Abstract
Most orally bioavailable drugs on the market are competitive inhibitors of catalytic sites, but a significant number of targets remain undrugged, because their molecular functions are believed to be inaccessible to drug-like molecules. This observation specifically applies to the development of small-molecule inhibitors of macromolecular interactions such as protein–membrane interactions that have been essentially neglected thus far. Nonetheless, many proteins containing a membrane-targeting domain play a crucial role in health and disease, and the inhibition of such interactions therefore represents a very promising therapeutic strategy. In this study, we demonstrate the use of combined in silico structure-based virtual ligand screening and surface plasmon resonance experiments to identify compounds that specifically disrupt protein–membrane interactions. Computational analysis of several membrane-binding domains revealed they all contain a druggable pocket within their membrane-binding region. We applied our screening protocol to the second discoidin domain of coagulation factor V and screened >300,000 drug-like compounds in silico against two known crystal structure forms. For each C2 domain structure, the top 500 molecules predicted as likely factor V-membrane inhibitors were evaluated in vitro. Seven drug-like hits were identified, indicating that therapeutic targets that bind transiently to the membrane surface can be investigated cost-effectively, and that inhibitors of protein–membrane interactions can be designed.
Keywords: computational chemistry, discoidin domain, surface plasmon resonance
The availability of thousands of genes potentially involved in disease has stimulated interest in the discovery of new drug targets (1). However, many such targets are underexploited, because their molecular functions are believed to be inaccessible to small drug-like molecules, and because the lead discovery costs are estimated to be too high, anywhere from $500,000 to $1,000,000 for screening 1 million compounds via high-throughput screening (HTS) experiments (2, 3). Along the same line of reasoning, the discovery of drug-like molecules acting outside catalytic sites is still considered an unattainable goal by many research scientists. However, with the advent of high-throughput technologies, we are witnessing a paradigm shift in drug discovery research. Potent inhibitors of protein–protein and protein–DNA interactions can be found, but the costs usually remain outrageous (4). Consequently, relatively few lead discovery campaigns against such targets have been performed, and even fewer studies have used in silico directed approaches, precluding cost-efficient discovery of active drug-like molecules against these macromolecular interactions. Although small nonpeptide inhibitors against macromolecular interactions are emerging, many cellular processes influencing the health and disease states depend on yet another kind of interaction, protein–membrane interactions. This interaction class has been largely neglected for conceptual and technical reasons, even though efficient and cost-effective protocols for the design of small inhibitors would represent a valuable new therapeutic approach for many disease indications. Indeed, with the availability of complete genome sequences for several different organisms and with structural genomics initiatives further supported by progress in homology modeling, an increasing number of potentially important therapeutic proteins that interact with the membrane surface are likely to be identified, indicating further that fast, inexpensive, and accurate protocols to target this molecular mechanism have to be developed.
Despite their wide and successful applications, HTS approaches often remain very costly for hit/lead identification purposes. Therefore, in silico techniques should be applied wherever possible prior and complementary to HTS experiments. For instance, if the 3D structure of a membrane-binding target is known, a rational approach to identify inhibitors is to use structure-based virtual ligand screening (SB-VLS) methods (5–9). However, it is important to note that SB-VLS methods are also expensive, because they usually require costly computer farms and several commercial software licenses (10, 11). In addition to the 3D structure of the target and a fast and accurate computational protocol, there is at least one other prerequisite for successful SB-VLS studies, the knowledge of the ligand-binding site. This is generally not known in detail for proteins interacting with the membrane surface, but binding site prediction methods can be applied to assist the identification of the most promising regions (12).
Next to the use of in silico experiments, appropriate in vitro protocols are required for the identification and validation of membrane-binding inhibitors. Traditionally, membrane-binding property assays are carried out by using different techniques, ranging from microtiter-plate based assays (ELISA-like) to direct binding experiments that make use of, for instance, surface plasmon resonance (SPR). The immobilization of a well defined phospholipid membrane surface and the stability and reproducibility of binding, along with a true quantitative and direct binding measurement character of the assay system, are of major importance for assay outcomes. We therefore suggest that the right functional assays coupled with SPR experiments appear to be an optimal combination for the identification of leads inhibiting protein–membrane interactions. Indeed, SPR is ideally suited for the identification of small molecular inhibitors (molecular mass >350 Da) in direct binding assays. Further, the use of SPR with liposomes captured to an L1-chip represents a general experimental approach to investigate inhibition of membrane binding at physiological temperature (13, 14). The method is extremely robust and reproducible and requires only minute amounts of the test compounds and the target protein. Although the SPR throughput is modest, it perfectly complements SB-VLS, because the number of molecules to be tested after in silico screening computations is usually small. Indeed, in our opinion, the combination of SB-VLS with SPR screening represents a generic approach enabling cost-effective identifications and developments of compounds that affect protein–membrane interactions.
In the present study, we investigated five proteins with known 3D structure that bind transiently to the membrane and performed a theoretical prediction of druggable pockets. We found that all these proteins possess a druggable pocket within the membrane-binding region. For our proof of concept, we selected the second discoidin domain (C2 domain) of coagulation factor V (FV) as a representative domain displaying calcium-independent membrane-binding properties (15). We used our hierarchical SB-VLS protocol (16), followed by functional and SPR-based assays, to identify and characterize membrane-binding inhibitors. We thereby demonstrate that membrane-binding inhibitors can be identified cost-effectively, and that this approach can be implemented and efficiently operated.
Results and Discussion
Membrane-Targeting Domains and Theoretical Prediction of Druggable Pockets.
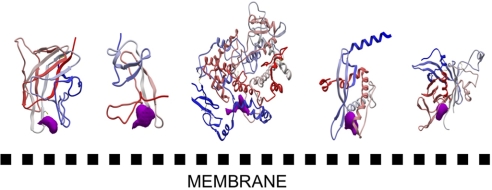
Structural analysis of proteins that bind to the membrane in a reversible manner via a membrane-targeting domain shows that these membrane-binding regions possess specific chemical and topological properties (e.g., a cationic patch surrounding aromatic and aliphatic clusters), whereas their overall fold varies, from all α through α/β to all β structures (17). Proteins or protein modules of therapeutic importance with a known 3D structure that belong to this membrane-binding class include the “tubby protein” (18), the PX domain (19), the plasma β2-glycoprotein I (20), cyclooxygenase (21), and coagulation FV (22). We investigated these five proteins and performed a theoretical prediction of druggable pockets (2, 23, 24). We found that the membrane-binding domains of all of the above-mentioned proteins not only display patches of basic and aliphatic/aromatic residues at the membrane-binding interface but, in the same region, also possess a cavity that could be potentially targeted by drugs (Fig. 1). These cavities are likely to play a major role in membrane binding and could participate in (stereo)-specific interactions with the phospholipid head groups. Interestingly, the sizes of the predicted binding pocket envelopes for our five membrane-binding proteins within the membrane-binding region range from 200 to 600 Å3, values commonly computed for proteins cocrystallized with small drug-like ligands (25). The pertinence of the pocket prediction algorithms used here has been reported (12, 25, 26) and is further validated in the case of FV. The C2 domain has been cocrystallized with the phenyl mercury molecule [a covalent bond is formed; Protein Data Bank (PDB) ID code 1CZS, open crystal form] (22), and the region surrounding this small compound has been proposed to form a key binding groove for specific interaction with acidic phospholipids. This site was readily predicted with both, the ICM PocketFinder utility (24) and with Q-SiteFinder (23).
Fig. 1.
Relative orientation of the predicted druggable pockets with respect to the membrane-binding surface. Five membrane-binding domains are oriented toward the membrane surface on the basis of previously reported biophysical and mutation studies. From left to right, the membrane binding domains of coagulation FV (22), plasma Beta2-glycoprotein I (20), cyclooxygenase (21), PX domain (19), and the “tubby protein” (18) are displayed. A color gradient is applied from the N-terminal region (blue) to the C-terminal region (red). The predicted druggable pockets are shown in magenta. This figure was prepared with ICM. The proteins are scaled differently to facilitate the reading.
Combined SB-VLS-SPR Screen to Identify FV–Membrane Interaction Inhibitors.
The C2 domain of coagulation FV.
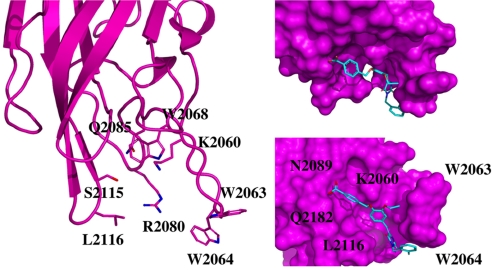
As proof of concept, we selected the C2 domain of coagulation FV as a representative protein domain involved in calcium-independent membrane binding. This domain is closely related, structurally and functionally, to the C2 domain of factor VIII (FVIII, a protein involved in hemophilia A) (27). Although this domain is named C2, this region of FV or FVIII is structurally unrelated to the so-called C2 domain of for instance cytosolic phospholipase A2. FV and FVIII are homologous molecules sharing the domain architecture A1-A2-B-A3-C1-C2. The binding of coagulation FV (via its C2 domain) to activated membranes of circulating platelets is essential for its functions in coagulation. Upon activation by thrombin, activated FV (FVa) acts as a cofactor of activated factor X (FXa) in the prothrombinase complex, which converts prothrombin to thrombin on an appropriate phospholipid surface (15, 28). Excess thrombin production can lead to thrombotic events, suggesting that small inhibitors of FV–membrane interactions could be the starting point for the development of a novel class of antithrombotic drugs. The FV C2 domain comprises a distorted jelly-roll β-barrel motif consisting of eight major antiparallel strands arranged in two β-sheets [supporting information (SI) Fig. 4]. Several loops, presenting hydrophobic/aromatic residues, facilitate immersion into the membrane. Specific interactions with phosphatidylserine (PS) head groups are expected to occur in the groove enclosed by these membrane-binding loops (22). This zone is surrounded by several basic residues, which facilitate the formation of an encounter complex with the negatively charged membrane phosphate groups via electrostatic steering, and that anchor the module into the membrane bilayer (22, 29–31). The FV membrane-binding loops can assume an open or closed conformation (SI Fig. 4) (22), and it is believed (but not proven) that the closed form has low membrane affinity, whereas the open form is suitable for membrane interaction. In the open crystal form, the exposed indoles of Trp 2063 and Trp 2064 contribute to the immersion of the module by interaction with the apolar membrane core (Fig. 2). The nearby-predicted druggable pocket is lined with polar/basic/aliphatic/aromatic side chains (Ser 2183, Gln 2085, Lys 2060, Trp 2068, Met 2120, Ser 2115, Leu 2116, and Arg 2080), ideally arranged for interactions with lipid headgroups.
Fig. 2.
Membrane-binding region of the FV C2 domain. (a) Ribbon plot of the C2 domain open form highlighting the membrane binding loops and displaying the amino acid side chains of the key membrane-binding players (Left). (b) Proposed docked pose for compound 001C07 (Upper Right, same orientation as in a). The small molecule after Surflex and LigandFit consensus docking occupies the phospholipids binding groove thereby impeding membrane interaction. Hydrophobic–aromatic interactions occur with W2063, W2064, and L2116 region. Hydrogen bonds between the side chain of K2060, N2089, and Q2182 and the compound carboxylate group are predicted, whereas the fluorobenzene group shows a favorable interaction with the side chains of either W2064 or R2080. (c) The domain was rotated to have a view from the membrane side (Lower Right). This figure was prepared with PyMol.
SB-VLS and SPR experiments on FV.
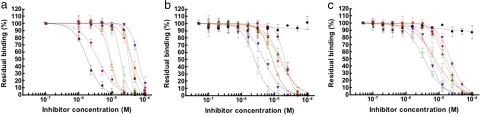
SB-VLS methods help to prioritize large compound collections before experimental testing and are complementary and often superior to massive experimental HTS (32). However, these computational approaches also suffer from limitations (9). We applied our validated multistep SB-VLS protocol (16, 33) to both the open and closed C2 crystal forms and selected the best 509 molecules for each receptor form (i.e., we decided to screen ≈1,000 compounds in total). These 1,018 molecules were tested in groups of four compounds (each at a final concentration of 100 μM) for their ability to inhibit prothrombin activation. Compound mixtures showing >95% inhibition of prothrombin activation were deconvoluted to determine the activity of the individual compounds. Nine molecules were identified that inhibited the prothrombinase assay by >99% at a concentration of 100 μM (Table 1 and SI Table 2). The IC50 of these molecules was determined by varying their concentrations in the functional assay (Fig. 3a). Control experiments showed that none of the nine hit compounds inhibited chromogenic substrate conversion by FXa. However, the two most active small molecules (001B03 and 010G06; Table 1) also inhibited prothrombin activation in the absence of FVa and/or phospholipids, indicating that the inhibitory effect of these compounds was not due to the impediment of the FVa–lipid interaction. Therefore, both molecules were excluded from further evaluation. To confirm that the remaining seven compounds inhibited association between the FV C2 domain and the membrane, we performed SPR experiments by using the isolated FVa light chain (FVa LC; this part of the molecule contains the A3 domain, and two discoidin domains) and a recombinant FV C2 domain. The compounds were tested for their ability to inhibit the binding of FVa LC or FV C2 to an immobilized phospholipid bilayer containing 20:80 PS:phosphatidylcholine (Fig. 3 b and c, respectively, and Table 1). We further verified that compounds failing to inhibit prothrombin activation (Fig. 3 b and c) were unable to impede membrane binding of the FVa LC and the FV C2 domain. The direct interaction of the hit compounds with FVa LC or the FV C2 domain was additionally confirmed by SPR analysis (data not shown). SPR control experiments were performed to show that none of the seven hit compounds significantly bound to the membrane [typically <25 resonance units (RU)], excluding the possibility that penetration and subsequent disruption/distortion of the lipid bilayer by these molecules were responsible for their inhibitory effect. Moreover, none of the seven hit compounds inhibited the calcium-dependent membrane-binding activity of the human vitamin K-dependent proteins, prothrombin and FXa. Furthermore, four of the seven hit compounds also inhibited membrane binding of the structurally related blood coagulation FVIII (Table 1), two of which (molecules 5B10 and 8A07) presented with an IC50 of 7.8 and 8.9 μM, respectively, for the inhibition of FVIII–membrane interaction, as measured by SPR. Although the small molecules described here show reasonable inhibition of FV C2 membrane binding and FVa cofactor activity in the activation of prothrombin by FXa, the compounds in their current form, at 100 μM final concentration, do not inhibit FVa procoagulant activity in a plasma-based assay system (data not shown). A likely explanation for this observation is that the active concentration of a given compound is markedly decreased when bound to plasma proteins (particularly albumin, α1 acid glycoprotein and lipoproteins). To verify this hypothesis, we tested the most potent compound (001C07) at a concentration of 50 μM in the functional assay in the presence of increasing albumin concentrations (0.5–80 mg/ml). As albumin concentration increases, the inhibitory activity of the compound gradually decreases to ≈25% at an albumin concentration of 40 mg/ml, which approaches the plasma albumin concentration (SI Fig. 5). The binding of hit compounds to albumin was further confirmed by SPR analysis (data not shown). Optimization procedures could be carried out to reduce affinity for albumin as shown for some cyclooxygenase 2 inhibitors (34) but are beyond the scope of the present investigation.
Table 1.
IC50 values for the identified hits
| ChemBrige ID and comments | IC50 Ptase, μM | IC50 Fva LC, μM | IC50 Fva C2, μM | Surflex ranking |
|---|---|---|---|---|
| 7364519 (molecule 001C07) FV-membrane specific | 18.6 ± 1.89 | 2.5 ± 0.18 | 3.51 ± 0.57 | Open form, position 502 |
| 6305867 (molecule 007H10) Acts on FV and FVIII membrane binding | 62.9 ± 2.56 | 4.80 ± 0.89 | 5.53 ± 1.04 | Open form, position 73 |
| 6043266 (molecule 006H08) FV-membrane specific | 9.19 ± 0.94 | 8.56 ± 1.14 | 6.71 ± 1.66 | Open form, position 305 |
| 5843746 (molecule 001D08) Acts on FV and FVIII membrane binding | 8.95 ± 1.02 | 8.55 ± 0.95 | 7.40 ± 1.05 | Open form, position 187 |
| 7688319 (molecule 005B10) Acts on FV and FVIII membrane binding | 32.4 ± 5.1 | 15.71 ± 2.26 | 14 ± 1.88 | Open form, position 476 |
| 7971347 (molecule 007D08) FV-membrane specific | 40.57 ± 2.42 | 14.07 ± 2.54 | 16.4 ± 1.38 | Open form, position 86 |
| 6446853 (molecule 008A07) Acts on FV and FVIII membrane binding | 38.05 ± 3.07 | 14.07 ± 2.54 | 21.92 ± 2.43 | Closed form, position 13 |
| 5169083 (molecule 001B03) Inhibition not due to membrane interference | 3.8 ± 0.61 | ND | ND | Open form, position 84 |
| 5870804 (molecule 010G06) Inhibition not due to membrane interference | 1.81 ± 0.17 | ND | ND | Closed form, position 115 |
The IC50 values represent the mean of two independent experiments with their corresponding 95% confidence intervals.
Fig. 3.
Titration curves for FV membrane-binding inhibitors identified by our multistep VLS procedure and in vitro screening. IC50 values of hit compounds were determined by using a functional assay (a) or SPR analysis by using FVa LC (b) or FV C2 domain (c), as described in Materials and Methods. Black triangle, 010G06; pink diamond, 001B03; green open diamond, 001C07; blue inverted triangle, 007H10; orange open circle, 006H08; open square, 001D08; green triangle, 005B10; orange diamond, 007D08; pink square, 008A07; black circle, 007A09 (Neg. control).
In silico structural analysis of the docked poses revealed that the active molecules were all partially buried in the PS-binding groove. Critical interactions with residues located in this cavity were identified. A consensus pose obtained by Surflex and LigandFit is shown in Fig. 2. In general, the compounds display favorable contacts with the side chains of FV C2 residues Gln 2085, Lys 2060, Trp 2068, Ser 2115, Leu 2116, Trp 2063, Trp 2064, Arg 2080, or Lys 2061. The compounds found by using the closed crystal form could also be docked in the open form, although with a different orientation. Further investigation of the precise interaction would require site-directed mutagenesis and/or NMR or x-ray crystallography experiments.
SB-VLS-SPR Generic Platform to Design Protein–Membrane Interaction Inhibitors.
To the best of our knowledge, lead discovery campaigns against membrane-targeting domains that include virtual ligand screening have not been carried out, and only one study has reported inhibition of FVIII–membrane interaction (35). In that report, a traditional HTS lead discovery approach was applied, leading to the initial identification of 10 best hits disrupting FVIII C2 domain–membrane interactions after screening 10,000 molecules from the ChemBridge collection. These 10 hits were grouped into three categories: weak inhibitors (five compounds with IC50 > 20 μM), moderate inhibitors (two compounds with IC50 between 10 and 20 μM), and strong inhibitors (three compounds with IC50 < 10 μM). The hit rate was 0.10, whereas we achieved a hit rate of 1.2 when using the FV C2 domain open crystal form (six inhibitors found after screening 500 molecules, four compounds with IC50 < 10 μM, and two compounds with IC50 < 20 μM). We obtained a hit rate of 0.7 when testing the 1,018 molecules (i.e., on the open and closed crystal structures). This value is still much better than when using HTS alone as in the case of FVIII.
After completion of our study, control in silico experiments were performed on the FVIII C2 domain to further validate our approach. We retrieved the x-ray structure of the FVIII C2 domain and applied the same VLS protocol as was used for FV, including the FVIII inhibitors of ref. 35 in our 300,000 ChemBridge absorption, distribution, metabolism, and excretion/tox filtered compound collection. Because the FVIII loops important for membrane binding are in an open conformation (27), we performed rigid body docking and flexible docking on this x-ray structure. We identified five compounds out of the 10 found by HTS in the top 1,000 Surflex list. In addition, we also found three molecules identified in the present study that cross-react with FVIII (see Table 1). Thus, we obtained an overall hit rate for FVIII of at least 0.8. These results indicate that for FVIII, our strategy is more efficient than the HTS-only approach. In fact, typical hit rates from SB-VLS are usually in the range of 1–5% (and more), depending on the knowledge about the binding pocket, its overall shape, and its physicochemical nature, compared with common hit rates from HTS of ≤0.1% (36). Higher hit rates than the ones calculated here using SB-VLS approaches can be obtained when some active molecules are already known (and when the binding pocket is well defined), but in such situations, the use of SB-VLS approaches in combination with ligand-based methods is highly recommended (37). In the present study, we initially ignored the data on FVIII, to evaluate the strengths and weaknesses of SB-VLS approaches. The advantages of using hierarchical SB-VLS protocols with rigid body docking before flexible ligand docking is that the computations run significantly faster (thus can be carried out on only one workstation in ≈2 weeks) than full flexible ligand docking of the entire compound collection (≈8 weeks on the same single workstation) while performing better or equally well (10, 16, 33). The hierarchical approach used here is also very interesting when several 3D structures of the receptor are available, because again, the computations can be performed on a few workstations in parallel.
Along with appropriate in silico protocols, the use of suitable experimental procedures is crucial to identify protein–protein or membrane-binding inhibitors. For example, when studying protein–protein interactions, ligand immobilization and regeneration are frequently a central problem. Whenever possible, capture assays are preferred to exclude ligand heterogeneity; however, proper capture reagents are not generally available. Protein–membrane interactions differ in this respect, because both native and artificial liposomes (13, 14) can be efficiently captured onto the surface of L1 sensorchips. The analyte (i.e., the membrane-binding protein) is used in its unmodified native form. Chemical modification and immobilization can obstruct the binding site, restrict conformational flexibility, and thus impair membrane binding. However, SPR enables real-time kinetics resolution even for fast and/or weak interactions. Highly reproducible binding data are generated because of precise control of experimental parameters. Captured liposomes are not affected by 5% DMSO, a property that is extremely useful in the early phase of drug development where the binding affinities of the compounds are low, and solubility is often a problem. Inhibition, but also stabilization of membrane binding due to the interaction of the analyte with a test compound, can be analyzed quantitatively. The assay is thus truly generic, because no specific detection reagents or biological assays are required, the only requirement being the membrane binding of the analyte. Consumption of the test compounds is minimal, and consumption of analyte is very favorable. For example, 1 mg of the C2 domain or of the LC segment is sufficient to screen ≈10,000 test compounds. Importantly, the lipid-binding properties of the test compound can be determined at the same time. Moreover, the binding of a drug to human serum albumin can be analyzed by SPR-based experiments (13, 38). These assays can thus also be used to generate early absorption, distribution, metabolism, and excretion data and to confirm the specificity and mode of action of the test compounds. A major disadvantage of the SPR assays is its modest throughput of ≈100 samples per day. However, this limitation can be elegantly overcome by combination with virtual screening approaches.
Conclusion
Experimental screening of difficult targets is time-consuming and cost-intensive. Therapeutic targets that bind transiently to the membrane have been neglected and remain among the most difficult challenges in contemporary drug discovery. The nature of protein–membrane interaction is not fully understood, success stories are extremely rare, and generic approaches are missing. Protein–membrane interactions are, however, crucial in many biological processes, because they localize key molecular factors on specific cell surfaces. In the present study, we combined SB-VLS experiments with in vitro assays and found seven active molecules that were able to disrupt FV membrane-binding activity in a timely and cost-effective fashion. These molecules are promising leads for the development of in vitro tools for hemostasis research and for the design of a novel class of anticoagulant drugs. It is remarkable that small molecules can impede membrane binding, because the binding interface between the membrane-binding domain and the phospholipids is relatively large, with immersion of the proteins several angstroms deep into the membrane bilayers. However, just as in the case of protein–protein interaction (39), the bulk of the binding energy appears to derive from contacts with just a small number of amino acid residues. We conclude that hierarchical virtual screening approaches in combination with SPR technology are an efficient and generally applicable approach for routinely identifying membrane-binding inhibitors. This strategy could therefore be applied to other relevant membrane-binding proteins in search of the next generation of therapeutics.
Materials and Methods
Computational Procedure.
Numerous membrane-binding domains from the RCSB Protein Data Bank (40) were investigated by using ICM (Molsoft, San Diego, CA). Five representative structures were selected to illustrate our study: the “tubby protein” (18) (PDB ID code 1I7E, resolution 1.95 Å), a PX domain (19) (PDB ID code 1H6H, resolution 1.70 Å), the plasma β2-glycoprotein I (20) (PDB ID code 1C1Z, resolution 2.87 Å), cyclooxygenase (21) (PDB ID code 1DIY, resolution 3.00 Å), and the C2 domain of coagulation FV (22) (PDB ID code 1CZT, resolution 1.87 Å). Binding pocket predictions were carried out with the ICM PocketFinder utility (24) and with Q-SiteFinder (23). For each structure, we analyzed pockets with a high druggability index (i.e., those with a pocket of appropriate size and chemical nature to bind a drug-like molecule). The best-ranked binding cavities were found at the expected protein–membrane interface.
We used an efficient multistep procedure that is both time- and cost-effective (16, 33) and studied the 500,000-molecule ChemBridge compound collection. Absorption, distribution, metabolism, and excretion/tox filtering was performed with Filter (OpenEye Scientific Software, Santa Fe, NM) and FAF-Drugs (41). The remaining 304,000 molecules were docked by using the rigid-body docking package FRED (OpenEye Scientific Software) into the closed (PDE ID code 1CZV) and open crystal (PDE ID code 1CZT or 1CZS) forms of the FV C2 domain. Up to 50 conformers were generated for each ligand with OMEGA (OpenEye Scientific Software). The top 60,000 unique molecules (Gaussian docking function) after FRED docking in the open and closed forms were selected for flexible docking with Surflex version 1.33 and the scoring function of version 1.31 (42). The top 2,000 docked poses were analyzed by using PyMOL (DeLano, San Carlos, CA), ICM (Molsoft) and Cerius2 (Accelrys, San Diego, CA). The hit compounds inhibiting FV-membrane binding were also redocked and investigated with LigandFit (43) to identify consensus poses. Protonation state definitions for the FV C2 domains, open and closed forms, were computed by using the PCE server (44) and the addition of hydrogen atoms was performed accordingly with InsightII (Accelrys). The binding pocket was defined theoretically by using Surflex, Q-SiteFinder and ICM PocketFinder. All water molecules were removed from the structures before docking. The hit rate was computed as the number of actives found divided by the number of molecules screened × 100.
Proteins and Reagents.
Human FVa and the FVa LC) were purified from human plasma as described (45). The recombinant FV C2 domain was obtained as follows. Phospholipid vesicles containing dioleoyl PS and dioleoyl phosphatidylcholine were prepared as described (45). Small molecules were purchased from ChemBridge (San Diego, CA). All other coagulation proteins were purchased through Kordia Lab Supplies (Leiden, The Netherlands).
Cloning, Expression, and Purification of the Human FV C2 Domain.
The pMT2-V expression vector (American Type Culture Collection no. 40515) containing the full-length human FV cDNA was used as a template for the construction of a PCR fragment containing the coding region of the hFV C2 domain (hFVC2), using the following forward and reverse primers: (FW: 5′-CTGGTCCCCCGGGGATGTTCCACACCCCTGGGTAT-3′; RV: 5′-TAGGATTGCCGTCAAGTTTGGCGCG-3′). After digestion with SmaI and SalI, the hFVC2 fragment was purified (Gel Extraction Kit, Qiagen, Valencia, CA) and subcloned into the pet43.1.a expression vector (Novagen, Madison, WI). The resulting pet43.1a-C2 plasmid was introduced into Escherichia coli DH5α cells, and positive clones were identified by DNA sequencing. The recombinant plasmid was then introduced into E. coli [Rosettagami(DE3)pLysS; Novagen] for expression of a NusA-hFV C2 fusion protein.
Bacteria were grown at 37°C in LB medium with 100 μg/ml carbenicillin/30 μg/ml kanamycin/34 μg/ml chloramphenicol/12.5 μg/ml tetracycline. At OD600∼0.6 cultures were induced with 1 mM isopropyl β-D-thiogalactoside and incubated overnight at room temperature (RT). Cells were pelleted, resuspended in cell lysis buffer [50 mM Tris/10 mM NaCl/0.1% (vol/vol) TritonX-100/2.5% (vol/vol) glycerol/1 μM PMSF/10 mM benzamidine/1 mg/ml lysozyme/0.1 units/ml benzonase (Novagen)/1 mM MgCl2, pH 8.5], incubated for 1 h at RT, and then sonicated twice (MSE Scientific Instruments, Beun de Ronde, The Netherlands) at 16 μm amplitude for 2 min. The cell debris were as removed by centrifugation and the supernatant containing the recombinant NusA-C2 fusion protein was applied to an anion exchange column (Q-Sepharose-FF, GE Healthcare, Uppsala, Sweden) equilibrated with 50 mM Tris/10 mM NaCl/2.5% glycerol, pH 8.5. After extensive washing with the same buffer, bound proteins were eluted with a linear gradient from 0.010 to 1 M NaCl. Peak fractions were analyzed for C2 antigen by ELISA and SDS/PAGE. Fractions from a major peak that eluted at 0.2 mM NaCl were pooled and applied to a Ni-NTA affinity chromatography column (HisTrap FF, GE Healthcare) equilibrated in 50 mM Tris/10 mM imidazole/300 mM NaCl/2.5% glycerol, pH 8.0. After extensive washing with the same buffer, bound proteins were eluted with a linear gradient from 0.01 to 1 M imidazole. Pooled fractions containing the NusA-C2 fusion protein were concentrated (Macrosep 3K centrifugal device, Pall Corporation, New York, NY) and applied to a PD10 column (GE Healthcare) to exchange the buffer to 50 mM Tris/100 mM NaCl, pH 8.0. The fusion protein was treated overnight at RT with 250 nM thrombin to cleave off the fusion tag. After 4-fold dilution in 50 mM Tris/10 mM NaCl/2.5% glycerol, pH 8.0, the protein suspension was loaded onto a Mono S column (GE Healthcare) in the same buffer. After washing, the bound native (untagged) hFV C2 domain was eluted with a linear gradient from 0.01 to 1 M NaCl. Fractions containing hFV C2 were concentrated and applied to a Superdex 200 column (GE Healthcare) equilibrated in 50 mM Hepes/100 mM NaCl/1 mg/ml PEG, pH 7.4. Peak fractions containing hFV C2 were identified by ELISA and SDS/PAGE. Peptide mass fingerprinting using MALDI-TOF MS was performed to further verify the identity of the purified protein. The concentration of purified hFV C2 was determined by measuring the absorbance at 280 nm.
In Vitro Screening of the Top-Scoring Molecules.
The 509 top-scoring molecules for the open and closed conformation of the FV C2 domain structure were screened in a functional assay, essentially as described (45). In brief, compounds were diluted from 10 mM stocks in DMSO to mixes of four compounds each at 2.5 mM per compound in DMSO. Next, an 100 μM concentration of each compound mixture was incubated for 5 min at 37°C with 20 pM FVa1/0.5 μM prothrombin/2 mM CaCl2 in 25 mM Hepes/150 mM NaCl/0.5 mg/ml ovalbumin, pH 7.5. Three minutes after the addition of phospholipid vesicles (5 μM 10:90 dioleoyl PS/dioleoyl phosphatidylcholine, mol/mol), prothrombin activation was started by the addition of FXa to a final concentration of 0.5 nM. After 2 and 4 min, aliquots were drawn from the reaction mixture, and rates of prothrombin activation were determined by using the chromogenic substrate S2238. Compound mixtures showing >95% inhibition were deconvoluted to identify the inhibitory individual compound(s) from the initial mixture. The screen resulted in nine compounds that inhibit prothrombin activation in the assay >99% at a concentration of 100 μM. The IC50 values of these hit compounds were determined by titration of variable concentrations of compounds (0.1–100 μM) in the functional assay described above. None of the inhibitors was found to inhibit prothrombin activation in the absence of phospholipids and/or FVa and also did not inhibit chromogenic substrate conversion by FXa.
SPR Measurements.
Experiments were performed on a Biacore T100 instrument (GE Healthcare) using a L1 sensor chip and PBS with 5% DMSO as running buffer. Phospholipid vesicles (500 μM 20:80 PS:phosphatidylcholine in 50 mM Hepes/150 mM NaCl, pH 7.6) were injected at a flow rate of 10 μl/min for 3 min resulting in capture levels of ≈5,000 RU. Captured vesicles were conditioned with a 30-s pulse of 50 mM NaOH. The resulting baseline was stable at 5,000 RU and allowed for >50 measurements. Vesicles were stripped off the chip with isopropanol:50 mM NaOH (40:60 vol/vol). A flowcell not covered with liposomes was used as reference cell. All experiments were performed at 37°C.
Injection of 200 nM C2 or 50 nM LC for 2 min resulted in binding signals of 400 and 1,000 RU, respectively. Both binding curves exhibited saturation toward the end of the injection phase. Inhibition of membrane binding was analyzed by using 2-fold serial dilutions of compounds in a range from 0.1 to 100 μM. Samples were prepared fresh in LoBind protein tubes (Eppendorf, Hamburg, Germany) and measured within 1 h. Blank subtracted binding levels were normalized, plotted against their corresponding compound concentrations, and fitted to the following equation by using GraphPad (San Diego, CA) Prism:
with y representing the relative binding signal, c the concentration of the compound, and c50 the concentration at which half-maximal inhibition is reached, and h the Hill coefficient.
Supplementary Material
Acknowledgments
This work was supported by Institut National de la Santé et de la Recherche Médicale (INSERM), the INSERM Avenir award (B.O.V.), the Dutch Organization for Scientific Research, Netherlands Organization for Scientific Research (NWO) VIDI Grant 916-046-330 (to G.A.F.N.), and INSERM-NWO Travel Grant 910-48-617 (to B.O.V. and G.A.F.N.).
Abbreviations
- HTS
high-throughput screening
- SB-VLS
structure-based virtual ligand screening
- SPR
surface plasmon resonance
- FV
Factor V
- FVa
activated FV
- FXa
activated Factor X
- PS
phosphatidylserine
- LC
light chain
- PDB
Protein Data Bank
- RU
resonance units.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701051104/DC1.
References
- 1.Hopkins AL, Groom CR. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 2.Hajduk PJ, Huth JR, Tse C. Drug Discov Today. 2005;10:1675–1682. doi: 10.1016/S1359-6446(05)03624-X. [DOI] [PubMed] [Google Scholar]
- 3.Davies JW, Glick M, Jenkins JL. Curr Opin Chem Biol. 2006;10:343–351. doi: 10.1016/j.cbpa.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 4.Whitty A, Kumaravel G. Nat Chem Biol. 2006;2:112–118. doi: 10.1038/nchembio0306-112. [DOI] [PubMed] [Google Scholar]
- 5.Jain AN. Curr Opin Drug Discov Dev. 2004;7:396–403. [PubMed] [Google Scholar]
- 6.Shoichet BK. Nature. 2004;432:862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abagyan R, Totrov M. Curr Opin Chem Biol. 2001;5:375–382. doi: 10.1016/s1367-5931(00)00217-9. [DOI] [PubMed] [Google Scholar]
- 8.Jalaie M, Shanmugasundaram V. Mini Rev Med Chem. 2006;6:1159–1167. doi: 10.2174/138955706778560157. [DOI] [PubMed] [Google Scholar]
- 9.Leach AR, Shoichet BK, Peishoff CE. J Med Chem. 2006;49:5851–5855. doi: 10.1021/jm060999m. [DOI] [PubMed] [Google Scholar]
- 10.Maiorov V, Sheridan RP. J Chem Inf Model. 2005;45:1017–1023. doi: 10.1021/ci050089y. [DOI] [PubMed] [Google Scholar]
- 11.Sperandio O, Miteva MA, Delfaud F, Villoutreix BO. Curr Protein Pept Sci. 2006;7:369–393. doi: 10.2174/138920306778559377. [DOI] [PubMed] [Google Scholar]
- 12.Laurie AT, Jackson RM. Curr Protein Pept Sci. 2006;7:395–406. doi: 10.2174/138920306778559386. [DOI] [PubMed] [Google Scholar]
- 13.Frostell-Karlsson A, Widegren H, Green CE, Hamalainen MD, Westerlund L, Karlsson R, Fenner K, van de Waterbeemd H. J Pharmacol Sci. 2005;94:25–37. doi: 10.1002/jps.20215. [DOI] [PubMed] [Google Scholar]
- 14.Abdiche YN, Myszka DG. Anal Biochem. 2004;328:233–243. doi: 10.1016/j.ab.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Dahlback B, Villoutreix BO. Arterioscler Thromb Vasc Biol. 2005;25:1311–1320. doi: 10.1161/01.ATV.0000168421.13467.82. [DOI] [PubMed] [Google Scholar]
- 16.Miteva MA, Lee WH, Montes MO, Villoutreix BO. J Med Chem. 2005;48:6012–6022. doi: 10.1021/jm050262h. [DOI] [PubMed] [Google Scholar]
- 17.Bhardwaj N, Stahelin RV, Langlois RE, Cho W, Lu H. J Mol Biol. 2006;359:486–495. doi: 10.1016/j.jmb.2006.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 19.Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJ, Lavenir I, Perisic O, Hawkins PT, et al. Mol Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- 20.Schwarzenbacher R, Zeth K, Diederichs K, Gries A, Kostner GM, Laggner P, Prassl R. EMBO J. 1999;18:6228–6239. doi: 10.1093/emboj/18.22.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malkowski MG, Ginell SL, Smith WL, Garavito RM. Science. 2000;289:1933–1937. doi: 10.1126/science.289.5486.1933. [DOI] [PubMed] [Google Scholar]
- 22.Macedo-Ribeiro S, Bode W, Huber R, Quinn-Allen MA, Kim SW, Ortel TL, Bourenkov GP, Bartunik HD, Stubbs MT, Kane WH, Fuentes-Prior P. Nature. 1999;402:434–439. doi: 10.1038/46594. [DOI] [PubMed] [Google Scholar]
- 23.Laurie AT, Jackson RM. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 24.An J, Totrov M, Abagyan R. Mol Cell Proteomics. 2005;4:752–761. doi: 10.1074/mcp.M400159-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.An J, Totrov M, Abagyan R. Genome Inform Ser Workshop Genome Inform. 2004;15:31–41. [PubMed] [Google Scholar]
- 26.Sotriffer C, Klebe G. Farmaco. 2002;57:243–251. doi: 10.1016/s0014-827x(02)01211-9. [DOI] [PubMed] [Google Scholar]
- 27.Pratt KP, Shen BW, Takeshima K, Davie EW, Fujikawa K, Stoddard BL. Nature. 1999;402:439–442. doi: 10.1038/46601. [DOI] [PubMed] [Google Scholar]
- 28.Esmon CT. Crit Care Med. 2001;29:S48–S51. doi: 10.1097/00003246-200107001-00018. discussion 51–52. [DOI] [PubMed] [Google Scholar]
- 29.Nicolaes GA, Villoutreix BO, Dahlback B. Blood Coagul Fibrinolysis. 2000;11:89–100. [PubMed] [Google Scholar]
- 30.Srivastava A, Quinn-Allen MA, Kim SW, Kane WH, Lentz BR. Biochemistry. 2001;40:8246–8255. doi: 10.1021/bi010449k. [DOI] [PubMed] [Google Scholar]
- 31.Kim SW, Quinn-Allen MA, Camp JT, Macedo-Ribeiro S, Fuentes-Prior P, Bode W, Kane WH. Biochemistry. 2000;39:1951–1958. doi: 10.1021/bi992256r. [DOI] [PubMed] [Google Scholar]
- 32.Mestres J. Biochem Soc Trans. 2002;30:797–799. doi: 10.1042/bst0300797. [DOI] [PubMed] [Google Scholar]
- 33.Cozza G, Bonvini P, Zorzi E, Poletto G, Pagano MA, Sarno S, Donella-Deana A, Zagotto G, Rosolen A, Pinna LA, et al. J Med Chem. 2006;49:2363–2366. doi: 10.1021/jm060112m. [DOI] [PubMed] [Google Scholar]
- 34.Mao H, Hajduk PJ, Craig R, Bell R, Borre T, Fesik SW. J Am Chem Soc. 2001;123:10429–10435. doi: 10.1021/ja015955b. [DOI] [PubMed] [Google Scholar]
- 35.Spiegel PC, Kaiser SM, Simon JA, Stoddard BL. Chem Biol. 2004;11:1413–1422. doi: 10.1016/j.chembiol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Harris NV, Clark DE. Glob Outsour Rev. 2004;6:27–31. [Google Scholar]
- 37.Hawkins PC, Skillman AG, Nicholls A. J Med Chem. 2007;50:74–82. doi: 10.1021/jm0603365. [DOI] [PubMed] [Google Scholar]
- 38.Rich RL, Day YS, Morton TA, Myszka DG. Anal Biochem. 2001;296:197–207. doi: 10.1006/abio.2001.5314. [DOI] [PubMed] [Google Scholar]
- 39.Clackson T, Wells JA. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 40.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miteva MA, Violas S, Montes M, Gomez D, Tuffery P, Villoutreix BO. Nucleic Acids Res. 2006;34:W738–W44. doi: 10.1093/nar/gkl065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain AN. J Med Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 43.Venkatachalam CM, Jiang X, Oldfield T, Waldman M. J Mol Graphics Model. 2003;21:289–307. doi: 10.1016/s1093-3263(02)00164-x. [DOI] [PubMed] [Google Scholar]
- 44.Miteva MA, Tuffery P, Villoutreix BO. Nucleic Acids Res. 2005;33:W372–W375. doi: 10.1093/nar/gki365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosing J, Bakker HM, Thomassen MC, Hemker HC, Tans G. J Biol Chem. 1993;268:21130–21136. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.