Abstract
Leukemia inhibitory factor (LIF) receptor is a cell surface receptor that mediates the actions of LIF and other IL-6 type cytokines through the formation of high-affinity signaling complexes with gp130. Here we present the crystal structure of a complex of mouse LIF receptor with human LIF at 4.0 Å resolution. The structure is, to date, the largest cytokine receptor fragment determined by x-ray crystallography. The binding of LIF to its receptor via the central Ig-like domain is unlike other cytokine receptor complexes that bind ligand predominantly through their cytokine-binding modules. This structure, in combination with previous crystallographic studies, also provides a structural template to understand the formation and orientation of the high-affinity signaling complex between LIF, LIF receptor, and gp130.
Keywords: gp130, IL-6 family, signaling
Leukemia inhibitory factor (LIF) is a pleiotropic cytokine that acts on many cell types including embryonic stem cells, megakaryocytes, osteoblasts, and neuronal cells (1). On the surface of these responsive cells, LIF first binds to the LIF receptor (LIFR) with low nanomolar affinity and then to gp130 to form a high-affinity (picomolar) functional signaling complex (2–4). The nonredundant functional importance of both LIF and LIFR has been demonstrated by gene knockout experiments in mice, with the ligand and receptor showing different phenotypes. Deletion of the LIF gene results principally in female infertility due to a failure of blastocyst implantation (5). The LIFR deletion, however, is more severe, causing perinatal lethal placental, skeletal, neural, and metabolic defects (6). The severity of the LIFR deletion reflects the fact that it (along with gp130) is required for signaling by many cytokines, including oncostatin M (OSM), ciliary neurotrophic factor, novel neurotrophin-1/cardiotrophin-like cytokine (NNT-1/CLC), and cardiotrophin-1 (CT-1).
Signaling by this family of cytokines can occur in two ways depending on the receptors used. Cytokines such as IL-6 and IL-11 signal through the formation of a hexameric complex with gp130 and a cytokine-specific nonsignaling receptor subunit (e.g., the [gp130:IL-6:IL-6Rα]2 complex) (7). On the other hand, LIF and related cytokines bind both gp130 and a second signaling receptor, such as LIFR or OSM receptor (OSMR), to form a trimeric complex (e.g., the LIF:LIFR:gp130 complex). In complexes of the second type some cytokines, such as ciliary neurotrophic factor, bind an additional nonsignaling receptor subunit (8).
Upon ligand-induced dimerization of the two receptor signaling chains, signal transduction occurs through activation of noncovalently associated Janus tyrosine kinases (Jaks), which are able to phosphorylate the cytoplasmic domains of gp130 and LIFR and thus recruit and activate the STAT family of transcription factors. STAT homo- or heterodimers translocate to the nucleus enabling them to regulate the transcription of a specific set of target genes (9, 10).
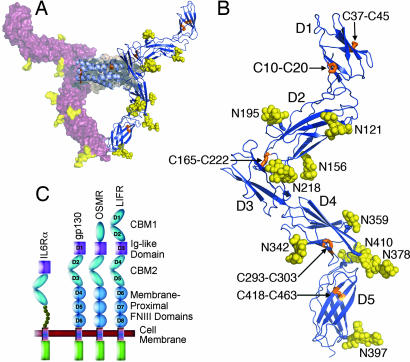
Sequence analysis indicates that the LIFR has an extracellular region with a modular structure containing two cytokine-binding modules (CBM) separated by an Ig-like domain and followed by three membrane-proximal fibronectin type-III (FNIII) domains (Fig. 1C). These FNIII domains both position the receptor on the cell surface and assist formation of the signaling complex through correct orientation of the receptor for activation (11). Mutagenesis studies have highlighted residues in both the LIF (12) and LIFR (13, 14) that are important for binding; however, a unique feature of the LIF:LIFR system is an unusual human:mouse species cross-reactivity that occurs despite high sequence identity between their ligands and receptors (78% and 76%, respectively). The binding and biological activity of mouse LIF (mLIF) is species restricted, whereas human LIF (hLIF) is able to bind both human and mouse receptors with high affinity (15). Here we describe the structure of the first five domains of mLIFR (Fig. 1) in complex with hLIF. The structure illustrates the unique binding arrangement between LIF and the Ig-domain of LIFR, aids in understanding species cross-reactivity and provides a structural template for modeling the high-affinity signaling complex between LIF:LIFR:gp130.
Fig. 1.
The crystal structure of the hLIF: mLIFR complex. (A) Side view of the complex illustrating the molecular surface for one complex, with receptor colored raspberry and ligand colored orange-gray. The NCS copy of the artificial dimer is shown in ribbons with receptor colored blue; ligand colored light blue; disulfide bonds colored orange, and N-linked carbohydrate colored yellow. (B) A detailed view of the receptor alone with domains and sites of posttranslational modifications labeled. Colors are as in A. (C) A domain representation of selected cytokine receptors illustrating differences in their modular domain structure.
Results and Discussion
Overall Structure.
For structural studies we expressed the first 5 extracellular domains of mouse LIFR (D1-D5 or CBM1-Ig-CBM2) and crystals of this fragment with human LIF contained two structurally similar but independent complexes. Despite the modest resolution of the data, a number of factors contributed significantly to the quality of the electron density maps, allowing clear placement of residues 1–180 of hLIF and 7–486 of mLIFR (see Materials and Methods). The extracellular fragment of mLIFR adopts an extended zigzag conformation (≈160 Å long and 56 Å wide) with the distinct domains lying in approximately the same plane (Fig. 1). In CBM1 (residues 7–202), the D1 and D2 domains are connected by a short linker and are at an angle of 80°. The D1 domain (residues 7–82) contains conserved disulfide bonds between Cys-10–Cys-20 and Cys-37–Cys-45 and is notably smaller than the corresponding domains from both gp130 and IL-6Rα (16, 17), due to shorter connecting β-strands (and a potential small N-terminal truncation of five to six residues). Superposition of LIFR D1 on these two structures results in a rmsd for Cα atoms of 1.82 Å and 1.95 Å, respectively. The D2 domain shows greater structural similarity to the corresponding CBM domains of both gp130 and IL-6Rα because it contains conserved structural features like the WSXWS motif, and the rmsd values for Cα atoms are 1.45 Å and 1.40 Å, respectively.
Following a short looped-out linker (residues 198–205), the Ig-like domain packs across the end of the D2 domain in a T-junction and is slightly twisted out of the plane of CBM1. Loops from the end of D2 form a relatively flat, hydrophobic interface with one sheet of the Ig-domain (residues 203–283). Central to this interface, there is an interdomain disulfide bond between Cys-165 and Cys-222 that is likely to fix the relative orientations of the D2 and Ig-like domains [supporting information (SI) Fig. 5B]. The positioning of this disulfide bond is unambiguous, despite weak density, as each cysteine is located near well defined N-linked glycosylation sites (Asn-156 and Asn-218). The weak density may be attributable to significant radiation damage during data collection. At the C-terminal end of the Ig-like domain there is no hinge and the polypeptide runs directly into D4 CBM2 (residues 284–380). These two domains lie collinearly with each other in an orientation similar to the first two domains of gp130 but larger loops in both domains of LIFR provide added rigidity for this interface (Fig. 1).
CBM2 is similar in size to the CBMs from other receptors with a characteristic angle of 74° between the D4 (residues 284–380) and D5 domains (residues 389–486). It contains conserved disulfide bonds in the D4 domain between Cys-293–Cys-303 and in D5 between Cys-418–Cys-463. CBM2 shares ≈20% sequence identity with the gp130 CBM and can be superposed with an rmsd of 1.88 Å for Cα atoms. However, conformational differences do occur between the two cytokine-binding modules, but are restricted mainly to the loop regions with the core secondary structure being well maintained.
Crystals of hLIF:mLIFR contained a dimer of complexes in the asymmetric unit where the predominant interactions occur between the N-terminal segments of two hLIF molecules, which were disordered in previous crystal and NMR structures of both human and mouse LIF (16, 18, 19). Additional dimer contacts occur between the D5 domains of NCS related mLIFR molecules. The presence of a hLIF:mLIFR dimer was unexpected because the formation of a 1:1 complex in solution has been previously demonstrated by native gel, cross linking, gel filtration, and sedimentation equilibrium analysis (3) as well as multiangle light scattering (16). The crystal packing also suggests that the dimer is likely to have little biological significance. In the dimer, hLIF and LIFR D3–5 form a closed loop, with two symmetry-related dimers linked in what could be described as magician's rings (SI Fig. 6); this would be highly inconsistent with a preformed dimer in solution and supports our argument that the dimer is a crystallization artifact.
Receptor:Ligand Interface.
The interface between hLIF and mLIFR is saddle-shaped, comprising 22 residues from the ligand and 25 from the receptor and burying a total of 1,890 Å2 of accessible surface area (Fig. 2). The ligand, hLIF, interacts via loops at one end of the four-helix bundle (site III, burying 945 Å2), whereas the receptor's contact region is predominantly formed from one sheet of the Ig-like domain (885 Å2), with the remainder coming from the N-terminal loops of D4 (residues 314–316 and 337–338, 160 Å2). Using the shape correlation statistic, Sc, to quantify interface complementarity (20), the value for hLIF:mLIFR is 0.70, consistent with a biologically relevant interaction.
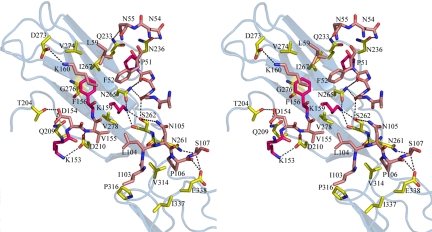
Fig. 2.
A stereo view of the hLIF:mLIFR binding interface. The receptor Ig-D4 domain secondary structure is shown as light blue ribbons. Interface residues are shown as sticks and colored yellow for receptor, salmon for ligand, and magenta for residues Pro-51, Lys-153, Phe-156 and Lys-159, previously demonstrated as essential residues for ligand binding (12).
Partial alanine scanning mutagenesis of hLIF identified Pro-51, Lys-153, Phe-156, and Lys-159 as important in LIFR binding with Phe-156 and Lys-159 mutations having the most dramatic impact (12). These two residues lie at the centre of the hLIF interface (Fig. 2) with Lys-159 surrounded by a hydrophobic patch containing Pro-51, Phe-52, Leu-59, Phe-156, Val-155, and Leu-104. The lysine side chain is tightly packed and hydrogen bonded to Ser-262 and Asn-265 which are both conserved in mammalian LIFR sequences. Although there is no compensating charge for Lys-159, the effects of the buried charge may be mitigated by the dipole from helix A. A significant hydrophobic interaction is made by a π-stacking arrangement of the Phe-156 side chain against the peptide bond of Gly-276 of mLIFR. The position of Phe-156 is maintained by interactions with Phe-52 and Leu-59 of hLIF and the phenyl ring sits between Asn-265, Ile-267, Val-274, and Val-278 on the receptor's β-sheet. Important putative hydrogen bonding interactions also occur toward the edge of the hydrophobic patch, including carbonyl groups of hLIF residues 48 and 50 with Asn-265 of mLIFR, and Lys-160 with Asp-273. Lys-153 of hLIF lies near the conserved Asp-210 of mLIFR and its side chain is partially enclosed by the D2/D3 link. Phe-52 has yet to be mutated in previous studies, but its proximity to Pro-51, a cis proline in all structures of LIF, suggest that it is likely to play a similar role to Pro-106 in hGH (21) and maintain a binding conformation for the Phe-52 loop. Pro-51 also makes favorable contacts to Ile-267 in the mLIFR Ig-like domain.
The association described for hLIF with mLIFR is quite unlike the primary binding of IL-6 to IL-6Rα or IL-6 and LIF to gp130. Yet, it is reminiscent of the contact of IL-6 with the Ig-like domain of gp130 in the IL-6:IL-6Rα:gp130 hexameric complex (7) (Fig. 3 A and D). For IL-6, this third receptor contact is crucial for signaling but it is not the major contributor to ligand binding. In the hLIF:mLIFR complex; however, the contact is more extensive (Fig. 3 C and E) with the loop between helices B and C (residues 103–108) interacting with an insert into the mLIFR Ig-like domain (residues 260–263) and two larger loops on domain D4 (Fig. 3E). Interactions include Pro-51 with Val-314 and the peptide bond of Glu-338, Ile-103 with Pro-316 and a putative hydrogen bond between Ser-107 and Glu-338 (Fig. 2). An additional hydrogen bonded interaction between Asn-105 and Ser-262 is not conserved interspecies because Ser-262 is a leucine in the human Ig-domain.
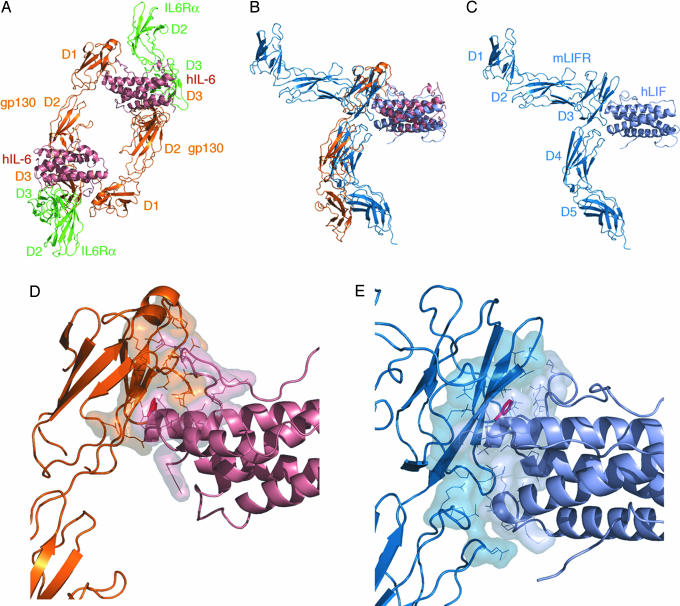
Fig. 3.
Comparison of the LIF:LIFR structure with the IL-6:IL-6Rα:gp130 hexameric complex. (A) Structure of the hexameric signaling complex of IL-6:IL-6Rα:gp130 (1P9M) (7) represented as ribbons. IL-6 is colored salmon, IL-6Rα is colored green, and gp130 is colored orange. (B) Structural superposition based upon the cytokines IL-6 from A and LIF from C. (C) Structure of the LIF:LIFR complex shown as ribbons and in the same orientation as in A and B. LIF is colored light-blue and LIFR is colored blue. (D) A detail of the interface between IL-6 and the Ig-domain (D1) of gp130. The buried surface residues calculated with a 1.4-Å probe radius are shown as a semitransparent surface to highlight the extent of the interaction. Also highlighted in red is Trp-157 the conserved hydrophobic residue at site III interactions. (E) A detail of the interface between LIF and LIFR. Buried surface calculated as in D is shown as a semitransparent surface with Phe-156 is highlighted in red.
Species Cross-Reactivity.
An unusual species cross-reactivity exists between human and mouse LIF and LIFR complexes despite >70% of the interface residues being conserved. The highest affinity complex is between hLIF and mLIFR with an affinity (equilibrium dissociation constant, KD) of ≈10–46 pM, one to two orders of magnitude higher than hLIF:hLIFR complex (300–900 pM) and mLIF:mLIFR complex (KD of 1–4 nM). Interestingly, mLIF cannot bind to hLIFR, or at least does so with very low affinity (15). Mapping the sequence differences onto the structure, it is clear that interactions at the center of the interface are maintained and that each difference appears to independently destabilize the structure of hLIF:mLIFR described here.
Mutation of six residues in mLIF to their human equivalent Glu57Asp, Thr107Ser, Gln112His, Val113Ser, Ala155Val, and Arg158Lys restores both binding to hLIFR and signaling activities comparable to hLIF (22). Of these residues, only Ser-107 and Val-155 are in direct contact with the receptor, with Val-155 improving the hydrophobic packing. Asp-57 and Lys-158 lie adjacent to interface residues and may slightly perturb them via the main chain, as could Ser-113, which appears to modify the start of the C helix by possibly replacing a main chain hydrogen bond.
Between mLIFR and hLIFR, there are 31 amino acid differences in the Ig-like domain (SI Fig. 7), and almost all of these are conservative hydrophobic changes to the Ig-domain core. In contrast, mLIFR:hLIFR mutations Gln233Leu, Asn236His, and Asn261Ser at the periphery of the interface would cause the loss of two to three hydrogen bonds to conserved LIF residues (Asn-55, Asn-54, and Ser/Thr-107, respectively) and contribute to the differences in the strength of interactions. Asp273Asn would mean the loss of a salt link, although the hydrogen bond would be maintained.
Other Ligands for LIFR.
Like LIF, CT-1 and OSM have the ability to induce signaling through the LIFR:gp130 complex. The significantly lower affinity of OSM for LIFR suggests that formation of a functional signaling complex occurs through OSM first forming a low-affinity gp130 complex before recruiting LIFR or OSMR to form a high affinity signaling complex (14, 23). Sequence alignment of LIF, CT-1, and OSM (SI Fig. 8) demonstrates that Phe-156 and Lys-159 in LIF are conserved in all of the ligands, whereas Pro-51 and Phe-52 are conserved in both CT-1 and LIF, and Phe-52 appears to be structurally replaced by a Leu-40 in OSM. Further differences between alternate ligands also occur through interaction of ligands with the D3/D4 region, although a similar to that observed for LIF:LIFR appears plausible.
Signaling Complex Model.
The formation of hLIF:mLIFR represents the first step toward cell signaling, and an interaction with the shared gp130 receptor is required to form a high-affinity signaling complex. The crystal structures, previously described, for unliganded gp130 (D2–D3) (24) and gp130 (D2–D3) in complex with hLIF (16) show minor conformational differences and gp130 can be superposed with an rmsd of 0.69 Å for Cα atoms. The hLIF:gp130 (D2–D3) complex (16) can also be superposed with the larger IL-6:gp130 (D1–D3) complex (7), giving an rmsd for gp130 of 0.84 Å. This finding illustrates that these receptor modules are fairly rigid structures, with negligible conformational changes occurring even upon ligand binding. Therefore, we now have structural templates to produce a highly accurate model of the hLIF:mLIFR:gp130 complex (Fig. 4), using a combination of our hLIF:mLIFR and available gp130 structures. The modeled complex has a buried surface of ≈3,500 Å2 with a contribution of ≈1,850 Å2 from the gp130:hLIF interface and ≈1,750 Å2 from the hLIF:mLIFR interface. Additional buried surface would also be expected through interaction between the membrane proximal FNIII domains. This large buried surface would be consistent with the very high affinity dissociation constant of the complex, although discreet hotspots are likely to provide the majority of the binding free energy through specific interactions.
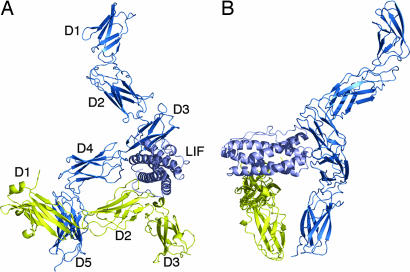
Fig. 4.
Structural model of the trimeric LIF:LIFR:gp130 complex oriented with the C-terminal domains of each receptor at the bottom. The spacing between these domains is consistent with known structural data (11, 25, 26). Membrane proximal domains would then extend downwards with an additional interaction between gp130 and LIFR before membrane. (A) View along LIF from the N-terminal end highlighting the gp130:LIF interface. The components of the complex are illustrated as ribbons, with LIFR D1-D5 shown in blue; LIF shown in light blue; and gp130 D1-D3 shown in yellow. (B) The same representation rotated ≈90° CCW to highlight the LIF:LIFR interface.
Positioning of the membrane proximal FNIII domains from both receptors would be at present quite speculative. Currently, the only structural information on the tall family of cytokine receptors is from recent EM reconstructions of human gp130:IL-6:IL-6Rα and murine gp130:IL-11:IL-11Rα receptor complexes, which highlight the interaction between two gp130 D6 domains (25, 26). In contrast, experiments using chimeric receptors indicate that the interaction of D5 of gp130 with D7 of LIFR occurs in the formation of a functional LIF:LIFR:gp130 signaling complex (11). Further structural studies of the full-length complexes will be therefore be required to unambiguously determine the arrangement of the membrane proximal “legs” on the cell surface.
Conclusion
For the four-helix cytokine:receptor complexes whose structures have been determined, the primary ligand binding interaction is usually via a cytokine-binding module (site I). In forming a signaling complex, a second CBM is sometimes used, as occurs in the growth hormone/prolactin system (site II). Although gp130 also uses a CBM to bind ligands at site II, it and a number of other cytokine receptors (e.g., leptin receptor, G-CSF receptor) also use an Ig-like domain to make a further contact with the ligand (site III) to form the signaling complex. In the LIF:LIFR:gp130 complex, the roles are reversed. More extensive site III contacts are formed between LIF and LIFR that comprise predominantly the Ig-like domain and loops of CBM2 in a noncanonical manner. To form the signaling complex, gp130 binds LIF via its CBM, and there is possibly another contact between FNIII domains nearer the membrane.
Thus in the IL-6 family, cytokines can generate signaling complexes in two ways. For IL-6, IL-11, CT-1, or ciliary neurotrophic factor, the cytokine binds a CBM from its specific, nonsignaling receptor via site I, and then binds two gp130 molecules via sites II and III to initiate activation. For LIF or OSM, only two sites are needed, with the cytokine binding the Ig-like domain of LIFR (or OSMR) via site III and the gp130 CBM via site II. The similarity in the interaction between gp130 (Ig):IL-6 and LIFR:LIF suggest an evolutionary link between binding modes and, although the type of interaction has been reused, the size of the interface has been modified for tighter or weaker binding as required.
Materials and Methods
Purification of Recombinant LIF and LIF Receptor.
Purified recombinant hLIF expressed in Escherichia coli was kindly provided by Zenyth Therapeutics (Melbourne, Australia). Recombinant mLIFR was purified from the conditioned medium of Chinese Hamster Ovary cell line CHO-K1 transfected with the construct pCHO1/mLIFR. This construct encodes the extracellular domains D1-D5 of mLIFR (residues 7–486, mature peptide) with an N-terminal IL-3 secretion signal and FLAG epitope (DYKDDDDK). Transfected CHO-K1 cells were grown in a fermentation apparatus with a working volume of 1.6 liters (Celligen Plus fermenter, New Brunswick Scientific, Edison, NJ). Conditioned media were concentrated 10-fold by ultrafiltration and mLIFR was then purified by Lentil Lectin-Sepharose (Amersham) followed by immunoaffinity chromatography using anti-FLAG M2 agarose (Sigma, St. Louis, MO), and eluted with FLAG peptide before being concentrated with a 10,000 MWCO centrifugal concentrator (Millipore) and purified further by size-exclusion chromatography using a Superdex 200 column (Amersham, Piscataway, NJ). The hLIF:mLIFR complex was formed by mixing equimolar amounts of receptor and ligand and concentrated to 10 mg/ml with a 10,000 MWCO centrifugal concentrator (Millipore) before crystallization trials.
Crystallization, Data Collection, and Structure Determination.
Several initial crystallization conditions for the hLIF:mLIFR complex were identified by using commercial grid and sparse matrix screens (Hampton Research). The best crystals grew by hanging drop vapor diffusion by equilibrating a 1-μl drop of protein with 1 μl of reservoir solution containing 1.4–1.6 M sodium malonate (pH 8.0). Small propeller shaped crystals, that grew to a maximum size of ≈75 × 20 × 20 μm over 1–2 weeks, were harvested into a solution containing 10% extra precipitant, then transferred to a cryoprotectant solution further supplemented with 10% (vol/vol) glycerol. They were also subjected to limited dehydration by suspension over a reservoir containing stabilization solution supplemented with 40% glycerol, then mounted in a nylon loop and flash frozen directly into liquid nitrogen.
Diffraction data were collected from several crystals using a Mar225 Mosaic CCD detector at the SouthEast Regional Collaborative Access Team (SER-CAT) beamline 22-ID, Advanced Photon Source (APS, Argonne National Laboratory, Chicago, IL). The best crystal showed diffraction to 4-Å resolution. Data for this crystal were collected and processed with HKL2000 (27) (for statistics see SI Table 1). The crystal belongs to the space group C2221 with two molecules of LIF and two molecules of LIFR in the asymmetric unit. The presence of a LIF:LIFR dimer was consistent with both peaks in the self-rotation function and a calculated Matthews coefficient of 5.3 Å3/Da corresponding to a solvent content of ≈76% (using the molecular mass of 19.7 kDa for hLIF and ≈90 kDa for glycosylated mLIFR from SDS/PAGE).
Initial phases were obtained by molecular replacement using the program MOLREP (28) using only the coordinates of human LIF derived from the crystal structure of the gp130: LIF complex (1PVH) (16) as a search model. Phases were improved by using density modification with solvent flipping implemented in CNS (29). The subsequent density modified map was of sufficient quality to allow an initial polyalanine trace of both LIFR molecules (SI Fig. 5A). Model building was hugely assisted by using the topology of the known structures of Ig and fibronectin domains from IL-6Rα and gp130. Several rounds of map improvement and manual rebuilding using DMMULTI (30) and COOT (31) were then carried out using multiple NCS operators between individual hLIF and receptor domains (D1–D5).
Initially all data between 20.0 and 4.0 Å were used in all map calculations. After setting aside 5% of the data for cross validation, refinement was commenced with data 20–4.31 Å resolution and strict NCS-weighted restraints for each domain. Multiple cycles of torsional simulated annealing refinement using CNS (29) were followed by manual rebuilding with COOT (31), where the presence of conserved disulfide bonds and numerous N-linked glycosylation sites clearly seen in the initial CNS solvent flipped map facilitated progress (SI Fig. 5A).
Side chains for which no density was observed modeled as alanine. Further refinement of the structure using REFMAC5 (32), with TLS refinement and NCS restraints, converged with Rcryst = 0.248 and Rfree = 0.298. Refinement with additional (weaker) data to 4.0 Å resolution and some more carbohydrate, converged with Rcryst = 0.237 and Rfree = 0.287.
The asymmetric unit contains an artificial hLIF:mLIFR dimer comprising residues 1–180 of hLIF, residues 7–486 of mLIFR and 21–24 sugar residues per receptor molecule. Validation of the final structure was performed with PROCHECK (33) and WHATCHECK (34), the final model having good geometry for a low-resolution structure with few Ramachandran outliers. Differences in LIFR interdomain angles are <5° except for D2-D3, which is 13°.
Modeling of LIF:LIFR:gp130 Complex.
The coordinates of LIF for the gp130 (D2–D3):LIF complex (1PVH) (16) were superposed upon the LIF coordinates of 1 molecule from our structure using LSQMAN (35) (rmsd, 0.86 Å). Further superposition of the larger gp130 receptor fragment (1I1R) containing the D1–D3 domains was performed with an rmsd of 0.82 Å for the D2–D3 domains. The final model generated of the high affinity complex (Fig. 3) contains the coordinates of LIFR (D1–D5) from this work and coordinates of gp130 (D1–D3) and LIF coordinates from published structures. The N-terminal “flap” of LIF is involved in gp130 binding and is in an altered conformation in our structure, due to dimer crystal contacts; thus, the final LIF coordinates in the model are derived from 1PVH as this is likely to be more representative of the actual complex. None of the glycosylation sites interfered with construction of the model.
Supplementary Material
Acknowledgments
We thank Clare McFarlane, Bronwyn Roberts, Tracy Willson, Wendy Carter, Phillip Morgan, Jason Corbin and Seth Masters for their excellent technical assistance, George Loverecz and Louis Lu (Commonwealth Scientific and Industrial Research Organization) for CHO cell fermentation and Peter Colman for his structural insight and continuous support. We are indebted to Michael Gorman (Walter and Eliza Hall Institute) for data collection, Andy Howard (SER-CAT beamline ID22) for his assistance, Masayuki Tsuchiya (Chugai Pharmaceuticals, Shizuoka, Japan) for providing the parental pCHO1 vector, and Zenyth Therapeutics (Melbourne, Australia) for the gift of recombinant hLIF protein. We also thank Robbie Joosten (Center for Molecular and Biomolecular Informatics, Nijmegen, Netherlands) for discussions on structure validation. The Australian Synchrotron Research Program, the National Institutes of Health National Center for Research Resources, and the U.S. Department of Energy supported use of the BioCARS sector and Advanced Photon Source. This work was supported by Australian Nation Health and Medical Research Council Grants 257500 and 280912, National Institutes of Health Grant R01-CA22556, and the Cooperative Research Centre for Cellular Growth Factors.
Abbreviations
- LIF
leukemia inhibitory factor
- LIFR
LIF receptor
- mLIF
mouse LIF
- hLIF
human LIF
- OSM
oncostatin M
- OSMR
OSM receptor
- CBM
cytokine-binding module
- FNIII
fibronectin type III.
Footnotes
The authors declare no conflict of interest.
Data deposition: Atomic coordinates and structure factors for LIF:LIFR complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2Q7N).
This article contains supporting information online at www.pnas.org/cgi/content/full/0705577104/DC1.
References
- 1.Metcalf D. Stem Cells. 2003;21:5–14. doi: 10.1634/stemcells.21-1-5. [DOI] [PubMed] [Google Scholar]
- 2.Gearing DP, VandenBos T, Beckmann MP, Thut CJ, Comeau MR, Mosley B, Ziegler SF. Ciba Found Symp. 1992;167:245–255. doi: 10.1002/9780470514269.ch15. and discussion, 255–249. [DOI] [PubMed] [Google Scholar]
- 3.Zhang JG, Owczarek CM, Ward LD, Howlett GJ, Fabri LJ, Roberts BA, Nicola NA. Biochem J. 1997;325:693–700. doi: 10.1042/bj3250693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilton DJ, Nicola NA. J Biol Chem. 1992;267:10238–10247. [PubMed] [Google Scholar]
- 5.Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Kontgen F, Abbondanzo SJ. Nature. 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- 6.Ware CB, Horowitz MC, Renshaw BR, Hunt JS, Liggitt D, Koblar SA, Gliniak BC, McKenna HJ, Papayannopoulou T, Thoma B, et al. Development (Cambridge, UK) 1995;121:1283–1299. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- 7.Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Science. 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 8.Bravo J, Heath JK. EMBO J. 2000;19:2399–2411. doi: 10.1093/emboj/19.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stahl N, Boulton TG, Farruggella T, Ip NY, Davis S, Witthuhn BA, Quelle FW, Silvennoinen O, Barbieri G, Pellegrini S, et al. Science. 1994;263:92–95. doi: 10.1126/science.8272873. [DOI] [PubMed] [Google Scholar]
- 10.Ernst M, Gearing DP, Dunn AR. EMBO J. 1994;13:1574–1584. doi: 10.1002/j.1460-2075.1994.tb06420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timmermann A, Kuster A, Kurth I, Heinrich PC, Muller-Newen G. Eur J Biochem. 2002;269:2716–2726. doi: 10.1046/j.1432-1033.2002.02941.x. [DOI] [PubMed] [Google Scholar]
- 12.Hudson KR, Vernallis AB, Heath JK. J Biol Chem. 1996;271:11971–11978. doi: 10.1074/jbc.271.20.11971. [DOI] [PubMed] [Google Scholar]
- 13.Bitard J, Daburon S, Duplomb L, Blanchard F, Vuisio P, Jacques Y, Godard A, Heath JK, Moreau JF, Taupin JL. J Biol Chem. 2003;278:16253–16261. doi: 10.1074/jbc.M207193200. [DOI] [PubMed] [Google Scholar]
- 14.Plun-Favreau H, Perret D, Diveu C, Froger J, Chevalier S, Lelievre E, Gascan H, Chabbert M. J Biol Chem. 2003;278:27169–27179. doi: 10.1074/jbc.M303168200. [DOI] [PubMed] [Google Scholar]
- 15.Owczarek CM, Zhang Y, Layton MJ, Metcalf D, Roberts B, Nicola NA. J Biol Chem. 1997;272:23976–23985. doi: 10.1074/jbc.272.38.23976. [DOI] [PubMed] [Google Scholar]
- 16.Boulanger MJ, Bankovich AJ, Kortemme T, Baker D, Garcia KC. Mol Cell. 2003;12:577–589. doi: 10.1016/s1097-2765(03)00365-4. [DOI] [PubMed] [Google Scholar]
- 17.Varghese JN, Moritz RL, Lou MZ, Van Donkelaar A, Ji H, Ivancic N, Branson KM, Hall NE, Simpson RJ. Proc Natl Acad Sci USA. 2002;99:15959–15964. doi: 10.1073/pnas.232432399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinds MG, Maurer T, Zhang JG, Nicola NA, Norton RS. J Biol Chem. 1998;273:13738–13745. doi: 10.1074/jbc.273.22.13738. [DOI] [PubMed] [Google Scholar]
- 19.Robinson RC, Grey LM, Staunton D, Vankelecom H, Vernallis AB, Moreau JF, Stuart DI, Heath JK, Jones EY. Cell. 1994;77:1101–1116. doi: 10.1016/0092-8674(94)90449-9. [DOI] [PubMed] [Google Scholar]
- 20.Lawrence MC, Colman PM. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 21.de Vos AM, Ultsch M, Kossiakoff AA. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 22.Layton MJ, Owczarek CM, Metcalf D, Clark RL, Smith DK, Treutlein HR, Nicola NA. J Biol Chem. 1994;269:29891–29896. [PubMed] [Google Scholar]
- 23.Heymann D, Godard A, Raher S, Bentouimou N, Blanchard F, Cherel M, Hallet MM, Jacques Y. Cytokine. 1996;8:197–205. doi: 10.1006/cyto.1996.0028. [DOI] [PubMed] [Google Scholar]
- 24.Bravo J, Staunton D, Heath JK, Jones EY. EMBO J. 1998;17:1665–1674. doi: 10.1093/emboj/17.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skiniotis G, Boulanger MJ, Garcia KC, Walz T. Nat Struct Mol Biol. 2005;12:545–551. doi: 10.1038/nsmb941. [DOI] [PubMed] [Google Scholar]
- 26.Matadeen R, Hon WC, Heath JK, Jones EY, Fuller S. Structure (London) 2007;15:441–448. doi: 10.1016/j.str.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. New York: Academic; 1997. [DOI] [PubMed] [Google Scholar]
- 28.Vagin A, Teplyakov A. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 29.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Cowtan K. CCP4/ESF-EACBM Newslett Protein Crystallogr. 1994;31:34–38. [Google Scholar]
- 31.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 34.Hooft RW, Vriend G, Sander C, Abola EE. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 35.Kleywegt GJ, Jones TA. CCP4/ESF-EACBM Newslett Protein Crystallogr. 1994;31:9–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.