Abstract
Many DNA-modifying enzymes act in a manner that requires communication between two noncontiguous DNA sites. These sites can be brought into contact either by a diffusion-mediated chance interaction between enzymes bound at the two sites, or by active translocation of the intervening DNA by a site-bound enzyme. EcoP15I, a type III restriction enzyme, needs to interact with two recognition sites separated by up to 3,500 bp before it can cleave DNA. Here, we have studied the behavior of EcoP15I, using a novel fast-scan atomic force microscope, which uses a miniaturized cantilever and scan stage to reduce the mechanical response time of the cantilever and to prevent the onset of resonant motion at high scan speeds. With this instrument, we were able to achieve scan rates of up to 10 frames per s under fluid. The improved time resolution allowed us to image EcoP15I in real time at scan rates of 1–3 frames per s. EcoP15I translocated DNA in an ATP-dependent manner, at a rate of 79 ± 33 bp/s. The accumulation of supercoiling, as a consequence of movement of EcoP15I along the DNA, could also be observed. EcoP15I bound to its recognition site was also seen to make nonspecific contacts with other DNA sites, thus forming DNA loops and reducing the distance between the two recognition sites. On the basis of our results, we conclude that EcoP15I uses two distinct mechanisms to communicate between two recognition sites: diffusive DNA loop formation and ATPase-driven translocation of the intervening DNA contour.
Keywords: imaging, nucleic acid, restriction-modification enzyme, scanning-probe
The action of a number of DNA-modifying enzymes requires communication between two spatially separated sites on the DNA molecule (for a review, see ref. 1). Two main mechanisms are used to accomplish this task (2). The protein can loop the intervening DNA by 3D diffusion, either by specific dimerization with a second site-bound protein or by specific or nonspecific binding to an unoccupied second site. Alternatively, the protein can translocate the intervening DNA while maintaining a contact with the initial binding site on the DNA. In this case, the translocated DNA will be extruded into an expanding loop.
The probability of diffusive looping depends on the spacing between the two sites, the stiffness (persistence length) of the DNA and any bend that the protein induces in the DNA (3). In the case of a specific interaction between two sites or site-bound proteins, the size of the loop will be determined simply by the distance between the recognition sequences on the DNA. However, in the case of a nonspecific interaction, the loop will have a distribution of sizes that can be predicted by modeling the DNA as a semiflexible polymer (3). In the absence of protein-induced bending, the most probable loop size occurs at ≈3.5 persistence lengths, or 550 bp. Shorter loops are strongly disfavored because of enthalpic contributions, but larger loops are less strongly disfavored as a result of entropic contributions, leading to an asymmetric probability distribution. Single molecule techniques have been used to observe diffusive looping by type II restriction enzymes using laser tweezers (4), tethered-particle motion assays (5), and electron microscopy (6). Similar behavior, using conventional atomic force microscopy (AFM), has also been seen for the Gal repressor (7) and for RNA polymerase and a transcription factor during transcriptional activation (8).
To be able to form extruded loops by DNA translocation, a protein must possess a “motor” or helicase domain. The unwinding of the DNA is inherent to the function of helicases such as RNA polymerase (9), the mismatch repair MutS proteins (10) and HSV-1 UL9 protein (11, 12), and hence these helicases must track the DNA. However, in the case of restriction enzymes (13) and the chromatin remodelling Snf2 proteins (14), there is no functional requirement to unwind the DNA. For instance, EcoR124I, a type I restriction enzyme and a superfamily 2 helicase, has been shown to track the DNA double helix without substantial strand separation. Tracking of the DNA helix will cause the generation of negative supercoiling in the expanding loop, at a rate of one superhelical twist per helical repeat (10.5 bp for B-form DNA) and should be sufficient to block translocation (2). Some supercoiled structures have been observed in AFM images of EcoR124I (15) and in electron microscope images of EcoKI and EcoBI (16). However, not all loops exhibited supercoiling. Supercoiling will pose the largest barrier to translocation for small loops, because the superhelical density will be largest immediately after translocation initiation. It has been proposed that this barrier may be overcome by melting the DNA during translocation initiation (15), or by the formation of a (relaxed) diffusive loop before translocation, which will reduce the superhelical density (17). Translocation rates of up to 550 bp/s have been reported for type I enzymes, such as EcoR124I (18, 19).
EcoP15I is a type III restriction enzyme (and a superfamily 2 helicase) that requires two copies of its asymmetric recognition site to be oriented in a head-to-head manner to cleave DNA efficiently. EcoP15I was previously thought to function solely by ATPase-dependent DNA translocation with concomitant extrusion of loops (20, 21), however, several studies have reached apparently contradictory conclusions. The ability of resolvase to concatenate plasmids in the presence of a cleavage-deficient mutant EcoPI (an enzyme highly homologous to EcoP15I) was taken as evidence that no loops, either extruded or diffusive, were formed (22), but experiments using this technique also suggested that the type II enzyme NaeI did not form diffusive loops (23). This has since been shown to be incorrect by experiments using the single-molecule tethered-particle motion assay (5). The anomalously low rates of ATP hydrolysis by EcoP15I, in comparison with type I restriction enzymes, make it unlikely that the entire inter-site distance is translocated (21). However, translocation is still thought to be involved, because the presence of a either a lac repressor specifically bound between the two sites or a nonspecific HU protein blocks DNA cleavage by EcoP15I (21).
AFM is a technique that relies on the mechanical deflection of a single-point probe that is raster scanned over a surface to gather an image. Unfortunately, conventional AFM requires at least 30 s (often much longer) to acquire an image. The frame rate of the technique is limited by two factors: the rate at which the tip can be scanned across the surface before the onset of resonant instabilities, and the time response of the cantilever; that is, how fast the tip is able to follow the surface topography. These limitations can be overcome in two ways. The recently introduced VideoAFM (Infinitesima, Oxford, U.K.) incorporates a microresonant scan stage which uses the resonant motion of a quartz tuning fork to provide the fast-scan motion (24). Alternatively, the small cantilever approach involves miniaturizing the sample stage, hence increasing the resonant threshold, and using a stiff, miniaturized cantilever with a faster response time (25). We adopted the latter approach, using a modified version of the microscope described by Ando et al. (25), and produced by Olympus (Tokyo, Japan). This system, operated under fluid, allowed us to gather 1,000 × 600 nm images at 192 × 144 pixels with rates of up to 10 frames per s.
Previous AFM-based studies of protein-DNA dynamics have examined transcription (26), restriction by the type I enzyme EcoKI (27), the action of DNase I on modified DNA (28), and the operation of photolyase (29). In these studies, maximum frame rates varied between 0.5 and 2 frames per min, which severely limited the time-resolution of the experiments. The only study yielding quantifiable data were that of Guthold et al. (26), which relied on the use of rate-limiting concentrations of nucleoside triphosphates to reduce the RNA polymerase velocity to a rate of 1.5 bp/s, which was observable with the conventional AFM instrument.
In this study, fast-scan AFM imaging showed that EcoP15I uses both translocation (and hence extruded looping) and diffusive looping to achieve communication between two distal DNA sites. The observation of dynamic protein-DNA interactions reported here are the fastest yet observed by AFM. This paper demonstrates that technological improvements in AFM instrumentation have increased the temporal resolution 300-fold from at best ≈30 s to 100 ms. Our results raise the possibility that many other biological processes might be observed in real time with previously unattainable temporal and spatial resolution.
Results
Imaging of EcoP15I-DNA Complexes Using Fast-scan AFM.
To image the dynamics of an EcoP15I-DNA complex, the complex must be stably adsorbed to the surface to allow imaging by the scanning AFM tip; however, the complex must also retain enough mobility to allow the enzyme to function. The fast-scan instrument is able to capture an entire image in the time normally taken to scan a few lines by conventional AFM, and therefore the requirement for stable adsorption is relaxed. Additionally, the tip scans more quickly and so contacts any point for a shorter time. Hence, the work done in displacing an adsorbed bio-molecule by the applied tip-sample force will be decreased in proportion to the reduced contact time, thus further reducing the requirement for stable adsorption. These factors allowed us to obtain images of nearly freely diffusing DNA on mica under fluid without having to pretreat the mica surface. The tips used were electron beam deposited, to produce a hydrophobic surface that is thought to have a minimal interaction with DNA and proteins (26). The mobility of the DNA on the surface varied considerably between experiments, most likely because of the particular dynamics of the tip being used and surface variations.
A two-site DNA template would allow the rapid formation of a stable precleavage complex consisting of two EcoP15I multimers. In contrast, on a one-site template the enzyme is unable to communicate with another enzyme, and so continues to act on the DNA ad infinitum, enabling the observation of its behavior at any time-point after the addition of ATP. For this reason, the DNA template used in this and subsequent experiments is 2,249 bp long and has a single EcoP15I recognition site 105 bp from one end.
Fig. 1 shows a series of consecutive images captured 100 ms apart from a movie of an EcoP15I-DNA complex imaged at a frame rate of 10 frames per s. [For the complete movie, see supporting information (SI) Movie 1]. In this case, the EcoP15I has formed a loop of DNA, and lies at the apex of the loop; however the EcoP15I appears inactive, despite being supplied with ATP, as no changes in the complex are observed. The motion of the complex on the surface is approximately equal in both x and y directions, even though the scanning tip velocity is much greater in the x axis than in the y axis. This indicates that the scanning tip is having a minimal effect, and that the DNA motion is primarily diffusive in nature. It is also noteworthy that the tip used to take this image had previously captured 9,860 images of 1,000 × 600 nm, demonstrating that tip wear is less of an issue than might have been expected. For comparison, a conventional atomic force microscope, operating at 2 frames per min, would take >3 days to collect the same number of frames.
Fig. 1.
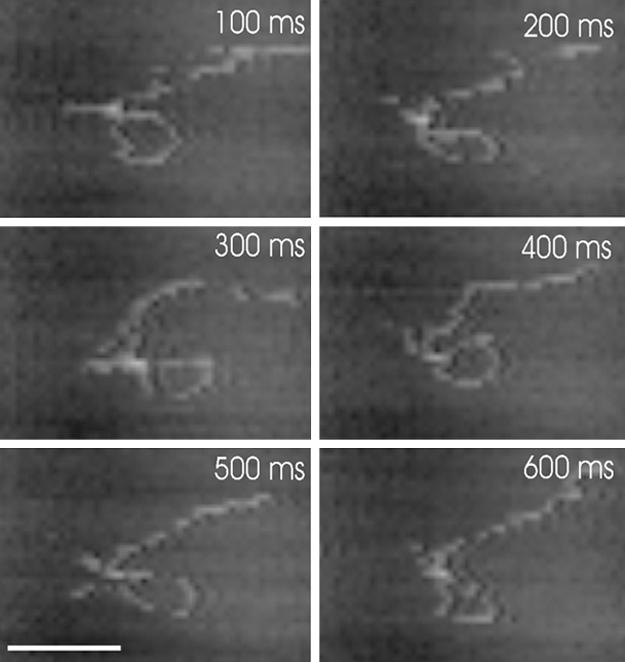
Consecutive time-lapse images of an EcoP15I-DNA complex obtained at 10 frames per s, using fast-scan AFM. The elapsed time is shown in each image. The image is cropped from an original scan size of 400 × 300 nm. (Scale bar, 100 nm.) See also SI Movie 1.
Translocation of DNA by EcoP15I.
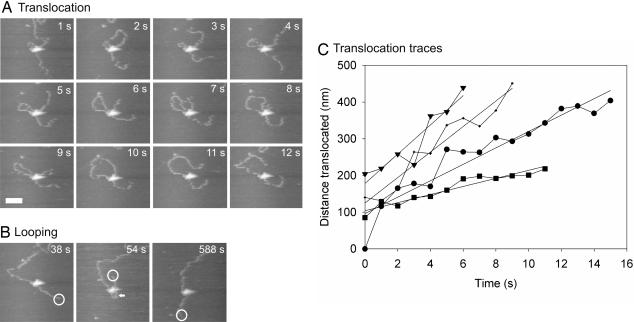
Fig. 2 shows a series of time-lapse AFM images of an EcoP15I-DNA complex that was observed to translocate, forming an extruded loop Fig. 2A), followed by the formation of a diffusive loop (Fig. 2B). (For the complete movie, see SI Movie 2). Note that the translocation does not initiate from the recognition site but from a region closer to the center of the DNA template. For this reason, we assume that the enzyme left its recognition site before the start of imaging. At the start of the imaging sequence, the enzyme began translocation, partitioning the translocated DNA into an expanding loop. The small loop present at t = 1 s was present from the start of the observation ≈1 min previously, so we cannot speculate on how it was formed. The loop expanded between t = 1 s and t = 10 s, before being released when the enzyme reached the end of the DNA template at t = 11 s. At t = 54 s, another loop was formed, before being released again at t = 588 s. This loop was ascribed to diffusive looping because the appearance of the loop was coupled with a large displacement of the DNA end (highlighted by the open circle at t = 38 s and t = 54 s), indicating that the DNA was severely bent by the formation of the loop and that the appearance of the loop was sudden. This result can be seen more clearly in SI Movie 2.
Fig. 2.
Translocation and extruded looping followed by diffusive looping by EcoP15I. Time-lapse images of an EcoP15I-DNA complex obtained at 1 frame per sec. The elapsed time is shown in each image. The image is cropped from an original scan size of 1,000 × 600 nm. (A) Translocation and formation of an extruded loop between 1 and 10 s, before release of the loop at 11 s. (Scale bar, 100 nm.) (B) Formation and release of a diffusive loop between 38 and 588 s. The loop is highlighted by an arrow. The presence of the loop can also be inferred from the large displacement of the DNA end, highlighted by an open circle. (C) Distance of DNA translocated by EcoP15I at various times, extracted from consecutive fast-scan AFM images. Each trace represents an independent experiment under identical conditions. The straight lines represent linear fits to each trace. See also SI Movie 2.
To demonstrate that the translocation events depicted in Fig. 2A were indeed due to uni-directional motion powered by ATP hydrolysis, the size of the expanding loop was measured at each time point for four individual complexes (Fig. 2C). Fitting each of these traces to a straight line gave translocation rates of between 32 and 117 bp/s, and an average translocation rate of 79 ± 33 bp/s, at a saturating ATP concentration of 1 mM. The variation in rates between experiments was likely due to variations in the specific tip dynamics and sample preparation. No translocation events were observed in the absence of ATP.
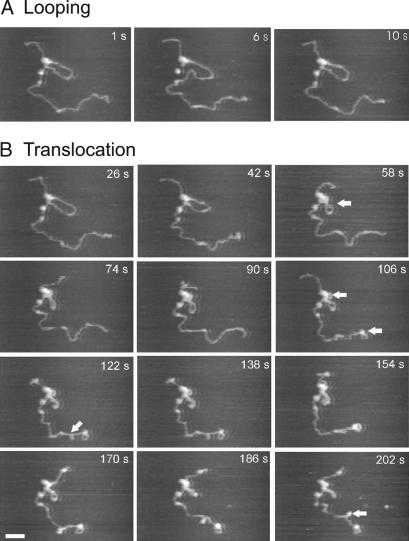
The translocation events illustrated in Fig. 2 appeared to occur when the DNA was free to move on the surface. This meant that supercoiling generated ahead of the translocating enzyme as a consequence of the enzyme tracking the DNA would have been dissipated from the nontethered DNA ends. In some cases, we observed complexes that appeared more tightly adsorbed to the surface, so that dissipation of supercoiling was blocked and translocation was inhibited either directly by the DNA-surface interaction, or indirectly by the accumulated supercoiling. Fig. 3 shows a series of time-lapse images of such a complex collected at 3 frames per s. (For the complete movie, see SI Movie 3). Notable features are indicated by the arrows. Initially the release and re-formation of a loop through diffusive looping were seen at t = 6 s and t = 10 s, respectively (Fig. 3A). During this process, the loop maintained a relaxed conformation. After loop re-formation, the loop became progressively more supercoiled between t = 26 s and t = 202 s (Fig. 3B). At t = 58 s the loop was seen to cross over itself as the increased negative supercoiling could no longer be contained through under-winding of the DNA helix and was instead partitioned in to writhe. One superhelical twist would be expected per helical repeat translocated (i.e., 10.5 bp or 3.5 nm). This was below the spatial resolution of the microscope, and thus the accumulation of moderate supercoiling need not be accompanied by translocation observable in the AFM images. Additionally, supercoiling accumulated in the linear DNA in the lower portion of the image presumably because the end of the DNA was firmly stuck on the surface. Of particular interest was the region of supercoiled DNA that formed a plectoneme at t = 122 s. We attribute this feature to supercoiling building up ahead of the enzyme, i.e., the enzyme was attempting to translocate along the longer section of DNA. Between t = 186 s and t = 202 s, the continued accumulation of positive supercoiling ahead of the enzyme (i.e., over-winding of the helix) caused the plectoneme to flip to the opposite side of the helix. This effect can be reproduced by winding a piece of flexible tubing in an anticlockwise fashion and watching the loop that is generated. A further control experiment (data not shown) performed immediately before the one illustrated in Fig. 3, using the same sample and tip, showed an inactive DNA-bound enzyme with no loop. In this case, no supercoiled structures were observed after prolonged scanning.
Fig. 3.
Diffusive looping followed by limited translocation by EcoP15I. Translocation was limited by a large DNA-surface interaction that prevents dissipation of accumulated supercoiling. Time-lapse images of EcoP15I-DNA complex obtained at 3 frames per s. The image is cropped from an original scan size of 1,000 × 600 nm. (A) Release and re-formation of a diffusive loop between 1 and 10 s. (B) Progressive accumulation of supercoiling as a result of translocation. Features of particular note are highlighted by arrowheads (see Results for details). (Scale bar, 100 nm.) See also SI Movie 3.
If both ends of an extruded DNA loop are held firmly by the enzyme then supercoiling buildup within the topologically constrained loop is expected irrespective of the DNA-surface interaction. However, comparison of Fig. 2A with Fig. 3B shows supercoiling in the expanding loop only when there is a strong DNA-surface, and presumably enzyme-surface, interaction. A possible explanation for this observation is that the enzyme has a mechanism to dissipate accumulated supercoiling and that the mechanism is blocked by a large surface interaction. This mechanism could allow one of the DNA contacts to swivel, transiently to release one of the DNA contacts, or to create a single-stranded nick in the DNA. Because EcoP15I has not been shown to possess a supercoiling activity in solution it seems likely such a mechanism may act to dissipate supercoiling in solution.
Dynamic Diffusive Looping of DNA by EcoP15I.
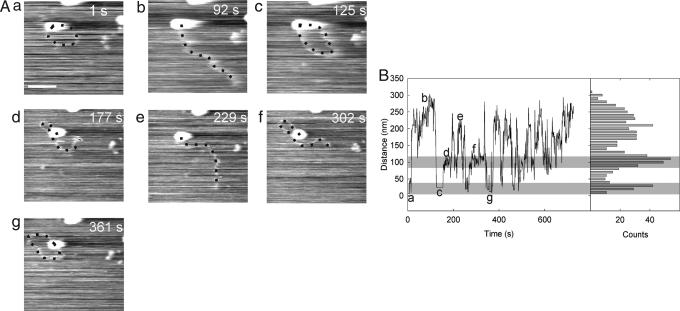
We next demonstrated that dynamic diffusive looping can be detected by fast-scan AFM (Fig. 4). An EcoP15I-DNA complex was observed bound close to the end of a DNA fragment, probably at the recognition site, which is located only 105 bp from one end (Fig. 4A). This complex was seen to form transient diffusive loops. The images were acquired at 1 frame per s. Because a number of consecutive images of a looped complex were required to distinguish a looped complex from a nonspecific association, we estimate a lower detection limit for loop lifetime of ≈5 s. Fig. 4B shows the variation with time of the distance between the center of the actively looping complex and the DNA end, for every frame collected in a 729 s interval (i.e., 729 images), together with a histogram of the measured distances. Inspection of the histogram revealed a tri-modal distribution. The peak at largest distance (≈ 250 nm) represents unlooped complexes, examples of which are shown in the images in Fig. 4 Ab and Ae. The peak at intermediate distance (≈100 nm) represents looped complexes in which EcoP15I associates with a nonspecific DNA sequence. The loops shown in the images in Fig. 4 Ad and Af had contour lengths of 480 bp and 550 bp, respectively. These compared favorably with the most probable loop size of 550 bp predicted by polymer theory (2). The model based on this theory treats the DNA as a homogeneous polymer and takes no account of topological constraints; hence, small loops, governed predominantly by the DNA stiffness, would always lie in a plane. For this reason, we could apply the 3D model to 2D looping on a surface. The peak at smallest distance (< 50 nm) represents the looped complex in which the EcoP15I associates with the other DNA end as shown in the images in Fig. 4 Aa, Ac, and Ag. In cases where the DNA end was not visible, the distance between the EcoP15I enzyme center and the exiting DNA arm was measured. Although statistics on the loop lifetime were limited by the temporal resolution of the technique, the lifetimes obtained in this particular experiment ranged between 12 and 119 s (n = 5).
Fig. 4.
Dynamic, diffusive looping by EcoP15I. (A) Time-lapse images of an EcoP15I-DNA complex obtained at 1 frame per s. The image is cropped from an original scan size of 600 × 400 nm. (Scale bar, 100 nm.) (B) For all 729 images, collected in a 729-s interval, the distance between the enzyme center and the end of the DNA was measured and is shown in the trace, along with the histogram. The two distinct looping states are seen as peaks in the histogram. The regions they correspond to in the trace are highlighted by gray rectangles. The letters in the trace refer to the corresponding AFM images.
Discussion
It has been observed in ensemble experiments that EcoP15I displays anomalously low rates of ATP hydrolysis, corresponding to a step size of many tens of bp per ATP molecule (20). The conclusion that EcoP15I is an extremely efficient motor appears to contradict the observation that EcoP15I is a member of the superfamily 2 helicases which usually hydrolyze ≈1 ATP/bp translocated. We suggest that this apparent paradox can be avoided if, when communicating between two DNA sites, EcoP15I only undergoes limited translocation in the manner of a conventional superfamily 2 helicase. The intervening DNA is also reduced by diffusive looping.
One aim of this paper was to determine the mechanism by which EcoP15I communicates between two noncontiguous DNA sites. The low rates of ATP hydrolysis suggest that translocation-coupled extruded looping cannot be the only mechanism and that diffusive DNA looping must also act to reduce the lengths of DNA that the enzymes must translocate. The single-molecule resolution of fast-scan AFM allows us to see that the same EcoP15I-DNA complex is capable of both looping via extrusion coupled to translocation and diffusive looping (Figs. 2 and 3). This raises questions about the order in which these two processes occur and the events that trigger them. The simplest model would involve the formation of a number of diffusive loops, followed by translocation of the reduced inter-site DNA distance. The number of diffusive loops formed would depend on the number of nonspecific DNA binding sites possessed by EcoP15I. On the basis of AFM images of EcoP15I-DNA complexes in air (data not shown) and the subunit stoichiometry of the enzyme, we believe this number to be two (20). Hence, a pair of enzymes will be capable of reducing the inter-site distance by four times the most probable loop size, or ≈2.2 kbp (4 × 550 bp). The remaining distance would then be covered by translocation of one or both enzymes. The initiation of translocation may require a large potential energy barrier to be overcome, making translocation initiation a slow process compared with diffusive looping. An attractive consequence of a model in which translocation follows diffusive looping is that the translocated DNA will be extruded into a preexisting relaxed loop, and hence the peak superhelical density will be much lower than if translocation initiation involved extrusion of a new loop (2). A similar model has been proposed for the type I enzyme EcoKI (16).
An average translocation velocity for EcoP15I of 79 ± 33 bp/s was obtained from our fast-scan AFM data. This value is most probably an underestimate of the true value because of the inhibitory affect of surface friction present in AFM experiments. Guthold et al. (26) estimated a frictional force of 1.5 pN would oppose an RNA polymerase that was pulling a 1,047-bp DNA fragment through itself with a translocation velocity of 0.5 bp/s. This estimate increases to 4 pN if the force required to both translate and rotate the DNA molecule on the mica surface is included (see Fig. 3 of ref. 26). This study used conventional AFM under identical buffer conditions to those used in this study (26). Guthold et al. (26) validated this value by comparing the expected translocation velocity from Michaelis–Menten kinetics with force-velocity measurements for RNA polymerase gained by using optical tweezers (9).
Using the diffusion coefficient obtained by Guthold et al. for DNA on mica under identical buffer conditions we can estimate the frictional force opposing translocation by EoP15I. The 2D diffusion coefficient, D, obtained by Guthold et al. for a 1,047-bp fragment of D = 1.4 ± 0.6 nm2/s can be scaled by using the scaling relationship D ≈ L−3/5 from the Zimm model (30), yielding D = 0.9 ± 0.3 nm2/s for the 2,249-bp DNA fragment used in our study. The measured translocation velocity is 79 ± 33 bp/s, which gives a frictional force of 115 ± 55 pN for linear motion only, and 330 ± 158 pN if rotational motion is included. These values appear particularly large. However, this simple model may overestimate the force as it does not take account of the complex DNA-mica interaction. The DNA and mica are both negatively charged in aqueous buffer, and the attractive interaction occurs through the correlation of adsorbed counterions surrounding the DNA contour and mica surface. If the DNA is moving fast enough to disrupt the counterion correlation then the frictional coefficient will be reduced and result in an overestimation of the force at large translocation velocities. Additionally the rapidly scanning AFM tip may also disrupt the interaction. For comparison the maximum force generation of EcoP15I is ≈240 pN. This is derived from the maximum force per ATP of 80 pN/nm and assumes a step size of 1 ATP/bp (31).
When DNA is stretched at a force of 65 pN, by tweezer methods, the DNA unwinds from B- to S-form (32). Whether this force is actually realized in our AFM experiments where a surface interaction is present is unclear. The presence of an attractive DNA-surface interaction is likely to prevent the unwinding of the DNA and hence raise the force at which this transition occurs.
This is the first study to measure the translocation velocity of EcoP15I, and no data exist on the force-velocity behavior of EcoP15I. However, it appears that EcoP15I is able to translocate against a large frictional force, which would imply an efficient coupling of the free energy of ATP hydrolysis to translocation. EcoR124I is a type I restriction enzyme also containing a superfamily 2 helicase domain; this enzyme has been shown to have a translocation velocity of 550 ± 30 bp/s against applied forces of up to 5 pN (the largest force used in the experiment) (19). We have no way of extrapolating our data to low force, however it seems likely that EcoP15I will achieve a substantially increased translocation velocity in the absence of a large friction force.
The data presented in Fig. 4 shows how fast-scan AFM can generate results similar to those obtained by using other single-molecule based techniques, such as tethered particle motion assays or tweezer experiments. Although these other methods are probably superior in terms of simplicity of experimental design and data analysis, fast-scan AFM has the great advantage that direct images are produced in addition to the conventional data trace. This means that distinct looping events (for example) can be identified not only by their particular data traces but also by the structure and morphology of the enzyme-DNA complexes determined from the corresponding AFM images. Our results on loop lifetime range between 12 and 119 s for EcoP15I, which is consistent with the distribution presented for the type II looping enzymes NaeI and NarI (5).
Application of fast-scan AFM to the type III restriction enzyme EcoP15I has provided new insights into the mechanisms underlying communication between two distal DNA sites. Based on our results, we believe that EcoP15I initially employs diffusive looping to reduce the inter-site DNA distance, followed by translocation-coupled extruded looping. This model circumvents the apparent paradox of why translocation is not blocked by supercoiling accumulated in the expanding loop: translocation does not partition DNA into a new loop but into a preexisting (relaxed) diffusive loop.
Materials and Methods
Fast-Scan AFM.
The fast-scan AFM system was based on the small cantilever apparatus of (25, 33). Small cantilevers, with dimensions (L×W×H) of 10 × 2×0.1 μm, were made from silicon nitride, using standard micromachining techniques (34). These cantilevers had a resonant frequency in water of 600–1,000 kHz and spring constants of 0.1–0.2 Nm−1, as determined by the thermal tune method (35). Typical free oscillation amplitudes during imaging were ≈4 nm, and the amplitude set-point was typically ≈70% of this value. A sharp probe was deposited on each cantilever, using electron beam deposition by Nanotools (Munich, Germany).
Sample Preparation.
The restriction endonuclease EcoP15I was purified, and its activity assayed, as described in ref. 36. A 2,249-bp DNA fragment containing a single EcoP15I recognition site 105 bp from one end was generated by digesting plasmid pTYB1 (New England Biolabs, Hitchin, U.K.) with SfoI and EcoRI. The fragment was gel purified and extracted by using a Qiaquick extraction kit (Qiagen, Crawley, West Sussex, U.K.). EcoP15I-DNA complexes were formed by incubating EcoP15I with 35 ng of DNA at a 1:1 recognition site:enzyme ratio in buffer (5 mM MgCl2/5 mM KCl/5 mM Tris·HCl, pH 7.9) supplemented with 80 μM S-adenosyl methionine in a final volume of 20 μl. After 2.5 min, ATP (1 mM; Sigma–Aldrich, Poole, Dorset, U.K.) was added and the reaction mix (3 μl) was immediately deposited onto 1 mm2 mica sample discs and incubated for 2 min. The sample was rinsed with 3 × 10 μl washes of buffer supplemented with 1 mM ATP, before being imaged in buffer, also supplemented with 1 mM ATP.
Image Analysis.
Video files were manipulated and compressed by using the open source program Virtual Dub (www.virtualdub.org) and Microsoft (Redmond, WA) video 1 codec. Individual frames were imported into ImageJ (http://rsb.info.nih.gov/ij/) and analyzed.
Supplementary Material
Acknowledgments
We thank Toshio Ando and members of the Olympus Corporation for helpful discussion and much technical advice. This work was supported in the United Kingdom by Biotechnology and Biological Sciences Research Council Grant BBS/B/1065X (to R.M.H., J.M.E., and D.T.F.D.) and in Japan by the Special Co-ordination Funds, the COE Research Grant and the Basic Research Grant (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Measurement and Intellectual Infrastructure Improvement Project for Small and Medium Enterprise from the Ministry of Economy, Trade and Industry of Japan. N.C. acknowledges travel support from the Biotechnology and Biological Sciences Research Council International Scientific Interchange Scheme.
Abbreviation
- AFM
atomic force microscopy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700483104/DC1.
References
- 1.Matthews KS. Microbiol Rev. 1992;56:123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halford SE, Welsh AJ, Szczelkun MD. Annu Rev Biophys Biomol Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]
- 3.Rippe K. Trends Biochem Sci. 2001;26:733–740. doi: 10.1016/s0968-0004(01)01978-8. [DOI] [PubMed] [Google Scholar]
- 4.Gemmen GJ, Millin R, Smith DE. Nucleic Acids Res. 2006;34:2864–2877. doi: 10.1093/nar/gkl382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Broek B, Vanzi F, Normanno D, Pavone FS, Wuite GJL. Nucleic Acids Res. 2006;34:167–174. doi: 10.1093/nar/gkj432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topal MD, Thresher RJ, Conrad M, Griffith J. Biochemistry. 1991;30:2006–2010. doi: 10.1021/bi00221a038. [DOI] [PubMed] [Google Scholar]
- 7.Virnik K, Lyubchenko YL, Karymov MA, Dahlgren P, Tolstorukov MY, Semsey S, Zhurkin VB, Adhya S. J Mol Biol. 2003;334:53–63. doi: 10.1016/j.jmb.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 8.Rippe K, Von Hippel PH, Langowski J. Trends Biochem Sci. 1995;20:500–506. doi: 10.1016/s0968-0004(00)89117-3. [DOI] [PubMed] [Google Scholar]
- 9.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 10.Jiang JJ, Bai L, Surtees JA, Gemici Z, Wang MD, Alani E. Mol Cell. 2005;20:771–781. doi: 10.1016/j.molcel.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Makhov AM, Boehmer PE, Lehman IR, Griffith JD. EMBO J. 1996;15:1742–1750. [PMC free article] [PubMed] [Google Scholar]
- 12.Makhov AM, Lee SSK, Lehman IR, Griffith JD. Proc Natl Acad Sci USA. 2003;100:898–903. doi: 10.1073/pnas.0237171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanley LK, Seidel R, van der Scheer C, Dekker NH, Szczelkun MD, Dekker C. EMBO J. 2006;25:2230–2239. doi: 10.1038/sj.emboj.7601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasad TK, Robertson RB, Visnapuu ML, Chi P, Sung P, Greene EC. J Mol Biol. 2007;369:940–953. doi: 10.1016/j.jmb.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Noort J, van der Heijden T, Dutta CF, Firman K, Dekker C. Nucleic Acids Res. 2004;32:6540–6547. doi: 10.1093/nar/gkh999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endlich B, Linn S. J Biol Chem. 1985;260:5720–5728. [PubMed] [Google Scholar]
- 17.Yuan R, Hamilton DL, Burckhardt J. Cell. 1980;20:237–244. doi: 10.1016/0092-8674(80)90251-2. [DOI] [PubMed] [Google Scholar]
- 18.Firman K, Szczelkun MD. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seidel R, Van Noort J, Van der Scheer C, Bloom JGP, Dekker NH, Dutta CF, Blundell A, Robinson T, Firman K, Dekker C. Nat Struct Mol Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 20.Janscak P, Sandmeier U, Szczelkun MD, Bickle TA. J Mol Biol. 2001;306:417–431. doi: 10.1006/jmbi.2000.4411. [DOI] [PubMed] [Google Scholar]
- 21.Meisel A, Mackeldanz P, Bickle TA, Kruger DH, Schroeder C. EMBO J. 1995;14:2958–2966. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peakman LJ, Szczelkun MD. Nucleic Acids Res. 2004;32:4166–4174. doi: 10.1093/nar/gkh762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milsom SE, Halford SE, Embleton ML, Szczelkun MD. J Mol Biol. 2001;311:515–527. doi: 10.1006/jmbi.2001.4893. [DOI] [PubMed] [Google Scholar]
- 24.Humphris ADL, Miles MJ, Hobbs JK. Appl Phys Lett. 2005;86 034106. [Google Scholar]
- 25.Ando T, Kodera N, Takai E, Maruyama D, Saito K, Toda A. Proc Natl Acad Sci USA. 2001;98:12468–12472. doi: 10.1073/pnas.211400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guthold M, Zhu XS, Rivetti C, Yang GL, Thomson NH, Kasas S, Hansma HG, Smith B, Hansma PK, Bustamante C. Biophys J. 1999;77:2284–2294. doi: 10.1016/S0006-3495(99)77067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis DJ, Dryden DTF, Berge T, Edwardson JM, Henderson RM. Nat Struct Biol. 1999;6:15–17. doi: 10.1038/4882. [DOI] [PubMed] [Google Scholar]
- 28.Abdelhady HG, Allen S, Davies MC, Roberts CJ, Tendler SJB, Williams PM. Nucleic Acids Res. 2003;31:4001–4005. doi: 10.1093/nar/gkg462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Noort SJT, van der Werf KO, Eker APM, Wyman C, de Grooth BG, van Hulst NF, Greve J. Biophys J. 1998;74:2840–2849. doi: 10.1016/S0006-3495(98)77991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith DE, Perkins TT, Chu S. Macromolecules. 1996;29:1372–1373. [Google Scholar]
- 31.Block SM. Trends Cell Biol. 1995;5:169–175. doi: 10.1016/s0962-8924(00)88982-5. [DOI] [PubMed] [Google Scholar]
- 32.Cluzel P, Lebrun A, Heller C, Lavery R, Viovy JL, Chatenay D, Caron F. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 33.Yokokawa M, Wada C, Ando T, Sakai N, Yagi A, Yoshimura SH, Takeyasu K. EMBO J. 2006;25:4567–4576. doi: 10.1038/sj.emboj.7601326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitazawa M, Shiotani K, Toda A. Jpn J Appl Phys, Part 1. 2003;42:4844–4847. [Google Scholar]
- 35.Hutter JL, Bechhoefer J. Rev Sci Instrum. 1993;64:1868–1873. [Google Scholar]
- 36.Raghavendra NK, Rao DN. Nucleic Acids Res. 2004;32:5703–5711. doi: 10.1093/nar/gkh899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.