Abstract
We report studies of the subcellular localization of the ClyA cytotoxic protein and of mutations causing defective translocation to the periplasm in Escherichia coli. The ability of ClyA to translocate to the periplasm was abolished in deletion mutants lacking the last 23 or 11 amino acid residues of the C-terminal region. A naturally occurring ClyA variant lacking four residues (183 to 186) in a hydrophobic subdomain was retained mainly in the cytosolic fraction. These mutant proteins displayed an inhibiting effect on the expression of the hemolytic phenotype of wild-type ClyA. Studies in vitro with purified mutant ClyA proteins revealed that they were defective in formation of pore assemblies and that their activity in hemolysis assays and in single-channel conductance tests was at least 10-fold lower than that of the wild-type ClyA. Tests with combinations of the purified proteins indicated that mutant and wild-type ClyA interacted and that formation of heteromeric assemblies affected the pore-forming activity of the wild-type protein. The observed protein-protein interactions were consistent with, and provided a molecular explanation for, the dominant negative feature of the mutant ClyA variants.
Cytolytic toxins are synthesized by a variety of gram-negative bacteria associated with diseases in humans and animals (1, 4, 27). Many of these toxins have been shown to represent important virulence factors, but their exact functions in the pathogenesis of infections are poorly understood. Most known cytolysins from gram-negative bacteria form pores in the cytoplasmic membranes of eukaryotic cells, but there are several different mechanisms of pore formation (5, 25). In Escherichia coli isolates associated with diseases, two pore-forming cytolysins have been identified so far, α-hemolysin and enterohemorrhagic E. coli (EHEC) hemolysin. α-Hemolysin (HlyA) is frequently produced by E. coli strains causing extraintestinal infections such as urinary tract infections, while EHEC hemolysin (Ehx) is found in EHEC strains of serogroup O157 (4, 32). The commonly used nonpathogenic E. coli laboratory strain K-12 contains neither the gene cluster required for the production and secretion of α-hemolysin nor the related EHEC hemolysin determinant and is nonhemolytic under normal laboratory conditions. However, recently it became evident that even derivatives of E. coli K-12 can express a hemolytic or cytolytic phenotype. A protein designated ClyA (for cytolysin A) has been shown to cause lysis of red blood cells from different species, e.g., human, horse, sheep, goat, and hen (23). Expression of ClyA also results in cytotoxicity towards cultured human and murine macrophages, human peripheral monocytes, and HeLa cells (17, 29). Activation of the clyA (hlyE, sheA) gene results in expression of the ClyA protein (10, 18, 20, 21, 29). For example, transcription of the clyA gene is derepressed in hns mutant E. coli (29, 33) and in bacteria carrying multicopy clones of the hlyX, slyA, or mprA gene (13, 18, 20). This results in a hemolytic phenotype on blood agar plates. In most wild-type E. coli strains there is a low level of ClyA protein produced, but it does not give a detectable phenotype (22) The mechanism by which E. coli K-12 may release cytolysin A has not been clarified, and it is not known whether there would be some similarities with HlyA and/or Ehx.
The ClyA cytolysin from E. coli is the prototype of what a novel type of pore-forming bacterial cytotoxins found in different enterobacteria (10, 11, 23, 24, 31). Recently, we showed that ClyA proteins from the human-pathogenic Salmonella enterica serovars Typhi and Paratyphi A have pore-forming activity and may be expressed at phenotypically detectable levels (23). The structural features of ClyA appear to be different from those of previously characterized cytolysins, and according to crystallographic studies, it represents a novel protein fold among cytotoxins (31). Earlier studies with overproducing strains suggested that ClyA accumulates in the periplasmic space (18, 21). However, the ClyA protein is made without cleavage of any N-terminal signal sequence, and it has remained unknown how it may reach the periplasm and be exposed on the bacterial cell surface. The C-terminal region was earlier suggested to be of importance for membrane targeting, pore formation, and translocation of ClyA, since mutant bacteria expressed a nonhemolytic phenotype (21). In this work, we have studied the subcellular localization of the wild type and some C-terminally mutant variants of ClyA, and we examined how the mutant proteins cause inhibition of expression of the hemolytic phenotype of wild-type ClyA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The relevant genotypes or characteristics of the bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown aerobically, with shaking, at 37°C in Luria-Bertani broth or on agar. Blood agar plates contained 5% (vol/vol) horse erythrocytes solidified with 1% (wt/vol) Columbia agar base (Oxoid). Antibiotics were added to Luria-Bertani broth or agar at 50 μg ml−1 (carbenicillin) and 30 μg ml−1 (kanamycin).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant genotype/character | Reference or source |
|---|---|---|
| Strains | ||
| BEU616 | MC1061 hns::cat | 33 |
| DH5α | endA1 hsdR17(rK mK+)supE44 thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 [φ80(ΔlacZ)M15] | 14 |
| MC1061 | araD139 Δ(ara-leu)7697 ΔlacX74 galU galK hsr hsm+strA | 8 |
| MC4100 | araD139 Δ(lacZYA-argF)U169 relA1 rpsL thi-1 | 7 |
| MWK11 | MC4100 ClyA+ (consensus CRP binding site) | 33 |
| YMZ19 | MC1061 clyA::kan | 20 |
| Plasmids | ||
| pUC18 | Cloning vector | 34 |
| pYMZ80 | Wild-type ClyA | 21 |
| pJON70 | A183D G184D ClyA mutant | 21 |
| pMWK16 | Δ(183-186) ClyA mutant | This laboratory (unpublished) |
| pJON63 | H292D Δ(293-303) ClyA mutant | 21 |
| pSNW168 | Δ(281-303) ClyA mutant | This work |
| pJON66 | A278P K279Q Δ(281-303) ClyA mutant | 21 |
Site-directed mutagenesis.
Mutants with mutations in the cloned clyA gene expressing alterations in the ClyA amino acid sequence were obtained by using the Quick Change site-directed mutagenesis kit (Stratagene) according to the instructions of the manufacturer. The plasmid pYMZ80 was used as the template with suitable oligonucleotide primers as described earlier (21). Mutations were verified by DNA sequencing with the ABI PRIM Dye Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA polymerase and an ABI PRIM 377 DNA sequencer.
Cell fractionation.
The bacteria were harvested by centrifugation. Periplasmic proteins were isolated by osmotic shock as follows. The bacteria from a defined culture volume (suspensions of 1011 cells/ml; usually 1 ml but up to 50 ml in cases of preparations to be used for purification of ClyA) were washed with 10 mM Tris-HCl (pH 8.0) and resuspended in 0.25 volume (compared to the starting volume) of a solution containing 20% sucrose, 20 mM Tris-HCl (pH 8.0), and 1 mM Na-EDTA. The mixture was incubated for 10 min at room temperature. Subsequently, the bacteria were pelleted and resuspended in 1 volume of ice-cold water. After incubation on ice for 10 min, the cells were removed by centrifugation at 12,000 × g. The supernatant was used as the periplasmic protein extract. The cell pellet was then disrupted by sonication in 1 volume of 10 mM Tris-HCl (pH 8.0) buffer. The cell debris and unbroken cells were removed by centrifugation at 5,000 × g for 10 min at 4°C, and the supernatant was fractionated into the membrane and cytoplasmic fractions by centrifugation at 10,000 × g for 30 min at 4°C. The supernatant was used as a cytoplasmic fraction. The sediment was resuspended with sterilized distilled water and was used as the membrane fraction. In order to separate the inner and outer membranes, the fraction was further treated with N-lauryl sarcosyl at a final concentration of 2% at room temperature and then centrifuged at 15,000 × g for 30 min. The resulting sediment was used as the outer membrane fraction, and the supernatant was used as the inner membrane fraction after dialysis and precipitation. Extracellular, periplasmic, cytoplasmic, and membrane-bound proteins were concentrated by precipitation with ice-cold trichloroacetic acid (final concentration, 10%), The precipitated proteins were collected by centrifugation at 12,000 × g, washed with acetone, dried under vacuum, and dissolved in sample buffer (50 mM Tris-HCl [pH 6.8], 10% glycerol, 5% β-mercaptoethanol, 2% sodium dodecyl sulfate [SDS], 0.05% bromophenol blue). Samples were neutralized by addition of saturated Tris solution and boiled for 5 min at 100°C. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) analyses of proteins were performed as described previously (16). The samples were applied to SDS-PAGE minigels and run at constant voltage (120 V for 17% gels and 100 V for 12.5% gels). Gels were stained with Coomassie blue and/or silver stain or analyzed by Western immunoblot analysis. Silver staining was carried out by using the SilverXpress silver staining kit according to the protocol of the manufacturer (Invitrogen).
Purification of ClyA proteins.
ClyA proteins from the periplasmic, cytoplasmic, and membrane fractions were further purified by fast protein liquid chromatography with a Mono Q ion-exchange column (Pharmacia, Uppsala, Sweden). The system was equilibrated with 20 mM Tris-HCl buffer (pH 8.0), and the protein was then eluted with a linear gradient of 0 to 1 M NaCl at a flow rate of 0.5 ml/min (2-ml fractions). The elution of the protein was monitored by measuring the extinction of the fractions at 280 nm. The proteins corresponding to eluted peaks were checked by SDS-PAGE and immunoblot analysis. The fractions containing ClyA protein were dialyzed against 20 mM Tris-HCl (pH 8.0) and stored at −20°C. Quantification of protein concentrations was carried out with the bicinchoninic acid protein assay kit according to the protocol of the manufacturer (Pierce).
Western blot analysis.
Cell extracts of E. coli strains were separated by SDS-PAGE and analyzed by immunoblotting with a polyclonal anti-ClyA antiserum (21), monoclonal anti-ClyA antibodies (S. N. Wai and B. E. Uhlin, unpublished data), polyclonal anti-DsbA antiserum (2), or polyclonal anti-H-NS antiserum (15). The separated proteins were transferred to a nitrocellulose filter by methods described earlier (28). Subsequently, the filters were probed with anti-ClyA antibodies. The monoclonal anti-ClyA antibodies were used at a final dilution of 1:5,000 with anti-mouse horseradish peroxidase-conjugated secondary antibody at a final dilution of 1:20,000. Polyclonal anti-ClyA antisera were used at final dilutions of 1:1,000 with biotinylated anti-rabbit secondary antibody and streptavidin-horseradish peroxidase conjugate at final dilutions of 1:1,000 and 1:1,500, respectively. Immunoreactive bands were visualized using the enhanced chemiluminescence Western blotting detection system of Amersham Pharmacia Biotech according to the instructions of the manufacturer.
Assay for cytolytic activity.
The cytolytic activities of strains expressing different mutant variants of ClyA were scored phenotypically after growth of the bacteria on blood agar plates as described before (33). For quantitative assessments of hemolytic activity, contact hemolytic assays were performed as previously described (21, 23, 26).
Analysis of single-channel formation in lipid bilayer membranes.
Black lipid bilayer membranes were formed as described previously (18). The instrumentation consisted of a Teflon chamber with two aqueous compartments connected by a small circular hole with a surface area of about 0.5 mm2. Membranes were formed across the hole by applying a 1% solution of diphytanoyl phosphatidylcholine (Avanti Polar Lipids, Alabaster, Ala.) in n-decane. The aqueous salt solutions (Merck, Darmstadt, Germany) were used unbuffered and had a pH of around 6. ClyA and its mutants were added from concentrated stock solutions either to the aqueous phase bathing a membrane in the black state or immediately prior to membrane formation. The temperature was kept at 25°C throughout. The membrane current was measured with a pair of Ag-AgCl electrodes switched in series with a voltage source and an electrometer. For the single-channel recordings, the electrometer was replaced by a homemade current amplifier. The amplified signal was monitored with a storage oscilloscope and recorded with a tape or a strip chart recorder.
Lipid monolayer crystallization of ClyA.
Purified wild-type and mutant ClyA proteins were crystallized into two-dimensional structures on planar lipid monolayer films by using a procedure described earlier (6). A brief description with specific details is given here. Crystallization was carried out in wells (1 mm high and 4 mm in diameter) made in a Teflon disk. The Teflon disks were stringently cleaned in 2% Hellmanex II for 1.5 h, after which they were rinsed with tap water for 1 h and with deionized water before excess water was removed. Approximately 18 μl of the purified wild-type or mutant ClyA proteins (30 μg/ml in 20 mM Tris-HCl buffer, pH 7.4) was placed in each well so that a meniscus was formed. In some experiments the wild-type and mutant ClyA protein were added together at a 1:1 ratio. A 75% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine-25% cholesterol mixture was diluted to 0.6 mM in chloroform, and 15 μl was placed on top of the protein solution meniscus. The preparation was incubated at room temperature for 3 days in a humidity chamber. The lipid-protein complexes were collected onto carbon-backed copper grids and immediately stained with 2% sodium phosphotungstate. The ClyA cytolysin assemblies were imaged under a JEOL 100B electron microscope.
RESULTS
Subcellular localization of wild-type and mutant ClyA proteins in E. coli.
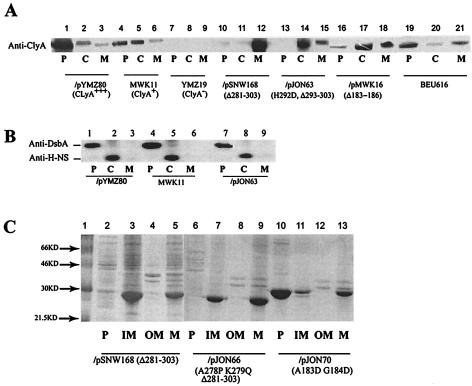
Intact E. coli cells expressing ClyA show cytotoxic effects upon direct contact with cultured mammalian cells, and the activity of ClyA appeared to be contact dependent (17, 21). Recent studies indicate that ClyA also is secreted into the extracellular space (12; our unpublished data). To further investigate the location of ClyA within bacterial cells, we examined the subcellular localization of ClyA in different E. coli strain derivatives. The bacterial cells were subjected to fractionation into cytoplasmic, periplasmic, and membrane fractions (see Materials and Methods for details). Derivatives of E. coli K-12 expressing different levels of ClyA protein were subjected to analysis as shown in Fig. 1. MWK11 is a derivative in which the chromosomal clyA locus has been altered in the promoter region such that it contains a consensus recognition site for the cyclic AMP receptor protein (CRP)-cyclic AMP regulatory complex and thereby expresses phenotypically detectable levels of ClyA (33). MC1061/pYMZ80 is an overexpressing derivative carrying a multicopy plasmid clone of the clyA structural gene. The chromosomal clyA::kan knockout mutant strain YMZ19 was the negative control strain. As shown by SDS-PAGE and Western immunoblotting analysis, large amounts of ClyA protein seemed to accumulate in the periplasmic fraction of the ClyA-overexpressing strain MC1061/pYMZ80 (Fig. 1A, lanes 1 to 3). ClyA localization to the periplasm was also readily detected in samples from strain MWK11 (Fig. 1A, lanes 4 to 6). We also detected ClyA protein in fractions representing the other cell compartments, i.e., in the cytoplasmic and the membrane fractions. Similar results were obtained with other strains expressing lower levels of ClyA from the chromosomal locus, e.g., strain BEU616 and the EHEC isolate 86-24 (Fig. 1A, lanes 19 to 21, and data not shown). In the case of the negative control strain (YMZ19), no anti-ClyA reactivity band was detected when it was tested by the immunoblot analysis (Fig. 1A, lanes 7 to 9). We verified that cytosolic proteins were absent from the periplasmic fraction and that periplasmic components were not present in the cytoplasmic fraction. Using immunoblot analysis, we employed the abundant nucleoid protein H-NS as a cytoplasmic marker and the periplasmic protein disulfide oxidoreductase (DsbA) as a periplasmic marker. The H-NS protein was readily detected in the cytoplasmic fractions but not in the periplasmic fractions (Fig. 1B). Accordingly, the DsbA protein was found in the periplasm, whereas it was not detected in the cytoplasmic fractions (Fig. 1B).
FIG. 1.
Subcellular localization of ClyA proteins. (A) Immunoblot analysis of ClyA proteins in periplasmic (P), cytoplasmic (C), and membrane (M) fractions from different derivatives of E. coli K-12 expressing wild-type or mutant alleles of the clyA gene. Lanes 1 to 3, strain MC1061/pYMZ80. Lanes 4 to 6, strain MWK11. Lanes 7 to 9, strain YMZ19. Lanes 10 to 12, strain MC1061/pSNW168. Lanes 13 to 15, strain MC1061/pJON63. Lanes 16 to 18, strain MC1061/pMWK16. Lanes 19 to 21, strain BEU616. The relevant characteristics of the strains (ClyA phenotype or mutation and plasmids encoding clyA variants) are shown below the lanes. For immunodetection, a mouse monoclonal antibody was used, as described in Materials and Methods. (B) Immunoblot analysis of the periplasmic and cytoplasmic marker proteins DsbA and H-NS, respectively. Subcellular fractions of strains MC1061/pYMZ80 (lanes 1 to 3), MWK11 (lanes 4 to 6), and MC1061/pJON63 (lanes 7 to 9) were subjected to analysis with anti-DsbA and anti-H-NS rabbit antisera. (C) SDS-PAGE analysis and Coomassie blue staining of proteins in periplasmic (P), inner membrane (IM), outer membrane (OM), and total membrane (M) fractions of strains DH5α/pSNW168 (lanes 2 to 5), BEU616/pJON66 (lanes 6 to 9), and BEU616/pJON70 (lanes 10 to 13). Molecular size markers were included in lane 1, and the sizes are indicated on the left.
Earlier results from studies of mutants with alterations in the ClyA C-terminal region suggested that this region may be important for surface expression of the ClyA protein (21). As shown here, the removal of 11 (pJON63) or 23 (pSNW168 and pJON66) amino acids from the C terminus resulted in ClyA proteins that were detected mainly in the cytoplasmic fraction and the membrane fraction, respectively (Fig. 1A, lanes 10 to 15; Table 2). These mutant proteins were not detected in culture supernatants (data not shown). Further separation of the membrane extract into outer membrane and inner membrane fractions showed that the mutant proteins appearing in the membrane fraction were localized to the inner membrane. In fact, in the case of the deletion mutant constructs pSNW168 and pJON66, the majority of the proteins were found in the inner membrane fraction (Fig. 1C).
TABLE 2.
ClyA subcellular protein localization and cytolytic expression
| Strain/plasmid | Relevant characteristics or alteration in ClyA amino acid sequencesa | Subcellular localizationb
|
Hemolysisc | Hemolysis when coexpressed with wild-type ClyAd | ||
|---|---|---|---|---|---|---|
| P | C | M | ||||
| MWK11 | Consensus CRP binding site in the clyA promoter regions ClyA+ | ++ | + | (+) | ++ | NAe |
| YMZ19 | clyA::kan ClyA− | − | − | − | − | NA |
| BEU616 | hns::cat | ++ | (+) | + | + | NA |
| MC1061/pYMZ80 | (1-266)-DDLMLSLLKEAAKKMINTCNEYQKRHGKKTLFFVPEV (wild type) | +++ | + | (+) | +++ | NA |
| MC1061/pJON63 | (1-266)-DDLMLSLLKEAAKKMINTCNEYQKRD (Δ293-303) | − | +++ | + | − | − |
| MC1061/pSNW168 | (1-266)-DDLMLSLLKEAAKK (Δ281-303) | − | − | +++ | − | − |
| MC1061/pJON66 | (1-266)-DDLMLSLLKEAPQK (Δ281-303) | − | + | +++ | − | − |
| MC1061/pJON70 | (1-182)-(A183D G184D)-(185-303) | ++ | + | + | − | (+) |
| MC1061/pMWK16 | (1-182)-Δ(AGVV)-(187-303) | + | +++ | ++ | − | − |
The wild-type and mutant variants of ClyA were expressed from plasmid-carried clyA alleles. Amino acid alterations in comparison with the wild-type ClyA protein are indicated by boldface.
Subcellular localization was monitored by immunoblot analysis of fractions (P, periplasm; C, cytoplasm; M, membrane) prepared as described in Materials and Methods (see also Fig. 1). Western blot signals, which reflect the relative amount of ClyA present in subcellular fractions, are reported as +++ (very strong), ++ (strong), + (moderately strong), (+) (weak), or − (none).
Hemolytic activity was scored on blood agar plates. Relative activity is reported as +++ (very strong), ++ (strong), + (moderately strong), (+) (weak), or − (none).
The dominant negative effect of different plasmid-encoded ClyA variants on expression of the hemolytic phenotype of wild-type ClyA was tested by use of the ClyA+ strains BEU616 and MWK11.
NA, not applicable.
A region in the middle of the ClyA polypeptide was, by X-ray crystallographic analysis, suggested to form a subdomain, designated the head domain and including the hydrophobic β tongue, which has to be maintained to allow its interaction with target membranes (31). Our earlier studies with mutants obtained by site-directed mutagenesis of residues 183 and 184 indicated that this region of ClyA was important for its cytolytic activity (21). The analysis of the subcellular localization of such a mutant ClyA (expressed by pJON70) showed that it did reach the periplasm to a large extent, although there was also some protein in the membrane fraction (Fig. 1B; Table 2). We also included a naturally occurring in-frame deletion variant of the clyA gene that expresses a ClyA protein lacking amino acids 183 to 186 due to a lack of the corresponding four codons. This Δ(183-186) variant was discovered in a strain from the ECOR (E. coli reference) collection (19). The protein expressed from this clyA allele (plasmid pMWK16) gave a nonhemolytic ClyA phenotype, and it was found primarily in the cytoplasmic fraction and at only a low level in the periplasm (Fig. 1A, lanes 16 to 18). The results from the analysis of subcellular localization correlated well with the degree of cytolytic activity as monitored by phenotypic tests (Table 2).
Effects of mutants altered in the ClyA C-terminal domain on phenotypic expression of the wild-type protein.
We decided to test whether mutant ClyA variants would affect the activity or localization of the wild-type protein if it also was expressed in the same bacterial cells. Therefore, a number of mutant clyA alleles cloned on plasmids were introduced into the ClyA-expressing strains MWK11 and BEU616, which carry the wild-type gene on the chromosome. There were clear effects on the phenotypes of the strains expressing both wild-type and mutant ClyA. Some of the mutants inhibited expression of the hemolytic phenotype of strains BEU616 and MWK11 (Table 2). The in-frame deletion variant pMWK16 (Δ183-186) and the C-terminal deletion constructs pJON63, pJON66, and pSNW168 displayed a clear inhibiting effect on the hemolytic phenotype, and the construct pJON70 (A183D G184D) showed a partial effect.
In vitro activities of wild-type and mutant ClyA.
We performed in vitro studies with purified proteins in order to test whether the observed effect of the mutant ClyA proteins could directly affect the cytolytic activity of the wild-type protein. For this purpose, we purified the wild-type ClyA and the A183G G184D, Δ(183-186), and Δ(281-303) mutant proteins, expressed by the clones pYMZ80, pJON70, pMWK16, and pSNW168, respectively. The cytolytic activity was monitored in a hemolysis assay with red blood cells. While the wild-type ClyA showed a clear hemolytic activity, the three mutant proteins showed very low activity in the assay, with levels only about 5 to 10% of the wild-type level (Table 3). The mutant proteins also displayed an inhibiting effect on the activity of the wild-type protein in this in vitro assay. When wild-type ClyA was mixed with an equal amount of the mutant proteins prior to the addition to the red blood cells, the activity was largely diminished; only about 10 to 20% of the activity remained (Table 3). Addition of smaller amounts of the mutant protein resulted in a less reduced level of activity; there was about 40% activity remaining when a 10-fold-lower concentration of mutant protein was included. These results provided additional evidence that the wild-type and mutant proteins could interact directly and that such interactions affected the function of wild-type ClyA.
TABLE 3.
In vitro analysis of ClyA protein activity
| ClyA proteina | Concnb (μg/ml)
|
Relative hemolytic activityc | |
|---|---|---|---|
| WT | Mutant | ||
| WT | 1 | 0 | 1.0 |
| Δ(183-186) | 0 | 1 | 0.13 |
| A183D G184D | 0 | 1 | 0.11 |
| Δ(281-303) | 0 | 1 | 0.03 |
| WT + Δ(183-186) | 1 | 1 | 0.14 |
| 1 | 0.1 | 0.39 | |
| WT + A183D G184D | 1 | 1 | 0.21 |
| WT + Δ(281-303) | 1 | 1 | 0.07 |
| 1 | 0.1 | 0.4 | |
Purified ClyA proteins from wild-type (WT) and different mutant alleles were obtained as described in Materials and Methods.
Final concentrations of proteins that were added in the erythrocyte assay. When wild-type and mutant proteins were tested together, the mixture was left at room temperature for 30 min prior to addition to the erythrocyte suspension.
The hemolytic activities of purified ClyA proteins were determined by the contact hemolytic assay performed for 90 min at 37°C. The relative activity was calculated by using the value for wild-type ClyA (0.295 at 545 nm), arbitrarily set to 1.0, as a reference. The data represent the means from three separate experiments.
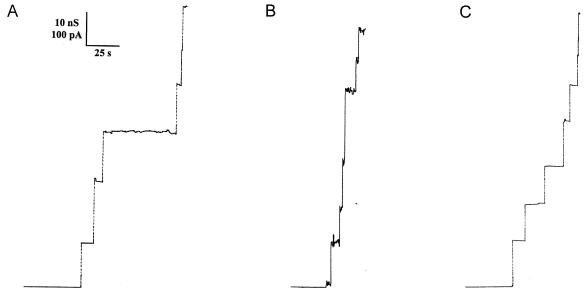
To further assess how the mutant proteins were affected with respect to functional properties, we studied their ability to form channels upon interaction with lipid bilayer membranes. The effect of wild-type ClyA has previously been studied in detail with membranes formed from diphytanoyl phosphatidylcholine-n-decane (18). In those experiments channel formation was observed, in agreement with the hemolytic activity of ClyA and its structure as revealed by electron microscopic analysis (31). As shown above, the Δ(183-186) and Δ(281-303) ClyA mutants had a very low hemolytic activity in comparison with wild-type ClyA. Measurements were performed with lipid bilayer membranes to check whether the ClyA mutants were able to form ion-permeable channels. The results were compared with the channels formed by wild-type ClyA. Figure 2 shows the results of single-channel conductance experiments with wild-type ClyA (Fig. 2A), the Δ(183-186) mutant (Fig. 2B), and the Δ(281-303) mutant (Fig. 2C) in 1 M KCl. The single-channel conductance is a measure of channel size. The somewhat smaller conductance of the mutant channels suggested that both mutants formed channels with a somewhat smaller size than the wild-type ClyA channel. The results also indicated that the mutants formed relatively stable channels with a lifetime similar to that observed previously with wild-type ClyA (18). Wild-type ClyA had the highest single-channel conductance under these experimental conditions, followed by the Δ(183-186) and Δ(281-303) mutants.
FIG. 2.
Single-channel recordings of diphytanoyl phosphatidylcholine-n-decane membranes in the presence of wild-type ClyA (trace A) and the Δ(183-186) (trace B) and Δ(281-303) (trace C) ClyA mutants. The aqueous phase contained 1 M KCl (pH 6) and 10 ng of ClyA wild-type or mutant protein per ml. The applied membrane potential was 20 mV (20°C). Note that the current noise for the single-channel recording of wild-type ClyA is similar to that for the mutants, indicating that the mutation did not induce a major change of the channel structure.
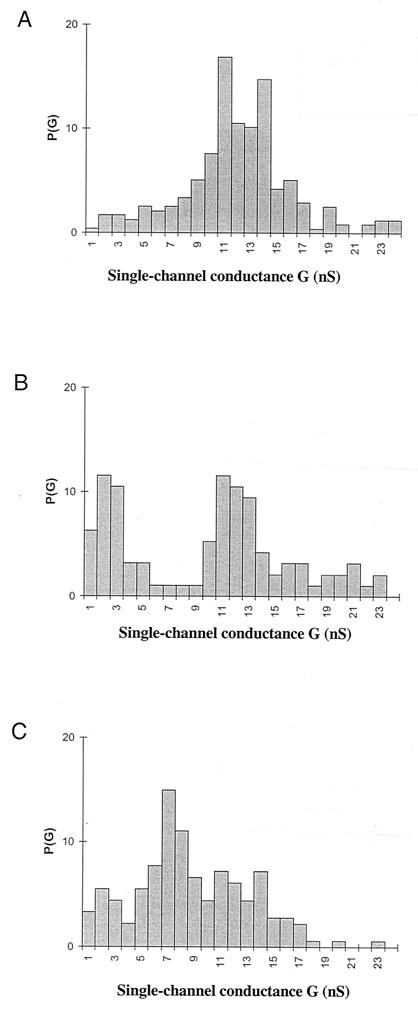
The small difference in the single-channel conductance could not account for the substantial difference in the hemolytic activity observed between wild-type ClyA and the mutants. Another possible effect of the mutations on the hemolytic activity would be a decrease in the channel-forming probability, which should also be detectable in lipid bilayer experiments. To check for such a possibility, multichannel measurements were performed. In these experiments ClyA was added in small quantities (50 to 500 ng/ml) to the aqueous solutions bathing a lipid bilayer membrane in the black state while stirring. Subsequently, the specific conductance of the membrane increased by several orders of magnitude. The ClyA-induced conductance increase was not sudden but needed some time to reach the maximum value. It was steep for about 10 to 20 min. Only a small further increase (compared with the initial one) occurred after that time. The increase of the membrane conductance was observed irrespective of whether wild-type ClyA was added to one side or both sides of the membrane. When the rate of conductance increase was relatively low (about 20 to 30 min after the addition of the toxin), it could be shown for different toxin concentrations that the membrane conductance was a linear function of the protein concentration in the aqueous phase. The addition of ClyA at 50 ng/ml to 1 M KCl resulted in a specific membrane conductance of 4 μS/cm2 after about 30 min. Increases of the ClyA concentration to 100, 200, and 500 ng/ml resulted in membrane conductivities of about 9, 19, and 45 μS/cm2 (means for three membranes), respectively. This means that differences in the ClyA concentration resulted in very similar variations of the specific membrane conductance, suggesting that the pore-forming oligomers did not show an association-dissociation reaction, which is in agreement with the single-channel data given above for wild-type ClyA and the mutants and previously for wild-type ClyA (18). Histograms of the channels formed by wild-type ClyA and the two tested mutant proteins in 1 M KCl solution are given in Fig. 3. Wild-type ClyA had the highest single-channel conductance under these experimental conditions followed by the Δ(183-186) and Δ(281-303) mutants.
FIG. 3.
Histogram of the probability P(G) for the occurrence of a given conductivity unit observed with membranes formed from diphytanoyl phosphatidylcholine-n-decane in the presence of 10 ng of wild-type ClyA (A) and the Δ(183-186) (B) and Δ(281-303) (C) mutants per ml. P(G) is the probability that a given conductance increment G is observed in the single-channel experiments. It was calculated by dividing the number of fluctuations with a given conductance increment by the total number of conductance fluctuations. The aqueous phase contained 1 M KCl. The applied membrane potential was 20 mV (20°C). The average single-channel conductances were 12 nS for 237 single-channel events (wild-type ClyA), 8.5 nS for 95 events [ClyA Δ(183-186) mutant], and 7.5 nS for 181 events [ClyA Δ(281-303) mutant]. Conductance is current divided by voltage.
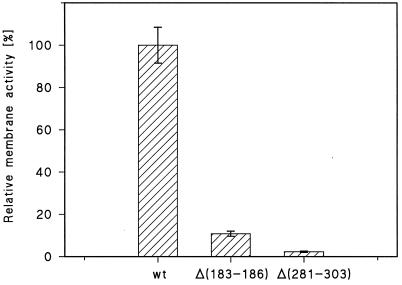
The measurement of the time course of the conductance increase allowed a meaningful comparison of the channel-forming activities of the two mutants compared to that of wild-type ClyA. The specific conductance of diphytanoyl phosphatidylcholine-n-decane membranes was measured at a defined concentration of the mutants and wild-type ClyA. The specific conductance caused by the mutants was much smaller than that caused by wild-type ClyA. Figure 4 shows the relative membrane (i.e., channel-forming) activities of the Δ(183-186) and Δ(281-303) ClyA mutants compared to that of wild-type ClyA. They accounted only for about 10% [Δ(183-186) mutant] and about 3% [Δ(281-303) mutant] compared to the wild type (100%). These values were in good agreement with the relative pore-forming activities determined by use of the hemolysis assay (Table 3).
FIG. 4.
Relative conductance caused by the ClyA Δ(183-186) and ClyA Δ(281-303) mutants in lipid bilayer membranes compared to wild-type ClyA (wt). Membranes were formed from diphytanoyl phosphatidylcholine-n-decane in 1 M KCl, and ClyA proteins were added at a concentration of 100 ng/ml to the aqueous phase. About 30 min after addition of ClyA proteins, the membrane conductance was measured and averaged for three membranes for the individual systems [wild-type ClyA, ClyA Δ(183-186) mutant, and ClyA Δ(281-303) mutant]. The membrane conductance of wild-type ClyA was set to 100%, and the conductances of the two ClyA mutants were calculated relative to that of wild-type ClyA. The membrane activity is given as the mean value ± standard deviation. T = 20°C; Vm = 20 mV.
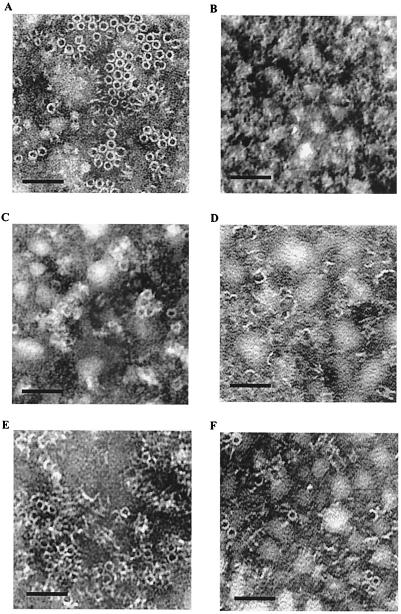
The cytolytic activity of the ClyA protein that causes hemolysis of erythrocytes includes pore formation, and results from protein structural analysis and electron microscopic studies suggested that there are eight ClyA molecules in each of the pore assemblies (31). In order to study how our mutant proteins might be affected in formation of such pore assemblies, we used the method of two-dimensional crystallization of the proteins on planar lipid films (see Materials and Methods for details). As shown in Fig. 5A, the wild-type ClyA protein formed easily recognized pore assemblies, and the pore structures were abundant and uniform in shape. However, in the case of the mutant proteins the ability to form such pore assemblies was impaired. The ClyA Δ(281-303) mutant protein did not result in any recognizable pore assemblies under these conditions (Fig. 5B). There were some pore assemblies found in the case of the ClyA Δ(183-186) mutant protein, but most of the protein seemed to be forming partial assemblies or altered structures (Fig. 5C). When the wild-type ClyA was mixed with an equal amount of mutant proteins, there were clear indications that the wild-type ClyA was impaired in its formation of pore assemblies (Fig. 5D to F). In particular, in the presence of the ClyA Δ(281-303) mutant protein there were mostly partial assemblies, and only a few complete pore structures typical of wild-type ClyA were observed (Fig. 5D [c.f. with Fig. 5A]). Similarly, the pore assembly formation by the wild-type ClyA protein was impaired when it was mixed with the ClyA Δ(183-186) mutant protein (Fig. 5E) or the ClyA (A183G G184D) mutant protein (Fig. 5F). The findings were in keeping with the above-described results and supported the conclusion that the mutant and wild-type ClyA proteins interacted and formed different complexes with altered cytolytic and pore-forming properties in comparison with those of the wild type.
FIG. 5.
Electron microscope images of formation of ClyA pore assemblies on planar lipid monolayer films. Purified wild-type and mutant ClyA proteins were allowed to assemble by the lipid monolayer crystallization method as described in Materials and Methods. Bars, 25 nm. (A) Wild-type ClyA protein. (B) ClyA Δ(281-303) mutant protein. (C) ClyA Δ(183-186) mutant protein. (D) Wild-type and ClyA Δ(281-303) mutant proteins in a 1:1 ratio. (E) Wild-type and ClyA Δ(183-186) mutant proteins in a 1:1 ratio. (F) Wild-type and ClyA (A183G G184D) mutant proteins in a 1:1 ratio.
DISCUSSION
We show here that some mutant ClyA polypeptides altered in the C-terminal part were unable to reach the periplasm (Fig. 1). The mutant Δ(281-303) protein was impaired in its translocation and was retained in the inner membrane. Furthermore, the mutant protein displayed a dominant negative effect on the wild-type ClyA protein. The C-terminal region was earlier suggested to be of importance for membrane targeting, pore formation, and translocation of ClyA, since mutant bacteria expressed a nonhemolytic phenotype (3, 21). According to the structural model based on X-ray crystallography, the C-terminal region includes a helix structure that is part of a tail domain in the folded protein and that may be somewhat mobile in relation to the rest of the protein (3, 31). How the lack of the C-terminal region might affect the overall structure of ClyA in the cell is not known, but we must consider that, e.g., the folding of the mutant Δ(281-303) protein could be quite different from that of the wild type. It was somewhat puzzling why ClyA with a 11-amino-acid deletion localized mainly to the cytoplasm whereas ClyA with a 23-amino-acid deletion localized to the membrane. However, it should be noted that there is also a substitution (H292D) in the former case, and it is feasible that the defective localization of ClyA expressed by pJON63 could be due to the substitution mutation rather than the deletion.
The naturally occurring ClyA variant lacking four amino acids in a hydrophobic region in the middle of the protein [Δ(183-186) mutant] retained the ability to be translocated to the periplasm although it had lost much of the pore-forming activity. It also displayed a dominant negative effect on wild-type ClyA.
Studies in vitro with purified proteins provided evidence that suggested an interaction between the wild-type and mutant ClyA proteins and that the activity of the wild-type polypeptide was affected negatively by the interaction (Table 3). The mutant proteins showed a low pore-forming activity as judged by the hemolysis assay with erythrocytes. Measurements of the single-channel conductance in the lipid bilayer membrane system showed that the mutant ClyA proteins retained some ability to form channels, albeit less well than the wild type. The results from electron microscopic studies of ClyA proteins assembled on planar lipid films by the lipid monolayer crystallization method fully supported the conclusion that mutant and wild-type ClyA proteins formed different complexes with altered cytolytic and pore-forming properties in comparison with those of the wild type.
The dominant negative phenotype of the ClyA mutants can therefore be explained as a consequence of the formation of dysfunctional mixed oligomers, comprised of both mutant and wild-type monomeric components. When the proteins are coexpressed in vivo, the formation of intracellular hetero-oligomers might interfere with the pool of monomeric wild-type ClyA. Such interference can presumably occur in either of the intracellular compartments where the mutant proteins primarily were found. Interestingly, a similar situation with formation of mixed oligomers was suggested as the more likely explanation for the dominant negative phenotype exerted by a mutant variant of the Helicobacter pylori VacA cytotoxin as well (30). In that case, the mutant protein retained the ability to be secreted from the bacteria and formed oligomeric complexes similar to those of the wild-type VacA. The ClyA Δ(281-303) mutant protein appeared to have lost the ability to form normal oligomeric complexes, and it clearly affected the ability of wild-type ClyA in its oligomerization in vitro (Fig. 5).
The ClyA protein is evidently not subject to any signal sequence cleavage, and there is no evidence of any other posttranslational modification (10, 21, 31). It remains to be elucidated whether the translocation of ClyA involves any accessory component(s) of some specific transport system. If so, the mutant proteins could possibly also affect the translocation of wild-type ClyA by competing for binding to such components of the transport system. In the case of the outer membrane lipoprotein XpsD of Xanthomonas campestris pv. Campestris, it was suggested that a particular mutant form of the protein could interfere with the secretion of the wild-type protein by competing for some other factor(s) (9). The wild-type XpsD protein forms a multimeric complex, and it was also shown that certain other mutant XpsD proteins caused secretion interference primarily by forming mixed nonfunctional multimers with the wild-type proteins (9). The ClyA protein represents a new type of cytolysin in enterobacteria with a novel protein fold according to the structural analysis (31). The dominantly negative ClyA mutants characterized in the present work should aid in further molecular analysis of the mechanisms by which this novel type of cytotoxin forms multimeric assemblies and how it is secreted by bacterial cells.
Acknowledgments
We thank Lenore Johansson for assistance with electron microscopy and K. Ito for kindly supplying anit-DsbA antiserum.
This work was supported by grants from the Swedish Research Council. S.N.W. was supported in part by a Visiting Scientist Fellowship from the Wenner-Gren Foundations, and J.O. was supported by a Research Associate Fellowship from the Faculty of Medicine and Odontology at Umeå University.
REFERENCES
- 1.Abrami, L., M. Fivaz, and F. G. van der Goot. 2000. Adventures of a pore-forming toxin at the target cell surface. Trends Microbiol. 8:168-172. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, Y., S. Kamitani, N. Kusukawa, and K. Ito. 1992. In vitro catalysis of oxidative folding of disulfide-bonded proteins by the Escherichia coli dsbA (ppfA) gene product. J. Biol. Chem. 267:22440-22445. [PubMed] [Google Scholar]
- 3.Atkins, A., N. R., Wyborn, A. J. Wallace, T. J. Stillman, L. K. Black, A. B. Fielding, M. Hisakado, P. J. Artymiuk, and J. Green. 2000. Structure-function relationships of a novel bacterial toxin, hemolysin E. The role of alpha G. J. Biol. Chem. 275:41150-41155. [DOI] [PubMed] [Google Scholar]
- 4.Bhakdi, S., H. Bayley, A. Valeva, I. Walev, B. Walker, M. Kehoe, and M. Palmer. 1996. Staphylococcal alpha-toxin, streptolysin-O, and Escherichia coli hemolysin: prototypes of pore-forming bacterial cytolysins. Arch. Microbiol. 165:73-79. [DOI] [PubMed] [Google Scholar]
- 5.Braun, V., R. Schonherr, and S. Hobbie. 1993. Enterobacterial hemolysins: activation, secretion and pore formation. Trends Microbiol. 1:211-216. [DOI] [PubMed] [Google Scholar]
- 6.Brisson, A., O. Lambert, and W. Bergsma-Schutter. 1999. Two-dimensional crystallization of soluble proteins on planar lipid films, p. 342-363. In A. Ducruix and R. Geige (ed.), Crystallization of nucleic acids and proteins: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 7.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 8.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 9.Chen, L.-Y., D.-Y. Chen, J. Miaw, and N.-T. Hu. 1996. XpsD, an outer membrane protein required for protein secretion by Xanthomonas campestris pv. campestris, forms a multimer. J. Biol. Chem. 271:2703-2708. [DOI] [PubMed] [Google Scholar]
- 10.Del Castillo, F. J., S. C. Leal, F. Moreno, and I. Del Castillo. 1997. The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted haemolysin. Mol. Microbiol. 25:107-115. [DOI] [PubMed] [Google Scholar]
- 11.Del Castillo, F. J., F. Moreno, and I. Del Castillo. 2000. Characterization of the genes encoding the SheA haemolysin in Escherichia coli 0157:H7 and Shigella flexneri 2a. Res. Microbiol. 151:229-230. [DOI] [PubMed] [Google Scholar]
- 12.Del Castillo, F. J., F. Moreno, and I. Del Castillo. 2001. Secretion of the Escherichia coli K-12 SheA hemolysin is independent of its cytolytic activity. FEMS Microbiol Lett. 204:281-285. [DOI] [PubMed] [Google Scholar]
- 13.Green, J., and M. L. Baldwin. 1997. The molecular basis for the differential regulation of the hlyE-encoded haemolysin of Escherichia coli by FNR and HlyX lies in the improved activating region 1 contact of HlyX. Microbiology 143:3785-3793. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 15.Johansson, J., and B. E. Uhlin. 1999. Differential protease-mediated turnover of H-NS and StpA revealed by a mutation altering protein stability and stationary-phase survival of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:10776-10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lai, X. H., I. Arencibia, A. Johansson, S. N. Wai, J. Oscarsson, S. Kalfas, K. G. Sundqvist, Y. Mizunoe, A. Sjöstedt, and B. E. Uhlin. 2000. Cytocidal and apoptotic effects of the ClyA protein from Escherichia coli on primary and cultured monocytes and macrophages. Infect. Immun. 68:4363-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig, A., S. Bauer, R. Benz, B. Bergmann, and W. Goebel. 1999. Analysis of the SlyA-controlled expression, subcellular localization and pore-forming activity of a 34 kDa haemolysin (ClyA) from Escherichia coli K-12. Mol. Microbiol. 31:557-567. [DOI] [PubMed] [Google Scholar]
- 19.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oscarsson, J., Y. Mizunoe, B. E. Uhlin, and D. J. Haydon. 1996. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol. Microbiol. 20:191-199. [DOI] [PubMed] [Google Scholar]
- 21.Oscarsson, J., Y. Mizunoe, L. Li, X. H. Lai, Å. Wieslander, and B. E. Uhlin. 1999. Molecular analysis of the cytolytic protein ClyA (SheA) from Escherichia coli. Mol. Microbiol. 32:1226-1238. [DOI] [PubMed] [Google Scholar]
- 22.Oscarsson, J., M. Westermark, L. Beutin, and B. E. Uhlin. 2002. The bacteriophage-associated ehly1 and ehly2 determinants from Escherichia coli O26:H- strains do not encode enterohemolysins per se but cause release of the ClyA cytolysin. Int. J. Med. Microbiol. 291:625-631. [DOI] [PubMed] [Google Scholar]
- 23.Oscarsson, J., M. Westermark, S. Löfdahl, B. Olsén, H. Palmgren, Y. Mizunoe, S. N. Wai, and B. E. Uhlin. 2002. Characterization of a pore-forming cytotoxin expressed by Salmonella enterica serovars Typhi and Paratyphi A. Infect. Immun. 70:5759-5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinold J., N. Starr, J. Maurer, and M. D. Lee. 1999. Identification of a new Escherichia coli She haemolysin homolog in avian E. coli. Vet. Microbiol. 66:125-134. [DOI] [PubMed] [Google Scholar]
- 25.Rowe, G. E., and R. A. Welch. 1994. Assays of hemolytic toxins. Methods Enzymol. 235:657-667. [DOI] [PubMed] [Google Scholar]
- 26.Sansonetti, P. J., A. Ryter, P. Clerc, A. T. Maurelli, and J. Mounier. 1986. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect. Immun. 51:461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanley, P., V. Koronakis, and C. Hughes. 1998. Acylation of Escherichia coli hemolysin: a unique protein lipidation mechanism underlying toxin function. Microbiol. Mol. Biol. Rev. 62:309-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uhlin, B. E., and Y. Mizunoe. 1994. Expression of a novel contact-hemolytic activity by E. coli. J. Cell. Biochem. Suppl. 18A:71.
- 30.Vinion-Dubiel, A. D., M. S. McClain, D. M. Czajkowsky, H. Iwamoto, D. Ye, P. Cao, W. Schraw, G. Szabo, S. R. Blanke, Z. Shao, and T. L. Cover. 1999. A dominant negative mutant of Helicobacter pylori vacuolating toxin (VacA) inhibits VacA-induced cell vacuolation. J. Biol. Chem. 274:37736-37742. [DOI] [PubMed] [Google Scholar]
- 31.Wallace, A. J., T. J. Stillman, A. Atkins, S. J. Jamieson, P. A. Bullough, J. Green, and P. J. Artymiuk. 2000. E. coli hemolysin E (HlyE, ClyA, SheA): X-ray crystal structure of the toxin and observation of membrane pores by electron microscopy. Cell 10:265-576. [DOI] [PubMed] [Google Scholar]
- 32.Welch, R. A., C. Forestier, A. Lobo, S. Pellett, W. Thomas, Jr., and G. Rowe. 1992. The synthesis and function of the Escherichia coli hemolysin and related RTX exotoxins. FEMS Microbiol. Immunol. 5:29-36. [DOI] [PubMed] [Google Scholar]
- 33.Westermark, M., J. Oscarsson, Y. Mizunoe, J. Urbonaviciene, and B. E. Uhlin. 2000. Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J. Bacteriol. 182:6347-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]