44 | — |
| V79-4/wt‡ | 1α,25-(OH)2D3 | 753 | 8.20 | 6.18 |
| V79-4/wt‡ | 1α,25-(OH)2D3 | 3 | 8.10 | — |
| V79-4/A326G‡ | 1α,25-(OH)2D3 | 753 | 0.12 | 0.12 |
| V79-4/A326G‡ | 1α,25-(OH)2D3 | 3 | 0.57 | — |
| V79-CYP24 | 25-OH-D3 | 10,003 | 4.60 | 2.83 |
| V79-CYP24 | 25-OH-D3 | 1,003 | 8.04 | — |
| OK | 25-OH-D3 | 5,003 | 0.08 | 0.10 |
| OK | 25-OH-D3 | 503 | 0.01 | — |
| V79-4/wt‡ | 25-OH-D3 | 753 | 4.83 | 3.70 |
| V79-4/wt‡ | 25-OH-D3 | 3 | 3.83 | — |
| V79-4/A326G‡ | 25-OH-D3 | 753 | 0.05 | 0.08 |
| V79-4/A326G | 25-OH-D3 | 3 | 0.70 | — |
All peak ratios for substrate 1α,25-(OH)2D3 refer to peaks 2/7. All peaks ratios for substrate 25-OH-D3refer to peaks 2a/7a. —, not applicable.
*Using [1β-3H]1α,25-(OH)2D3or [26,27-3H]25-OH-D3 (sp. activities in text).
†Ratio after summation of weighted products from either C24- or C23-hydroxylation pathways by using the method described in Materials and Methods.
‡V79-4 cells transfected with wild-type (wt) or A326G-mutated hCYP24A1.
A substrate docking study was undertaken with the CYP24A1 homology model (21), to determine the proximity of the Ala-326 residue in relation to the 25-hydroxylated side chain of the substrate. As shown in Fig. 5, Ala-326 is located in the I-helix that abuts the substrate binding pocket. Furthermore, substrate docking using the average hydroxylation-susceptible carbon approach (Fig. 5A) places the side chain of the vitamin D substrate in juxtaposition to the I-helix with the C26 carbon only 4.00 Å from the Ala-326 methyl carbon in wild-type enzyme and leaving the C24R-hydrogen in an ideal hydroxylation orientation only 3.55 Å from the heme Fe atom (Fig. 5B). Using the same substrate docking approach for the A326G mutant places the side chain of the vitamin substrate close to the I-helix with the C26 carbon still only 3.48 Å from the Gly-326 α-carbon but now leaving the C23S-hydrogen only 3.94 Å from the heme Fe atom (Fig. 5C). We thus believe that the substitution of glycine for alanine at position 326 in hCYP24A1 permits the side chain of the substrate to slide further into the substrate binding pocket, placing C23 rather than C24 into an optimal position for hydroxylation.
Discussion
This study has demonstrated the critical juxtaposition of the I-helix alanine 326 residue in the active site of CYP24A1 with the side chain of 25-hydroxylated vitamin D substrates. The studies reported here show that, when this alanine 326 in the hCYP24A1 is changed to a glycine, as it is in opossum and guinea pig, the 24-hydroxylation pattern primarily giving rise to calcitroic acid is dramatically altered to a 23-hydroxylation pattern, in which the terminal metabolite is 1α,25-(OH)2D3-26,23-lactone, a pattern that closely resembles that observed with the opossum CYP24A1. Such a radical change in hydroxylation pattern does not occur when other residues in hCYP24A1 are mutated (21), making a strong case for the fact that the side chain of Ala-326 is a major determinant of the depth of substrate penetration into the substrate-binding pocket and the alignment of the hydroxylation site above the heme Fe. Indeed, homology modeling of hCYP24A1 (21), and its related CYP, CYP27A1 (25), places the I-helix closely abutting the substrate binding pocket and the residue Ala-326 facing the terminal methyl groups of the vitamin D side chain, thus independently supporting the conclusion from our mutagenesis studies that Ala-326 occupies a critical position. Although our results are clear-cut, at first glance these findings seem to be counter to the work of Hamamoto et al. (23), which argues a role for Met-416 and Ile-500 in controlling C23- and C24-hydroxylation in rat CYP24A1 function. However, these authors demonstrated only modest changes in the C24-/C23-hydroxylation ratio and did not discern the amino acid residues contacting the side chain, arguing that the residues that they mutated are in contact with other regions of the substrate. Accordingly, our results do not exclude a role for other amino acid residues in CYP24A1 that could further modulate enzyme hydroxylation/regioselectivity by altering the orientation of the substrate within the active site.
Why a putative catabolic enzyme such as CYP24A1 catalyzes two independent pathways from the 25-hydroxylated forms, 1α,25-(OH)2D3 or 25-OH-D3, resulting in totally different end products is an intriguing question. The issue has been apparent because in vivo and in vitro studies collectively showed that calcitroic acid and 1α,25-(OH)2D3-26,23-lactone are both 1α,25-(OH)2D3-induced products, both result from recombinant CYP24A1 action, and both are absent in VDR-null and CYP24A1-null mice in vivo (8, 9). These products are physiologically different because the calcitroic acid is a rapidly cleared catabolite with no discernible biological activity and 1α,25-(OH)2D3-26,23-lactone belongs to a family of known VDR antagonists (14, 15) with better vitamin D-binding protein-binding properties and greater metabolic stability (26). The fact that the dehydration products of 1α,25-(OH)2D3-26,23-lactone are strong VDR antagonists raises the possibility that metabolic products of the lactone pathway provide a redundant or fail-safe mechanism to more rapidly attenuate an environmental vitamin D challenge. Moreover, species with CYP24A1 containing Gly-326 might be better able to adapt to excessive 1α,25-(OH)2D3 activation or excessive Ca or PO4 intakes. Interestingly, this hypothesis is supported by the work of Yamada's group (24), which compared the time course of induction of CYP24A1 in OK (opossum) and COS-7 (green monkey) cells and concluded that OK cells turn off 1α,25-(OH)2D3-induced CYP24A1 mRNA expression faster. Clearly, future studies need to compare the two CYP24A1 variants with equivalent VDR expression and appropriate vitamin D-responsive element-dependent reporter gene constructs to evaluate the full physiological implications of the two pathways.
As a result of our studies, we examined the frequency of polymorphisms at the 326 position in the species orthologs of 14 P450 isoforms (353 sequences). Although alanine and glycine are conserved in 70 and 17% of the human P450 proteome, respectively, only CYP24A1 displayed a species-based polymorphism. The alanine/glycine polymorphism, a transversional mutation at the second nucleotide position of the GCN and GGN codons, occurs as alanine in 18 of 31 species (58%, mostly mammalian), and glycine (42%) in guinea pig, bushbaby, three opossums (M. domesticus, Didelphis virginiana, and Trichosurus vulpecula), Xenopus, and seven bony fish species including lamprey (Fig. 2; ref. 21; data not shown). It remains to be seen whether the CYP24A1 orthologs with glycine use the C23–lactone pathway and benefit their hosts with added protection against harmful effects of excessive vitamin D action.
hCYP24A1 exhibits no known deficiency phenotype, unlike other vitamin D-related P450s, such as CYP27A1 (27), CYP27B1 (28), and CYP2R1 (29). At this time, no single-nucleotide polymorphisms (Entrez build 127) localize to the alanine/glycine polymorphism site in humans (196 entries) or mouse (103 entries), and the four and one nonsynonymous mutations, respectively, which have been identified are uncharacterized but unlikely to significantly affect activity or metabolic pathway utilization. Thus, CYP24A1 is physiologically essential and potentially an important clinical target for inhibition to extend the action of 1α,25-(OH)2D3. Inhibitors designed on azole chemistry (e.g., ketoconazole) (30) and a series of competitive inhibitors based on natural substrates (31) have previously had successes as topical agents in human clinical trials for hyperproliferative conditions such as psoriasis. It remains to be seen whether characterization of the lactone pathway and better understanding of the CYP24A1 active site will facilitate new design strategies for CYP24A1 inhibitors.
One of the important implications of this study is the growing sophistication of homology models for the vitamin D-related CYPs including CYP24A1 (21–23, 25) to accurately predict substrate binding. The model (21) used here had previously defined the important roles of specific residues surrounding the substrate binding domain, these being the following: Ile-131 in A-ring and cis-triene contacts; Trp-134 and Gly-499 in substrate access; Leu-148 in side-chain contact; Met-246 in modulating regioselectivity. In this study, we further enhanced our substrate docking strategy by measuring the average distance of the hydroxylation-susceptible carbons and heme-ligating heteroatoms from 67 CYP crystal structures in the available databases and using these distances to position the vitamin D substrate within CYP24A1. By this process, which brought the hydroxylation-susceptible carbon to within 4 Å of the Fe atom, the terminal methyls were brought into juxtaposition with residue 326 of the I-helix. In the case of the wild-type hCYP24A1 where the C24R hydrogen was 3.55 Å, the C26-Ala-326 distance was 4.00 Å; whereas in the mutant A326G hCYP24A1 where the C23S hydrogen was 3.94 Å, the C26-Gly-326 distance was 3.48 Å (Fig. 5 A and B). Of course, further tests of these models will be their ability to predict subtleties of the substrate binding pocket that might explain some of the anomalous experimental hydroxylation patterns observed with various calcitriol analogs (21). Ultimately, the models will also be judged by their similarity to crystal structures of the vitamin D-related CYPs, which currently remain undetermined.
Materials and Methods
Cell Culture.
V79-4 Chinese hamster lung fibroblast cells [American Type Culture Collection (ATCC), Manassas, VA; CCL-93] and V79-hCYP24 were cultured as described previously (7). OK cells (ATCC; CCL-1840) were cultured in Eagle's Minimal Essential Medium (Sigma, Burlington, ON, Canada) supplemented with 10% (vol/vol) FBS, 1% (vol/vol) antibiotic/antimycotic (Invitrogen, Carlsbad, CA), and 1% (vol/vol) sodium pyruvate.
Preparation of Plasmid Constructs.
Full-length hCYP24A1 (21) was subcloned as an NheI–XhoI fragment into a pcDNA5/FRT construct (Invitrogen), which was modified to include the C-terminal V5-His epitope from pcDNA3.1 V5-HisB. Site-directed mutations were introduced by using a QuikChange kit (Stratagene, La Jolla, CA) per the manufacturer's protocol, and oligonucleotides were synthesized by Cortec (Kingston, ON, Canada) (SI Table 3). Mutant plasmids were transformed into Gold XL-1 Blue supercompetent cells and grown on LB agar plates containing ampicillin. Plasmids were purified for transfection by using Plasmid Maxi kit (Qiagen, Mississauga, ON, Canada). Full-length CYP24A1 from D. virginiana was cloned from OK cells by RT-PCR.
Transfection and Incubation.
Incubations with V79-CYP24 or OK cells used medium containing 1% BSA (Roche Molecular Biochemicals, Laval, PQ, Canada) and 100 mM N′,N-diphenyl-p-phenylenediamine (Sigma) (7). V79-CYP24 cells were incubated with 1 or 10 μM [1β-3H]1α,25-(OH)2D3 (32) [specific (sp.) activity, 150 or 15 mCi/mmol] (1 Ci = 37 GBq) and 1 or 10 μM [26,27-3H]25-OH-D3 (Amersham, Bucks, U.K.) (sp. activity, 90 or 9 mCi/mmol). OK cells were pretreated with 10 nM 1α,25-(OH)2D3 to induce CYP24A1 expression before use in subsequent experiments. OK cell were then incubated with 500 nM or 5 μM [1β-3H]1α,25-(OH)2D3 (sp. activity, 300 or 30 mCi/mmol, respectively) and 500 nM or 5 μM [26,27-3H]25-OH-D3 (sp. activity, 180 or 18 mCi/mmol, respectively).
Transient transfection experiments used V79-4 cells, seeded at a density of 1.5 × 105 cells per well in six-well plates and allowed to recover overnight before being transfected with 2 μg of either wild-type or mutant hCYP24A1 expression constructs by using Lipofectamine and Plus reagents (Invitrogen) for 3 h, per the manufacturer's instructions. The transfected cells were cultured in the appropriate medium for 18 h before being incubated with 3 or 750 nM [1β-3H]1α,25-(OH)2D3 (sp. activity, 50 Ci/mmol or 200 mCi/mmol); 3 or 750 nM [26,27-3H]25-OH-D3 (sp. activity, 30 Ci/mmol or 120 mCi/mmol) in medium containing 1% BSA and N′,N-diphenyl-p-phenylenediamine.
All incubations with cell lines or transiently transfected cells were conducted for 30 h, in triplicate wells, in parallel with no-cell, and/or nontransfected (V79-4) cell controls. Two micrograms of 1α-OH-D3 or vitamin D3 (Sigma) was used as internal standard.
Modeling and Substrate Docking.
We based our model of hCYP24A1 (21) on a previously described sequence alignment and model of CYP27A1 (25). Notable characteristics in these models are the extensive hydrogen bonding networks associated with heme-binding, the ERR triad, the β-sheet structures, the structural arrangement of the E-, G-, and H-helices, and the presence of a fully formed active-site cavity capable of easily accommodating ligand. An extensive set of distance constraints was used to preserve conserved hydrogen bonds in the structure during geometry minimization and constrained molecular dynamics using the Discover module in InsightII, version 2000 (Accelrys, San Diego, CA). These were set to vary between 1.43 and 1.76 Å. Ligand docking structures were prepared as previously described (21). An examination of substrate-heme geometry in 49 crystal structures and inhibitor-heme geometry in 18 crystal structures revealed the preferred positioning of hydroxylation-susceptible carbons and heme-ligating heteroatoms adjacent to the heme iron. The locations of these atoms were triangulated by calculating the distances to each of the heme methyls, CMA, CMB, and CMD, using the PDB coordinates and a spreadsheet (SI Table 4). The average triangulation distances (±SD) used to constrain C23 or C24 during docking of 1α,25-(OH)2D3 were as follows: D-CMA = 6.139 ± 0.14 Å; D-CMB = 7.642 ± 0.13 Å; D-CMD = 6.421 ± 0.17 Å.
The stereochemical configuration of the docked C23 (pro-S) or C24 (pro-R) was established by constraining the corresponding aliphatic hydrogen atom toward the heme iron atom, because P450-mediated hydroxylation does not invert the stereochemistry of the target atom. A pro-R configuration at C25 was established by rotation of the C24–25 bond to position the C26 methyl toward the heme iron. A heavily constrained molecular dynamics simulation was subsequently used to refine substrate docking with C23 or C24 in the average “sweet spot” of a productive ligand orientation.
Lipid Extraction and HPLC.
The medium was extracted using methanol/methylene chloride (7). The aqueous phase was acidified with 0.1% (vol/vol) glacial acetic acid (GAA) (Fisher Scientific, Houston, TX) and reextracted with 2.5 ml of methylene chloride (Caledon Laboratories, Georgetown, ON, Canada). Lipid extracts from incubations involving 1α,25-(OH)2D3 or 25-OH-D3 were subjected to HPLC analysis using 91:7:2 or 96:3:1 (%vol/vol/vol) hexane/isopropanol/methanol (Caledon) on Zorbax-SIL (3 μm; 6.2 × 80 mm) (Agilent, Santa Clara, CA) at 1 ml/min using an Alliance 2695 separations module (Waters, Milford, MA). Detection used either a photodiode array detector or an on-line radiochromatography detector (Berthold, Bad Wildbad, Germany) running Ready Flow III liquid scintillation mixture (Beckman, Fullerton, CA) at a flow rate of 2 ml/min, calibrated with known amounts of [1β-3H]1α,25-(OH)2D3 (50 Ci/mmol) or [26,27-3H]25-OH-D3 (30 Ci/mmol). The aqueous phase reextracts were subjected to reverse-phase HPLC on Zorbax SB-C18 (5 μm; 4.6 × 150 mm) (Agilent) using a linear gradient solvent system starting at 69:30:1 (%vol/vol/vol) water/acetonitrile/10% GAA in water and ending at 0:99:1 water/acetonitrile/10% GAA in water over 30 min, at 1 ml/min, using 3 ml/min scintillation mixture.
Periodate Cleavage and LC-MS Analysis.
Purified 1α,23,25-(OH)3D3 (800 ng) from incubations of OK cells/or A326G-transfected V79-4 cells or control 1α,24,25-(OH)3D3 in methanol was subjected to treatment with 5% (wt/vol) sodium periodate (BDH, Poole, U.K.) for 1 h (33), the reaction products purified on Zorbax-SIL and subjected to LC-MS using a Quattro Ultima triple-quadrapole mass spectrometer (Waters) in electrospray-positive (ES+) mode with a Zorbax SB-C18 (3.5 μm; 2.1 × 150 mm) (Agilent) column eluted as described above.
Identification of Metabolites.
Products of 1α,25-(OH)2D3 and 25-OH-D3 were identified based on the following criteria: (i) cochromatography with the synthetic standards 1α,24,25-(OH)3D3, 24,25-(OH)2D3, 24,25,26,27-tetranor-1α,23-(OH)2D3 (Leo Pharma, Ballerup, Denmark), or 1α,25-(OH)2D3-26,23-lactone (Dr. S. Ishizuka, Teijin, Japan); (ii) presence of vitamin D chromophore (λmax = 265 nm; ε = 18 300); (iii) sensitivity to sodium m-periodate cleavage; (iv) mass spectrometry data.
Quantitative Assessment of Hydroxylation.
The ratio of 24-hydroxylation to 23-hydroxylation activity was expressed by (i) using a weighted activity method (21) based on the sum of all products weighted for their position in either the C23- or C24-hydroxylation pathway, or (ii) determining the ratios of 1α,24,25-(OH)3D3 to 1α,23,25-(OH)3D3 or 24,25-(OH)2D3 to 23,25-(OH)2D3 when using substrate concentrations >500 nM.
Supplementary Material
Acknowledgments
Shelly West provided technical assistance in some of these studies. This work was supported by Canadian Institutes of Health Research Operating, Major Equipment, and Maintenance Grants MA-9475, MMA-38116, and MMA-69106 (to G.J.). M.K. is the recipient of an Ontario Graduate Scholarship.
Abbreviations
- 1α,25-(OH)2D3
1α,25-dihydroxyvitamin D3
- 25-OH-D3
25-hydroxyvitamin-D3
- CYP
cytochrome P450
- VDR
1α,25-dihydroxyvitamin D3 receptor
- OK
opossum kidney
- LC
liquid chromatography
- sp.
specific
- GAA
glacial acetic acid.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702093104/DC1.
References
- 1.Jones G, Strugnell SA, DeLuca HF. Physiol Rev. 1998;78:1193–1231. doi: 10.1152/physrev.1998.78.4.1193. [DOI] [PubMed] [Google Scholar]
- 2.Haussler MR, Whitfield GK, Haussler CA, Hsieh JC, Thompson PD, Selznick SH, Dominguez CE, Jurutka PW. J Bone Miner Res. 1998;13:325–349. doi: 10.1359/jbmr.1998.13.3.325. [DOI] [PubMed] [Google Scholar]
- 3.Masuda S, Jones G. Mol Cancer Ther. 2006;5:797–808. doi: 10.1158/1535-7163.MCT-05-0539. [DOI] [PubMed] [Google Scholar]
- 4.Prosser DE, Jones G. Trends Biochem Sci. 2004;29:664–673. doi: 10.1016/j.tibs.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Beckman MJ, Tadikonda P, Werner E, Prahl J, Yamada S, DeLuca HF. Biochemistry. 1996;35:8465–8472. doi: 10.1021/bi960658i. [DOI] [PubMed] [Google Scholar]
- 6.Sakaki T, Sawada N, Komai K, Shiozawa S, Yamada S, Yamamoto K, Ohyama Y, Inouye K. Eur J Biochem. 2000;267:6158–6165. doi: 10.1046/j.1432-1327.2000.01680.x. [DOI] [PubMed] [Google Scholar]
- 7.Masuda S, Gao M, Zhang A, Kaufmann M, Jones G. Recent Results Cancer Res. 2003;164:189–202. doi: 10.1007/978-3-642-55580-0_14. [DOI] [PubMed] [Google Scholar]
- 8.Masuda S, Byford V, Arabian A, Sakai Y, Demay MB, St-Arnaud R, Jones G. Endocrinology. 2005;146:825–834. doi: 10.1210/en.2004-1116. [DOI] [PubMed] [Google Scholar]
- 9.Endres B, Kato S, DeLuca HF. Biochemistry. 2000;39:2123–2129. doi: 10.1021/bi9923757. [DOI] [PubMed] [Google Scholar]
- 10.St-Arnaud R, Arabian A, Travers R, Barletta F, Raval-Pandya M, Chapin K, Depovere J, Mathieu C, Christakos S, Demay MB, et al. Endocrinology. 2000;141:2658–2666. doi: 10.1210/endo.141.7.7579. [DOI] [PubMed] [Google Scholar]
- 11.Esvelt RP, Schnoes HK, DeLuca HF. Biochemistry. 1979;18:3977–3983. doi: 10.1021/bi00585a021. [DOI] [PubMed] [Google Scholar]
- 12.Harnden D, Kumar R, Holick MF, Deluca HF. Science. 1976;193:493–494. doi: 10.1126/science.941020. [DOI] [PubMed] [Google Scholar]
- 13.Siu-Caldera ML, Clark JW, Santos-Moore A, Peleg S, Liu YY, Uskokovic MR, Sharma S, Reddy GS. J Steroid Biochem Mol Biol. 1996;59:405–412. doi: 10.1016/s0960-0760(96)00134-3. [DOI] [PubMed] [Google Scholar]
- 14.Toell A, Gonzalez MM, Ruf D, Steinmeyer A, Ishizuka S, Carlberg C. Mol Pharmacol. 2001;59:1478–1485. doi: 10.1124/mol.59.6.1478. [DOI] [PubMed] [Google Scholar]
- 15.Ishizuka S, Kurihara N, Reddy SV, Cornish J, Cundy T, Roodman GD. Endocrinology. 2005;146:2023–2030. doi: 10.1210/en.2004-1140. [DOI] [PubMed] [Google Scholar]
- 16.Engstrom GW, Reinhardt TA, Horst RL. Arch Biochem Biophys. 1986;250:86–93. doi: 10.1016/0003-9861(86)90704-6. [DOI] [PubMed] [Google Scholar]
- 17.Pedersen JI, Hagenfeldt Y, Björkhem I. Biochem J. 1988;250:527–532. doi: 10.1042/bj2500527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simboli-Campbell M, Jones G. J Nutr. 1991;121:1635–1642. doi: 10.1093/jn/121.10.1635. [DOI] [PubMed] [Google Scholar]
- 19.Ohyama Y, Noshiro M, Okuda K. FEBS Lett. 1991;278:195–198. doi: 10.1016/0014-5793(91)80115-j. [DOI] [PubMed] [Google Scholar]
- 20.Chen K-S, Prahl JM, DeLuca HF. Proc Natl Acad Sci USA. 1993;90:4543–4547. doi: 10.1073/pnas.90.10.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda S, Prosser DE, Guo YD, Kaufmann M, Jones G. Arch Biochem Biophys. 2007;460:177–191. doi: 10.1016/j.abb.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 22.Annalora AJ, Bobrovnikov-Marjon E, Serda R, Pastuszyn A, Graham SE, Marcus CB, Omdahl JL. Arch Biochem Biophys. 2007;460:262–273. doi: 10.1016/j.abb.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamamoto H, Kusudo T, Urushino N, Masuno H, Yamamoto K, Yamada S, Kamakura M, Ohta M, Inouye K, Sakaki T. Mol Pharmacol. 2006;70:120–128. doi: 10.1124/mol.106.023275. [DOI] [PubMed] [Google Scholar]
- 24.Horiuchi N, Saikatsu S, Akeno N, Abe M, Kimura S, Yamada S. Horm Metab Res. 1995;27:83–89. doi: 10.1055/s-2007-979914. [DOI] [PubMed] [Google Scholar]
- 25.Prosser DE, Guo Y-D, Jia Z, Jones G. Biophys J. 2006;90:3389–3409. doi: 10.1529/biophysj.105.069369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bishop JE, Collins ED, Okamura WH, Norman AW. J Bone Miner Res. 1994;9:1277–1288. doi: 10.1002/jbmr.5650090818. [DOI] [PubMed] [Google Scholar]
- 27.Gallus GN, Dotti MT, Federico A. Neurol Sci. 2006;27:143–149. doi: 10.1007/s10072-006-0618-7. [DOI] [PubMed] [Google Scholar]
- 28.Miller WL, Portale AA. Endocr Dev. 2003;6:156–174. doi: 10.1159/000072775. [DOI] [PubMed] [Google Scholar]
- 29.Cheng JB, Levine MA, Bell NH, Mangelsdorf DJ, Russell DW. Proc Natl Acad Sci USA. 2004;101:7711–7715. doi: 10.1073/pnas.0402490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuster I, Egger H, Herzig G, Reddy GS, Vorisek G. Recent Results Cancer Res. 2003;164:169–188. doi: 10.1007/978-3-642-55580-0_13. [DOI] [PubMed] [Google Scholar]
- 31.Kahraman M, Sinishtaj S, Dolan PM, Kensler TW, Peleg S, Saha U, Chuang SS, Bernstein G, Korczak B, Posner GH. J Med Chem. 2004;47:6854–6863. doi: 10.1021/jm040129+. [DOI] [PubMed] [Google Scholar]
- 32.Makin G, Lohnes D, Byford V, Ray R, Jones G. Biochem J. 1989;262:173–180. doi: 10.1042/bj2620173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller BE, Chin DP, Jones G. J Bone Miner Res. 1990;5:597–608. doi: 10.1002/jbmr.5650050609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.