Abstract
The polytene chromosomes of Drosophila melanogaster consist of condensed heterochromatin regions most of which are in the chromocenter, telomeres, and the fourth chromosome. Whereas suppressor of variegation 3-9 [SU(VAR)3-9], a histone methyltransferase, is mainly responsible for lysine 9 of histone H3 (H3K9) methylation of the chromocenter and consequent binding of the heterochromatin-protein HP1, the enzyme for painting of the fourth chromosome by H3K9 methylation has been elusive. We show here that dSETDB1, the Drosophila ortholog of the mammalian SETDB1, is an authentic H3K9 methyltransferase and a pleiotropic regulator of the fly's development. Drosophila mutants hypomorphic or null in dSETDB1 expression lose most of the H3K9 methylation as well as HP1-binding on the fourth chromosome. We also show that binding of painting of fourth (POF), a known fourth chromosome-specific protein, and the dSETDB1-controlled H3K9 methylation of this chromosome are interdependent. Furthermore, POF and dSETDB1 interact with each other in vivo. The deregulation of H3K9 methylation, HP1-binding, and POF-binding resulted in, on the average, a global reduction of gene expression from the fourth chromosome but not the other chromosomes. Deficiency of dSETDB1 also up-regulated the expression of HP1. These results have suggested an interactive network, as controlled in part by dSETDB1, regulating the epigenetic modification and gene expression of Drosophila chromosome 4.
Keywords: epigenetics, gene regulation, polytene chromosomes, heterochromatin, histone methylation
The eukaryotic chromosomes are composed of two types of chromatins, euchromatin and heterochromatin, regulated by epigenetic marks such as DNA methylation and histone modification (1, 2). In general, the heterochromatin consists of highly compacted DNA and repetitive DNA sequences that are present in transcriptionally silent regions around the centromeres and telomeres. In vertebrates, the heterochromatin, in contrast to euchromatin, is usually enriched in methylated CpG and hypoacetylated histones. More-recent studies have demonstrated the importance of histone methylation in the formation and maintenance of heterochromatin (1–3). In particular, methylation at lysine 9 of histone H3 (H3K9) creates the binding sites and subsequent recruitment of chromodomain proteins, such as the heterochromatin protein HP1 (4–6), to condense the chromatin (7, 8). Heterochromatin plays a central role in the developmental stage- and differentiation state-specific regulation of gene expression through epigenetic processes such as the X-chromosome inactivation in mammals, telomeric and mating-type silencing in yeast, and the position effect variegation in Drosophila (9–11).
Like in the mammals (12), multiple histone methyltransferases (HMTases) have been described in Drosophila melanogaster (13–15). For H3K9, the HMTases that have been well characterized include suppressor of variegation 3-9 [SU(VAR)3-9] and dG9a (4, 16). Of the two, SU(VAR)3-9 mainly associates with and is responsible for histone H3K9 methylation of the chromocenter heterochromatin, although weak signals of SU(VAR)3-9 binding could also be detected on the fourth chromosome, the telomeres and a few euchromatic sites (4, 14). In SU(VAR)3-9-deficient mutants, however, H3K9 methylation and HP1-binding were greatly reduced in the chromocenter, but they remained unchanged on the fourth chromosome of the polytene chromosomes (4). Interestingly, the localization of SU(VAR)3-9 and HP1 to the chromocenter are interdependent (4). More recently, SU(VAR)3-9 was also shown to act in vitro as an auxiliary factor and facilitate the maintenance of HP1 on the heterochromatin (17). SU(VAR)3-9 appeared to act synergistically with dG9a for methylation and heterochromatin formation of the chromocenter (16).
An interesting connection with the apparent lack of an effect by the SU(VAR)3-9 mutation on H3K9 methylation and HP1-binding of the fourth chromosome is the existence of a chromosome 4-specific binding protein, painting of fourth (POF). POF is a chromosome-specific protein containing an RNA-binding motif. Like HP1, POF-binding on the fourth chromosome was not affected by the SU(VAR)3-9 mutation, either (4, 18). Moreover, the bindings of HP1 and POF to the fourth chromosome were interdependent (18). Gene-expression profiling by microarray analysis showed that expression of genes on the fourth chromosome, but not on other chromosomes, was globally coregulated by POF and HP1, with HP1 being a transcription repressor while POF behaved like an activator (18, 19). Based on the above, Johansson et al. (18) have suggested that there may exist a Drosophila chromosome 4-specific H3K9 methyltransferase.
The fly dSETDB1 was initially identified as an ortholog of the mammalian SETDB1/ESET, and it consists of a methyl-CpG-binding domain, a PreSET [pre-SU(VAR)3-9, Enhancer of Zeste, Trithorax]/SET domain, and two tudor motifs (20). In the following, we present biochemical data that dSETDB1 is indeed a histone H3K9 methyltransferase. We then show by genetic analysis that dSETDB1 is essential for the survival and proper development of the flies. Remarkably, dSETDB1 is mainly responsible for H3K9 methylation of the fourth chromosome as well as painting of this chromosome by HP1- and POF-binding. Based on these results, the previously known properties of HP1 and POF, and our gene-expression profiling analysis of the dSETDB1 mutants in comparison to the wild type, we outline a model in which dSETDB1 functions cooperatively with HP1 and POF for the epigenetic regulation of the fourth chromosome in Drosophila.
Results
Gene Structure and Expression of dSETDB1.
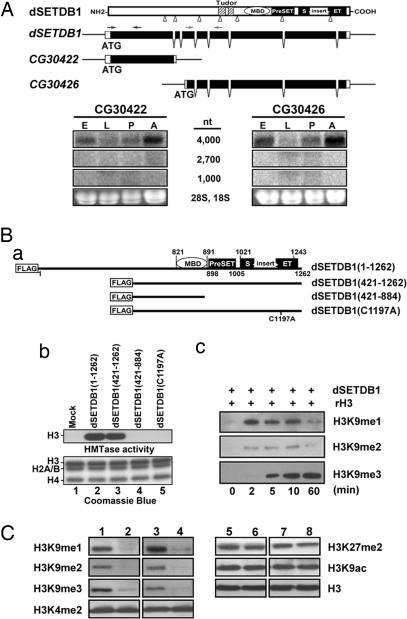
dSETDB1 was initially identified as a 3,948-bp open reading (CG12196) in the Fly Genomic Annotation (release no. 2.0), the sequence of which suggests that it is the ortholog of the mammalian H3K9 methyltransferase SETDB1/ESET containing a methyl-CpG-binding domain, a PreSET/SET domain, and two tudor motifs (20). More recent annotations listed dSETDB1 as encoding two transcripts. One of the transcripts is a 1,049-nt exon (CG30422), and other is 2,716 nt long (CG30426), consisting of six exons (top maps, Fig. 1A).
Fig. 1.
Characterization of dSETDB1 as an H3K9 histone methyltransferase. (A) Structure and expression of dSETDB1. The schematic illustrations of the gene structure, the domains/motifs of the protein, and the two annotations of dSETDB1 are shown (Upper). The approximate locations of the primers used to generate the probes for Northern blot analysis are indicated above the gene. The probes made from the CG30422 and CG30426 regions both gave a single 4,000-nt band on the Northern blots (Lower) throughout the embryonic (E), larval (L), pupal (P), and adult (A) stages. (B) Histone methyltransferase assay of dSETDB1. (Ba) Schematic illustration of the inserts of plasmids expressing different dSETDB1 polypeptides tagged with FLAG: full-length dSETDB1 (1–1,262), N-terminal deletion mutant (421–1,262), N-terminal deletion mutant (421–884), and the single amino acid mutant dSETDB1 (C1197A). (Bb) Histone methyltransferase activities of dSETDB1. The HMT activities in the extracts of cells transfected with plasmids expressing different forms of dSETDB1 were assayed in vitro after immunoprecipitation with the anti-FLAG M2 agarose. The core histone substrates were stained with Coomassie blue (Bb Lower), and the radioactively labeled histone H3 was identified by autoradiography (Bb Upper). Similar to human hSETDB1 (23), recombinant dSETDB1 purified from bacteria exhibited no activity in vitro (T.-Y.T., data not shown). (Bc) Sequential methylation of H3 by dSETDB1 in vitro. Two micrograms of the recombinant histone H3 (rH3) were used as the substrate for in vitro HMTase assay with anti-Flag purified dSETDB1 from transfected SL2 cells. At different times, the reaction products were analyzed by Western blotting using antibodies against different forms of methylated H3K9. (C) Western blot analysis of the expression levels of different modification forms of the histones. The levels in the total extracts of the third instar larvae of the wild type (lanes 1 and 5), the homozygotes of dSETDB1GS7132 (lanes 2 and 6) and dSETDB1null (lanes 4 and 8), and dSETDB1GS7132; UAS-dSETDB1/arm-GAL4 (lanes 3 and 7) were analyzed by Western blotting. The epitopes or proteins against which the antibodies were made are indicated at the side of the gel panels.
Northern blotting with two cDNA probes from the CG30422 and CG30426 regions (one probe from each region), detected only one transcript of the size of ≈4,000 nt (Fig. 1A). This finding was consistent with the results of others (21, 22). The data of Fig. 1A also showed that dSETDB1 transcription is developmentally regulated, with higher levels at the embryonic and adult stages than those at the larval and pupal stages (Fig. 1A).
dSETDB1 Has a Methyltransferase Activity for Histone H3K9.
We first tested whether dSETDB1 possessed intrinsic HMTase activity by transfection of Drosophila SL2 cells with Drosophila actin5C (AC) promoter-based expression plasmids containing the full-length or portions of the dSETDB1 cDNA tagged with FLAG (Fig. 1Ba). Indeed, a robust histone H3-specific methyltransferase activity was detected when the core histones were used as the substrate (compare lane 2 with lane 1, Fig. 1Bb). On the other hand, only dSETDB1 (421–1,262) but not dSETDB1 (421–884), which lacked the PreSET/SET domain, was capable of methylating the H3 histone in vitro (lanes 3 and 4, Fig. 1Bb). Single amino-acid substitution C to A at the highly conserved C1197 within the SET domain of hSETDB1 (23) abolished its ability to methylate H3 (lane 5, Fig. 1Bb). These data supported that the PreSET/SET domain of dSETDB1 was required for its HMTase activity on H3. Furthermore, as its human ortholog, dSETDB1 specifically methylated H3K9, but not H3K4 or H3K27 [supporting information (SI) Fig. 6].
Finally, histone H3 could be methylated at lysine 9 by mono-, di-, or trimethylation (24). As exemplified in Fig. 1Bc, dSETDB1 methylated H3 to generate K9me1, K9me2, and K9me3 in a sequential manner (me1, me2, and me3 standing for mono-, di-, and trimethyl, respectively). This data demonstrated that dSETDB1 could generate all three methylations on K9 of H3 and, thus, could globally affect the H3K9 methylation in Drosophila cells.
Generation and Characterization of dSETDB1 Mutants.
To address the function of dSETDB1 in normal development of the flies, we first acquired a P-element insertion line, GS7132, generated by the Drosophila Gene Search Project (DGSP), the P-element of which was inserted 110 bp downstream of the dSETDB1 transcription start site (gel in SI Fig. 7A). This insertion caused a decrease of dSETDB1 expression by ≈80% (SI Fig. 7A). The homozygous dSETDB1GS7132 flies exhibited significantly reduced viability with only 57% of the adult flies enclosed, and more than half of them dying within 10 days. The deficiency Df(2R)ED4071, which covered dSETDB1, failed to complement the lethality phenotype caused by dSETDB1GS7132. The mutant flies had spread-out wings (SI Fig. 7B) and disorganization of the interommatidial bristles (SI Fig. 7C). To assure the phenotypes observed above for the homozygous dSETDB1GS7132 came from single gene disruption, we used the flipase–flipase-recombination-target (FLP–FRT)-based deletion approach (refs. 25 and 26; also see SI Materials and Methods) to generate dSETDB1 knockout lines. A single line, dSETDB1null, with the entire dSETDB1-coding region removed, was isolated. Homozygotes of dSETDB1null as well as its transheterozygotes with the dSETDB1GS7132 flies also had the wing phenotype, and they exhibited more severe lethality. Finally, the phenotypes of the homozygous dSETDB1GS7132 or its transheterozygotes with dSETDB1null could be rescued by expression of the full-length dSETDB1 protein (SI Fig. 7 B and C).
dSETDB1 Is a Histone H3K9 Methyltransferase in Drosophila.
To test whether dSETDB1 functions as an authentic HMTase in Drosophila, we extracted the total histones from the nuclei of cells derived from the third instar larvae of the wild type, the homozygous dSETDB1GS7132, and the homozygous dSETDB1null flies. The extracts were analyzed by Western blotting with the use of antibodies against different H3 modification. Whereas the global levels of H3 itself, H3K9ac, H3K4me2, and H3K27me2 remained unchanged (compare lanes 6 and 8 with lane 5, Fig. 1C), those of the H3K9me1, H3K9me2, and H3K9me3 all decreased significantly in the two mutant lines (compare lanes 2 and 4 with lane 1, Fig. 1C). The levels of all three methylated H3K9 species could be rescued by overexpression of dSETDB1 (lane 3, Fig. 1C). These findings established dSETDB1 as one of the major histone H3K9 methyltransferases in Drosophila.
dSETDB1 Is a Fourth Chromosome-Specific Histone Methyltransferase.
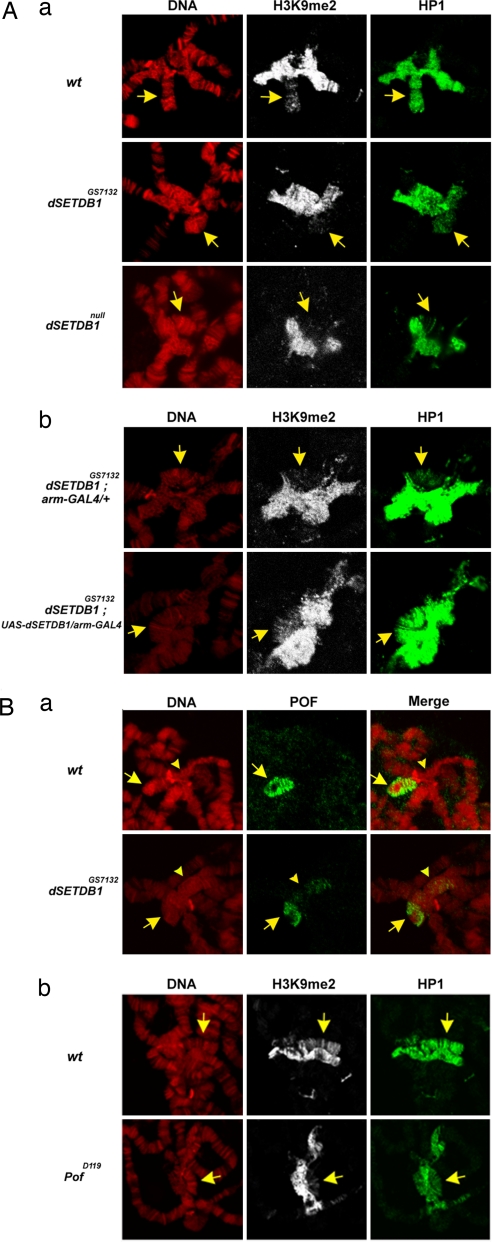
We have carried out immunostaining to analyze the distribution patterns of methylated H3K9 on the polytene chromosomes from the salivary gland cells of the third instar larvae. With use of the H3K9me2 antibody, staining of both the chromocentral heterochromatin and, to a less extent, the fourth chromosome of the wild type was observed (Fig. 2Aa Top Center). This finding is similar to those observed in the previous studies, e.g., refs. 4 and 14. Significantly, we found that decoration by H3K9 di-methylation on the fourth chromosome, but not on the chromocenter, was almost abolished in either the dSETDB1GS7132 homozygotes or the null mutant (compare Middle Center and Bottom Center with Top Center in Fig. 2Aa). Similarly, despite its relatively low amount (14), the level of H3K9-trimethylation was also reduced by the dSETDB1 mutation (SI Fig. 8). Notably, the reduction of the level of H3K9me2 on the fourth chromosome could be rescued by arm-GAL4-directed overexpression of the dSETDB1 protein in the dSETDB1GS7132 background (compare the two central panels of Fig. 2Ab). The above data suggest that dSETDB1 regulates the H3K9me2 and H3K9me3 painting of the fourth chromosome in the salivary gland cells of Drosophila.
Fig. 2.
Immunostaining analysis of polytene chromosomes. (Aa) Immunostaining analysis of the distributions of H3K9me2 and HP1 in the polytene chromocenter and on the fourth chromosome. The patterns of the wild type (Aa Top), homozygous dSETDB1GS7132 (Aa Middle), and homozygous dSETDB1null (Aa Bottom), respectively, are shown. (Aa Left) DNA patterns as stained by DAPI. Note the reduced staining signals of both H3K9me2 and HP1 on the fourth chromosome (arrows) in both mutant lines. (Ab) Rescue of the HP1-binding and H3K9 methylation by overexpressing the dSETDB1 protein. The immunostaining patterns of the dSETDB1GS7132; UAS-dSETDB1/ arm-GAL4 (Lower) are shown in comparison to dSETDB1GS7132; arm-GAL4/+ (Upper). (Ba) Immunostaining patterns of POF on the polytene chromosomes. The pattern in the wild type (wt) and homozygous dSETDB1GS7132 are compared. Note the reduction of POF-binding on the fourth chromosome (arrows) and the spread of the signals into the chromocenter (arrowheads) in the mutant (Middle Lower). (Bb) Immunostaining pattern of H3K9me2 in PofD119 flies. The distribution patterns of H3K9me2 and HP1 in the polytene chromosomes of the POF-deficient line PofD119 are compared with the wild type. Note the reduced signals of both H3K9me2 and HP1 on the fourth chromosome (arrows) in the mutant.
HP1- and POF-Binding on the Polytene Fourth Chromosome Are Both dSETDB1-Dependent.
In view of the reduced signals of H3K9me2 and H3K9me3 on the fourth chromosome in the dSETDB1 mutant background, we have analyzed the pattern of association of HP1 on chromosome 4. Indeed, immunocytological analysis showed that the staining of HP1 on the fourth chromosome was also greatly decreased in both dSETDB1 mutant lines (compare Fig. 2Aa Middle Right and Bottom Right with Top Right in SI Fig. 8). Similar to H3K9me2, the decrease of the HP1 painting on the fourth chromosome could be reversed by transgenically expressing the dSETDB1 protein (compare Fig. 2Ab Right and Center).
POF is the only known chromosome-specific protein painting an autosome, e.g., chromosome 4, in Drosophila. We thus also examined the immunostaining pattern of POF by using a specific anti-POF antibody (27). As seen, the painting of this chromosome by POF was also significantly decreased by the dSETDB1 mutation (Fig. 2Ba). Interestingly, some spread of the POF binding into the nearby chromocenter was observed (arrowhead, Fig. 2Ba Lower Center). These findings indicated that the restricted binding of POF on the Drosophila polytene chromosomes 4 as well as the recruitment of HP1 to this chromosome were tightly controlled by dSETDB1.
Painting of the Fourth Chromosome by H3K9me2 Is Also POF-Dependent.
To gain further insight of the upstream–downstream signals for control of H3K9 methylation of the fourth chromosome, we analyzed the chromosomal distribution of H3K9me2 and HP1 in a POF mutant line by immunostaining (Fig. 2Bb). As observed previously (18), HP1 binding on the polytene chromosome 4 but not the chromocenter of PofD119 was significantly reduced (compare Fig. 2Bb Lower Right with Upper Right). Remarkably, the H3K9me2 signal of the fourth chromosome was also reduced in the mutant (compare Fig. 2Bb Upper and Lower). This data indicated that painting of the fourth chromosome with HP1 and H3K9 methylation was regulated by dSETDB1 as well as POF.
dSETDB1 Physically Interacts with POF in Vivo.
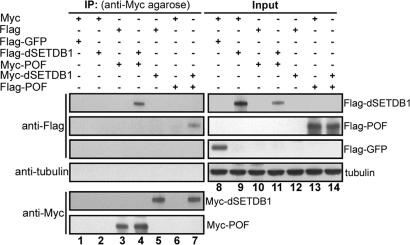
In view of the coregulation of H3K9 methylation by dSETDB1 and POF in a chromosome 4-specific manner, we tested the possibility of physical interaction between dSETDB1 and POF. With the lack of available dSETDB1 antibody or transgenic flies carrying tagged dSETDB1, we carried out immunoprecipitation experiments with SL2 cells transfected with plasmids expressing Myc- or Flag-tagged dSETDB1 or POF. Indeed, as shown in Fig. 3, interaction in vivo between either the Flag-dSETDB1/ Myc-POF pair (lane 4, Fig. 3) or the Myc-dSETDB1/ Flag-POF pair (lane 7, Fig. 3) could be detected.
Fig. 3.
dSETDB1 association with POF in vivo. Drosophila SL2 cells were transfected with different combinations of the pAC-based expression plasmids. The whole-cell extracts were then prepared, immunoprecipitated with anti-Myc agarose, blotted (lanes 1–7), and probed with anti-Flag, anti-tubulin, or anti-Myc, as indicated to the left of the panels. Five percent of the whole-cell extracts used for the individual immunoprecipitated reactions were loaded as the “Input” controls and probed with the same antibodies (lanes 8–14). The identities of bands detected on the blots are indicated to the right of the panels. The interaction in vivo between dSETDB1 and POF is evidenced by the presence of Flag-dSETDB1 in lane 4 and Flag-POF in lane 7. IP, immunoprecipitate.
dSETDB1 Contributes to a Global Regulation of Gene Expression from the Fourth Chromosome.
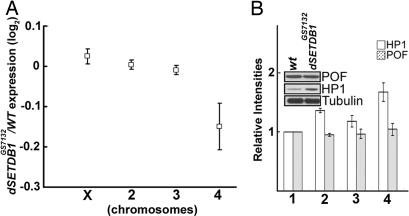
Because dSETDB1 mutations specifically affected the painting of the fourth chromosome by H3K9me2, H3K9me3, HP1-binding, and POF-binding (Fig. 2 A and B and SI Fig. 8), we analyzed the expression levels of genes of the fourth chromosome in comparison to those on the other chromosomes in homozygous dSETDB1GS7132 and in the wild type. Three biological replicates of mRNA samples were prepared from the third instar larvae for synthesis of cRNAs used as probes for an Affymetrix (Santa Clara, CA) microarray analysis (Genome 2.0). As shown in SI Fig. 9, the lack of dSETDB1 caused reduction of the expression levels of a majority (73%) of the 92 genes of the fourth chromosome analyzed. Although the extents of reduction where small for the individual genes, the overall mean reduction of gene expression on the fourth chromosome was apparent when compared with chromosomes X, 2, and 3 (Fig. 4A).
Fig. 4.
Gene expression comparison of wild type and dSETDB1 mutants. (A) Mean changes of the levels of gene expression of the individual chromosomes of homozygous dSETDB1GS7132 in comparison to the wild type. Each square indicates the log2 value of the ratio of the average of the median expression levels of all genes on the particular chromosome of the mutant to that of the wild type. (B) Western blot analysis of the levels of HP1, POF, and tubulin. The total protein extracts of the third instar larvae from the wild type (bar set 1), homozygous dSETDB1GS7132 (bar set 2), dSETDB1GS7132; UAS-dSETDB1/arm-GAL4 (bar set 3), and homozygous dSETDB1null (bar set 4), respectively, were analyzed by Western blotting with different antibodies. The relative intensities derived from two sets of experiments are indicated by the bar histographs, with the open bars representing those of HP1 and the shaded bars representing POF. The Western blot of the wild type in comparison to the homozygous dSETDB1GS7132 is shown as an example.
dSETDB1 Mutations Lead to Increase of HP1 Expression.
After the immunostaining study of Fig. 2, we also examined the effects of the dSETDB1 mutations on the expression levels of the HP1 and POF proteins. As shown by Western blotting analysis of the total protein extracts from the third instar larvae, the levels of the HP1 protein, but not those of POF, were elevated, instead of decreased by ≈40–70% in the two mutant lines (compare bar sets 2 and 4 with bar set 1, Fig. 4B). Transgenic expression of dSETDB1 in the mutant partially restored the level of HP1 (bar set 3, Fig. 4B). This result suggested that, contrary to what might be expected from the anti-HP1 immunostaining data of Fig. 2A, dSETDB1 in fact played a repressive role in HP1 expression (see Discussion).
Discussion
dSETDB1 Is an Authentic H3K9 Methyltransferase Regulating Drosophila Development.
Our combined biochemical and genetic data (Fig. 1 and SI Figs. 6 and 7) have demonstrated that dSETDB1 indeed is an H3K9 methyltransferase regulating the early development of D. melanogaster. The fact that the hypomorphic (dSETDB1GS7132) and the null (dSETDB1null) mutations resulted in similar phenotypes (SI Fig. 6 and data not shown) suggested that a threshold of the expression level of dSETDB1 was critical for fly development. Our data on the importance of dSETDB1 in the regulation of fly development were consistent with two other recent reports. In one of them (22), GAL4-driven knock-down of the dSETDB1 expression by RNAi caused various defects in the development of the flies. In another study consisting of the genetic screening and rescue experiments of EMS-induced mutants, dSETDB1 was shown to be essential for oogenesis of Drosophila and mutation of the dSETDB1 gene of the sterile flies led to a reduction of H3K9 trimethylation in the germ and somatic cells of the germanium (21).
dSETDB1 Is Mainly a Chromosome 4-Specific H3K9 Methyltransferase in the Salivary Gland Cells, but It Also Regulates H3K9 Methylation Globally.
Histone H3K9 methylation is critical for heterochromatin formation and transcription regulation (1, 28, 29). Our in vitro characterization of the H3K9 methyltransferase activity of dSETDB1 (Fig. 1B) in combination with the in vivo analysis of its epigenetic properties (Figs. 1C and 2) have demonstrated that this enzyme, like the well known SU(VAR)3-9, is also an essential chromatin modifier for gene regulation. Remarkably, the epigenetic effect of dSETDB1 in the salivary gland cells appeared to be exerted mainly on the fourth chromosome (Fig. 2). Our immunostaining data of dSETDB1GS7132 and dSETDB1null homozygotes in comparison to the wild type (Fig. 2A) is highly suggestive that dSETDB1 is the long-sought-for enzyme responsible for H3K9 methylation on the fourth chromosome of the polytene chromosomes. It is important to note here that, after submission of this manuscript, Seum et al. (30) reported that HA-tagged dSETDB1 proteins were mainly localized on the polytene chromosome 4 in a transgenic fly line.
Notably, mutations in dSETDB1 also globally significantly affected the levels of H3K9 mono-, di-, and trimethylation of the flies (Fig. 1C). Previous mutation analysis has suggested that SU(VAR)3-9 is the main H3K9 methyltransferase in Drosophila (4). Furthermore, it was recently shown that another H3K9 methyltransferase, dG9a, interacted with SU(VAR)3-9 to affect the global H3K9 methylation level and the development of Drosophila (16). Our data thus indicate that dSETDB1, while being the major enzyme for H3K9 methylation of the polytene chromosome 4, also functions synergistically with other H3K9 methyltransferases including SU(VAR)3-9 and dG9a to accomplish the global H3K9 methylation patterns and the consequent regulation of the developmental program of Drosophila.
Interdependence of dSETDB1, HP1, and POF in the Epigenetic Painting of Chromosome 4.
Our immunostaining results showed that in vivo binding of HP1 on the fourth chromosome, but not that in the chromocenter, depended on H3K9 methylation as directed by dSETDB1 (Fig. 2A). The dSETDB1-dependent binding of HP1 on the fourth chromosome (this study; ref. 30) and the transgenic fly analysis by Seum et al. (30) are highly suggestive that the endogenous dSETDB1, similar to the effect of SU(VAR)3-9 on the chromocenter (4) and its interaction with/recruitment of HP1 (4, 17), modifies the chromatin structure of the polytene chromosome 4 through chromosome 4-specific binding, followed by H3K9 methylation and recruitment of HP1.
By immunostaining analysis, we have further shown that the chromosome 4-restricted binding of the protein POF also depended on dSETDB1 and/or H3K9 methylation (Fig. 2Ba), although the expression level of POF was not altered in the dSETDB1 mutants (Fig. 4B). Because the chromosome 4-specific binding of POF, mainly within the gene bodies, was interdependent with that of HP1 (18), our data in Fig. 2 A and B could be explained by a scenario in which dSETDB1 generates methylated H3K9 residues on the fourth chromosome, which provides the docking sites for HP1. These chromosome 4-bound HP1 molecules in turn help to stabilize the binding of POF proteins that are recruited to the fourth chromosome through an independent mechanism(s).
On the other hand, POF also appeared to facilitate H3K9 methylation of the fourth chromosome and, by inference, the binding of dSETDB1 on the chromosome (Fig. 2Bb). This latter possibility was strengthened by the capabilities of the two proteins to interact with each other in vivo (Fig. 3). The above together suggest the existence of multiway interactions, functionally as well as physically, among dSETDB1, POF, and HP1 (see further discussions and modeling below).
Interactive Network for Epigenetic Regulation of the Polytene Chromosome 4.
Our microarray analysis has shown that dSETDB1 mutation affected mainly the expression of genes of the fourth chromosome (SI Fig. 9) but not those of the other chromosomes (Fig. 4A). This change in gene expression, mostly reduction, of the fourth chromosome is an interesting correlation with the reduced binding of POF, an activator of transcription (18), on this chromosome as affected by the same dSETDB1 mutation (Fig. 2Ba). Johansson et al. (18) have proposed a feedback model in which HP1 and POF counteract each other with respect to their chromosome 4-binding and function as a transcription activator (POF) or repressor (HP1) to globally regulate the expression of genes from the fourth chromosome.
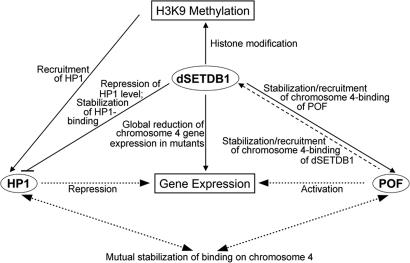
Based on our data and the discussion above, we suggest a “balancing” model for the epigenetic regulation of the polytene chromosome 4, in which dSETDB1, HP1, and POF function cooperatively and counteractingly with respect to their binding to the fourth chromosome as well as their effects on the histone modification and gene expression of this chromosome (Fig. 5). It is proposed that both dSETDB1 and POF bind specifically to the fourth chromosome, although whether one of them binds first or both proteins bind together as a complex is unknown at the moment. Furthermore, the two proteins stabilize each other's binding, mainly within the genes, on the fourth chromosome. The chromosome 4-bound dSETDB1, as a H3K9 methyltransferase and possibly as an auxilatory factor for chromatin binding of other factors, then generates methylated H3K9 residues that serve as the recognition marks to recruit HP1 to the fourth chromosome, which in turn also helps to stabilize the binding of POF. With respect to gene expression, through its physical interaction with the gene body-bound POF, dSETDB1 could contribute to the activation function of POF. In addition, dSETDB1 actively recruits HP1 as mentioned above, and it also represses the expression level of the HP1 protein, thus providing another “force” counteracting the HP1 effects on heterochromatin formation/maintenance and transcription repression. The details of the molecular and cellular basis of the different steps of this model await investigation.
Fig. 5.
A “balancing” model for heterochromatin formation and gene regulation of chromosome 4. The initial feedback model of gene regulation of chromosome 4 by HP1 and POF was suggested by Johansson et al. (18), in which HP1 and POF affect each other with respect to their chromosome 4-restricted binding and effects on gene expression from this chromosome (dotted lines). In this balancing model, the balancing effects among dSETDB1, H3K9 methylation, and HP1 expression are included. In particular, the dSETDB1-binding to chromosome 4 generates methylated H3K9 for the recruitment of HP1 to this chromosome, but at the same time dSETDB1 also represses the level of the HP1 protein, thus limiting the available pool of HP1. dSETDB1 and POF also cooperatively interact with each other physically for their recruitment to and/or stabilization of binding on the fourth chromosome. For more descriptions of the model, see Discussion.
Materials and Methods
Fly Stocks and Genetic Crosses.
The flies were grown and crossed on standard medium at 25°C unless indicated otherwise. The different fly stocks/lines used and the establishment of transgenic flies are described in SI Materials and Methods.
Transgenic Flies.
The plasmid pUAST-dSETDB1 (see SI Materials and Methods for its construction) was used in germline transformation as described previously (31) to generate the UAS-dSETDB1 lines. Twelve independent transgenic lines were generated with the DNA inserted on X, second, and third chromosomes (two lines on the X, six lines on the second, and four lines on the third chromosomes).
Generation of dSETDB1 Mutants.
Generation of the flipase–flipase recombination (FLP-FRT) target deletion was performed as previously described (25, 26). A single line, dSETDB1null, with the dSETDB1 genomic region removed was used for subsequent analysis.
Immunostaining of the Polytene Chromosomes.
Flies for immunostaining analysis of the polytene chromosomes were grown at 18°C. Immunostaining of the polytene chromosomes were performed essentially as described previously (32). Details of the processes and image analysis are described in SI Materials and Methods.
Antibodies.
The antibodies against histone H3 and the modified forms of histones were obtained from Upstate Biotechnology (Lake Placid, NY). The anti-HP1 antibody C1A9 was from the Developmental Studies Hybridoma Bank, University of Iowa (Iowa City, IA). Anti-POF was a kind gift from Jan Larsson (Umeå University, Umeå, Sweden). The anti-tubulin was from Sigma (St. Louis, MO). The secondary antibodies, goat anti-rabbit or anti-mouse antibodies conjugated with Alexa Fluor 488 (1:200) or Alexa Fluor 647 (1:100), used in the immunostaining experiments were from Molecular Probes (Eugene, OR).
Cell Culture.
Drosophila SL2 cells were maintained at 24°C in Schneider's Drosophila medium (Sigma) containing 10% FBS (Invitrogen, Carlsbad, CA) and 1% antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin; Invitrogen).
Plasmids.
The procedures for construction of different expression plasmids are described in SI Materials and Methods.
DNA Transfection and Immunoprecipitation.
Details are described in SI Materials and Methods.
In Vitro Assay of Histone Methyltransferase Activity.
Histone methyltransferase assays were performed as previously described (15). The reaction products were analyzed by 15% SDS/PAGE and by x-ray autoradiography. See SI Materials and Methods for more details.
Northern Blot Analysis.
The Northern blotting was carried out following the standard procedures (33). See SI Materials and Methods for the details.
Western Blot Analysis.
Total histones extract was prepared from isolated nuclei of 25 third instar larvae as previous described (34). See SI Materials and Methods for more details.
Gene Profiling by Microarray Hybridization.
The details of the microarray hybridization are described in the SI Materials and Methods. For microarray data analysis, the intensity values were first normalized and summarized by using the MAS 5.0 software (Affymetrix). The data treatment and statistical analysis were carried out by using GeneSpring (Agilent Technologies, Palo Alto, CA) to calculate the median intensities, the average of the median intensities, etc. (see the legends of Fig. 4 and SI Fig. 9).
Supplementary Material
Acknowledgments
We thank J.-T. Wu and C.-K. Chen in C.-T. Chien's laboratory, C.-C. Yang in D.-H. Huang's laboratory, and H. Sun's laboratory for their help and suggestions on experiments involving Drosophila genetics; Mr. T.-L. Lin in the Electron Microscope laboratory of the Institute of Cellular and Organismic Biology for help with the use of their facilities; Dr. Jan Larsson's laboratory for their generosity in sharing the PofD119 line and the anti-POF antibody; members of C.-K.J.S.'s laboratory, in particular W.-P. Wang, G.-S. Huang, S.-Y. Pai, and Dr. J.-L. Chao, for contributing to sample collection and helpful discussions; and the Microarray Core and Bioinformatics Core of the Institute of Molecular Biology for their help. This research has been supported by a Frontier of Science Grant from the National Science Council, Taipei, Taiwan. C.-K.J.S. is an Academia Sinica Awarded Investigator and T.-Y.T. is an Academia Sinica postdoctoral Fellow.
Abbreviations
- POF
painting of fourth
- SU(VAR)3-9
suppressor of variegation 3-9
- HMTase
histone methyltransferase
- H3K9
lysine 9 of histone H3.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705534104/DC1.
References
- 1.Grewal SI, Jia S. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 2.Quina AS, Buschbeck M, Di Croce L. Biochem Pharmacol. 2006;72:1563–1569. doi: 10.1016/j.bcp.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 3.Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 4.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 6.Eissenberg JC, Elgin SC. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 7.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 8.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 9.Cavalli G. Curr Opin Cell Biol. 2002;14:269–278. doi: 10.1016/s0955-0674(02)00324-1. [DOI] [PubMed] [Google Scholar]
- 10.Grewal SI, Moazed D. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 11.Weiler KS, Wakimoto BT. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 12.Bannister AJ, Kouzarides T. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez-Venegas R, Avramova Z. Gene. 2002;285:25–37. doi: 10.1016/s0378-1119(02)00401-8. [DOI] [PubMed] [Google Scholar]
- 14.Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mis J, Ner SS, Grigliatti TA. Mol Genet Genomics. 2006;275:513–526. doi: 10.1007/s00438-006-0116-x. [DOI] [PubMed] [Google Scholar]
- 17.Eskeland R, Eberharter A, Imhof A. Mol Cell Biol. 2007;27:453–465. doi: 10.1128/MCB.01576-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson AM, Stenberg P, Bernhardsson C, Larsson J. EMBO J. 2007:1–10. doi: 10.1038/sj.emboj.7601604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu LP, Ni JQ, Shi YD, Oakeley EJ, Sun FL. Nat Genet. 2005;37:1361–1366. doi: 10.1038/ng1662. [DOI] [PubMed] [Google Scholar]
- 20.Hung MS, Shen CK. Eukaryotic Cell. 2003;2:841–846. doi: 10.1128/EC.2.5.841-846.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clough E, Moon W, Wang S, Smith K, Hazelrigg T. Development. 2007;134:157–165. doi: 10.1242/dev.02698. [DOI] [PubMed] [Google Scholar]
- 22.Stabell M, Bjorkmo M, Aalen RB, Lambertsson A. Hereditas. 2006;143:177–188. doi: 10.1111/j.2006.0018-0661.01970.x. [DOI] [PubMed] [Google Scholar]
- 23.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., III Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bannister AJ, Schneider R, Kouzarides T. Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 25.Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 26.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 27.Larsson J, Chen JD, Rasheva V, Rasmuson-Lestander A, Pirrotta V. Proc Natl Acad Sci USA. 2001;98:6273–6278. doi: 10.1073/pnas.111581298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 29.Schotta G, Ebert A, Reuter G. Genetica (The Hague) 2003;117:149–158. doi: 10.1023/a:1022923508198. [DOI] [PubMed] [Google Scholar]
- 30.Seum C, Reo E, Peng H, Rauscher FJ, Spierer P, Bontron S. PLoS Genet. 2007;3:e76. doi: 10.1371/journal.pgen.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubin GM, Spradling AC. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 32.Huang DH, Chang YL. Methods Enzymol. 2004;377:267–282. doi: 10.1016/S0076-6879(03)77016-5. [DOI] [PubMed] [Google Scholar]
- 33.Roder K, Hung MS, Lee TL, Lin TY, Xiao H, Isobe KI, Juang JL, Shen CJ. Mol Cell Biol. 2000;20:7401–7409. doi: 10.1128/mcb.20.19.7401-7409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner BM, Birley AJ, Lavender J. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.