Abstract
In natural photosynthesis, the two photosystems that operate in series to drive electron transport from water to carbon dioxide are quite similar in structure and function, but operate at widely different potentials. In both systems photochemistry begins by photo-oxidation of a chlorophyll a, but that in photosystem II (PS2) has a 0.7 eV higher midpoint potential than that in photosystem I (PS1), so their electronic structures must be very different. Using reaction centers from 15N-labeled spinach, these electronic structures are compared by their photochemically induced dynamic nuclear polarization (photo-CIDNP) in magic-angle spinning (MAS) NMR measurements. The results show that the electron spin distribution in PS1, apart from its known delocalization over 2 chlorophyll molecules, reveals no marked disturbance, whereas the pattern of electron spin density distribution in PS2 is inverted in the oxidized radical state. A model for the donor of PS2 is presented explaining the inversion of electron spin density based on a tilt of the axial histidine toward pyrrole ring IV causing π-π overlap of both aromatic systems.
Keywords: photosynthesis, photosystem I, solid-state NMR, electron transfer, redox potential
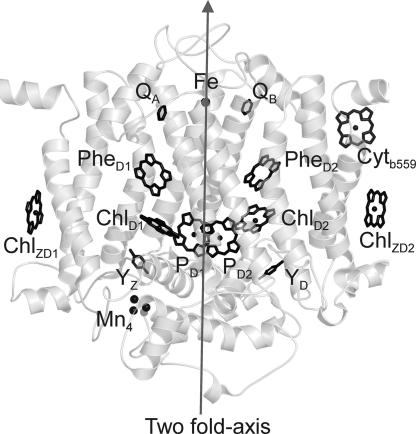
In photosynthesis, the two photosystems that operate in series to drive electron transport from water to carbon dioxide are similar in structure and function, but operate at widely different potentials. In both photosystems, photochemistry begins by photo-oxidation of a chlorophyll a (Chl). The oxidized electron donor of photosystem II (PS2) is the strongest oxidizing agent known in living nature, having a redox potential of + 1.2V (1), required for water oxidation. The electronically excited donor of photosystem I (PS1), probably the most reducing compound in living nature (2), initiates the dark reaction. The question arises what factors tune those electronic properties. The spatial structure of PS2 (Fig. 1) shows two inner Chls (PD1 and PD2), two accessory Chls (ChlD1 and ChlD2), two pheophytin a (Phe) cofactors and two quinones in an arrangement similar to that in bacterial reaction centers (RCs) of purple bacteria.
Fig. 1.
Spatial arrangement of cofactors in PS2. Schematic structure of the PSII core complex of Thermosynechococcus elongatus (3) obtained with the program PYMOL (DeLano Scientific, South San Francisco, CA) is shown. On the donor side, two Chls (PD1 and PD2) and two accessory Chls (ChlD1 and ChlD2) are localized. At the acceptor side, two Phe cofactors and two quinones are localized. The cofactors are nearly arranged in a C2 symmetry perpendicular to the membrane plane as indicated by the axis.
Photochemically induced dynamic nuclear polarization (photo-CIDNP) magic-angle spinning (MAS) NMR is an optical solid-state NMR method using the high electron polarization in the correlated electron pair and allows for strong increase of sensitivity and selectivity (4, 5) allowing to study the electronic structure of photosynthetic cofactors in great detail. The solid-state photo-CIDNP effect, discovered in 1994 (6), relies on different mechanisms called three-spin mixing (7), differential decay (8), and differential relaxation (9, 10). Recently, the contribution of these three mechanisms has been analyzed by field-dependent measurements on unlabeled RCs of the purple bacterium Rhodobacter sphaeroides (11, 12). The chemical shift refers to the electronic ground-state after the photocycle, the photo-CIDNP signal intensity is linked to the intermediate radical state. Analytical expressions imply that for short lifetimes of the radical pair the nuclear polarization created by the electron–electron–nuclear three spin mixing mechanism is proportional to the square of the pseudosecular hyperfine coupling and thus to the square of the hyperfine anisotropy (13). The hyperfine anisotropy in turn is approximately proportional to the spin density ρp in the 2pz orbital on the aromatic atom under consideration. In related work, we have performed numerical computations considering both the three spin mixing and differential decay mechanisms (11) of 13C photo-CIDNP for eight carbon atoms of both the donor and acceptor in bacterial RCs. At a field of 4.7 T, we found that the polarization Δp0 generated by a single photocycle is given by Δp0 = (−0.216 ± 0.086)ρp2 (41). We thus expect that also for 15N in plant RCs the amplitude of signals of the same cofactor is approximately proportional to the square of the spin density in the pz orbital of each aromatic atom. Until now, plant photosystems had been investigated only by 13C photo-CIDNP MAS NMR and without isotope enrichment (14–16). As origin of the high redox power in PS2, a local electrostatic field causing the asymmetric electron distribution has been proposed (14), however, the origin of this field remained unclear. The possibilities of local matrix involvement (16) and of general charge effects of the D1D2 protein on all inner Chls (17) have been discussed. The 13C signal patterns of unlabeled RCs are complex and difficult to interpret. On the other hand, 15N photo-CIDNP MAS NMR of isotope labeled RCs can be interpreted straightforwardly, as each signal corresponds to one of the four pyrrole subunits. Until now, studies of plant RCs were hampered by the difficulty to label plants. Here we present the 15N photo-CIDNP MAS NMR data of uniformly 15N labeled plant RCs of spinach (Spinacia oleracea).
Results and Discussion
15N Photo-CIDNP MAS NMR on PS1.
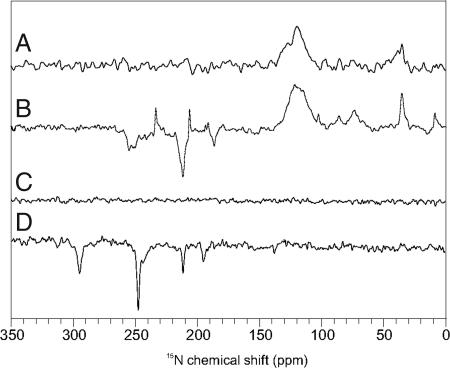
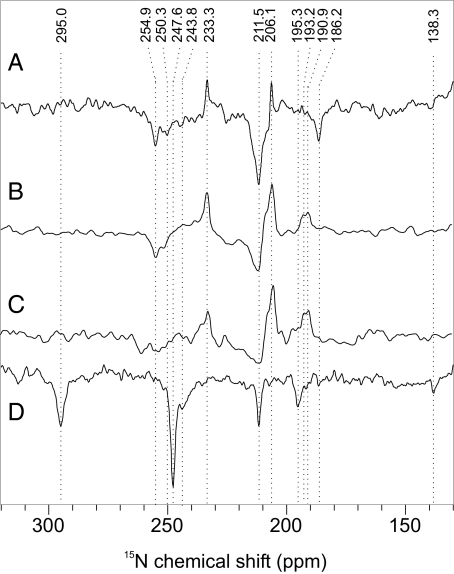
Spectrum A in Fig. 2 has been obtained from PS1 in the dark. Only absorptive (positive) signals occur, a broad hump between 120 and 130 ppm from the protein backbone as well as some weak features <100 ppm which can be assigned to well-known signals of arginine and lysine residues (18). Under illumination (Fig. 2, spectrum B), enhanced absorptive (positive) and emissive (negative) signals occur. The ratio of enhanced absorptive and emissive signals depends on the magnetic field (Fig. 3, spectra A and B). Both the two emissive as well as the enhanced absorptive set of signals can be assigned conveniently to Chl cofactors (Table 1) based on assignment obtained on Chls in solution (19). At the donor site, which has been shown to be very rigid (15, 20), slow signal recovery is expected. Hence, an experiment with very short cycle delay of 0.4 s (Fig. 3, spectrum C), showing a strong decay of the emissive compared with the enhanced absorptive signals, suggests that the emissive signals originate from the donor Chl. The emissive signal at 211.5 ppm is broadened by a shoulder at ≈215 ppm (Table 1), indicating involvement of a second donor Chl cofactor, which can be due to the epimerization at the C-132 and the different hydrogen-bonding at the 131-keto group (21), causing also the split of the N-IV signal (250.3 and 254.9 ppm), whereas no effect is observed on the remote N-I position (186.2 ppm). The resonance of N-III is observed at 193.2 ppm. The four absorptive signals (Fig. 3, spectrum C) can be assigned to a single Chl cofactor, probably the primary electron acceptor A0 (15). The enhanced absorptive signal at 233.3 ppm indicates a strong local disturbance at a pyrrole ring IV in the electronic ground state. On the other hand, the intensity patterns demonstrate that PS1 in the radical-pair state is assembled by Chl cofactors with nuclear polarization patterns similar to that of isolated Chls having their maximum on pyrrole ring II (22–24) (Fig. 4A). Hence, the assignment of the emissive signals to the donor implies a dimeric donor having an asymmetric electron spin distribution between two undisturbed Chl cofactors, as it also has been found with ENDOR (25–27) and by quantum-chemical calculations (28).
Fig. 2.
15N MAS NMR spectra of PS1 and PS2. Overview spectra of PS1 (A, dark; B, light) were obtained at 9.6 T (400 MHz proton frequency). Spectra of PS2 (C, dark; D, light) were measured at 4.7 T (200 MHz proton frequency). PS1–110 and D1D2 sample preparations were studied at 240 K and with a cycle delay of 4 s.
Fig. 3.
15N photo-CIDNP MAS NMR spectra of PS1 and PS2. Detailed spectra of PS1 (A–C) and PS2 (D) obtained at 9.6 T (A) and 4.7 T (B–D). Cycle delay was 4 s (A, B, and D) and 0.4 s (C).
Table 1.
15N chemical shifts of the photo-CIDNP signals in comparison with published chemical shift data
| Assignment |
Solution data |
PS1 |
PS2 |
|
|---|---|---|---|---|
| Cofactor | Atom | σliq* | σsolid† | σsolid† |
| Chl a | N-I | 186.0 | 186.2 (e) | |
| 190.9 (a) | ||||
| N-II | 206.5 | 206.1 (a) | 211.5 (e) | |
| 211.5 (e) | ||||
| N-III | 189.4 | 193.2 (a) | 195.3 (e) | |
| N-IV | 247.0 | 233.3 (a) | 247.6 (e) | |
| 250.3 (e) | ||||
| 254.9 (e) | ||||
| Phe a | N-I | 125.5 | — | |
| N-II | 241.5 | — | ||
| N-III | 133.9 | — | 138.3 (e) | |
| N-IV | 295.8 | — | 295.0 (e) | |
All chemical shifts are referenced to liquid ammonia with use of an external standard of solid 15NH4NO3 (δ = 23.5). a, absorptive (positive); e, emissive (negative).
*Chemical shift in ppm. Measured in CDCl3. Source: ref. 19.
†Chemical shift in ppm. Source: this work.
Fig. 4.
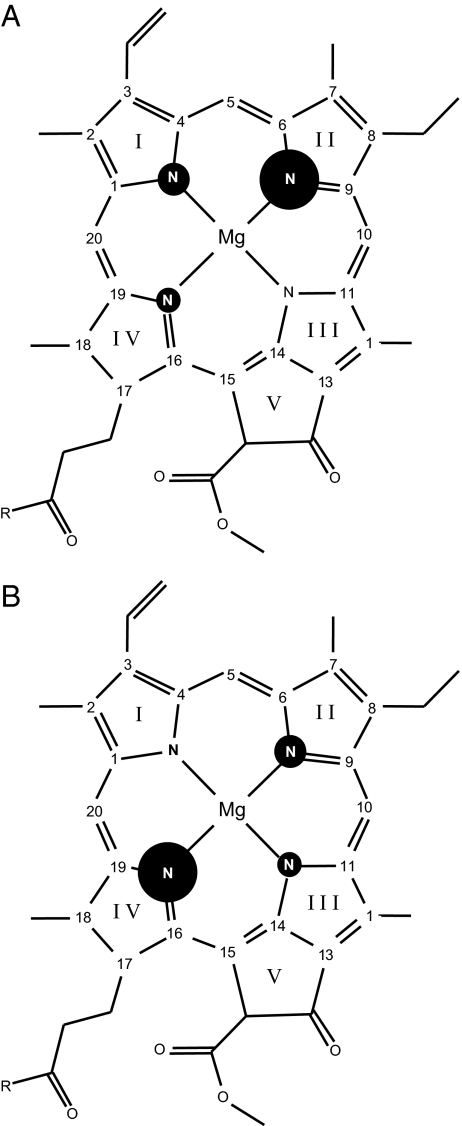
Electron spin density patterns. Based on the 15N photo-CIDNP intensities, electron spin density pattern of the donor cofactors of PS1 (A) and PS2 (B) are shown. The size of the circles refers to the relative signal intensity.
15N Photo-CIDNP MAS NMR on PS2.
In the dark, due to low concentration, no signal is detected from the PS2 sample (Fig. 2, spectrum C). Upon illumination, several emissive signals appear (Fig. 2, spectrum D, and Fig. 3, spectrum D). The pattern demonstrates clearly that the radical pair is formed by a Chl and a Phe having well separated signals (Table 1). The strongest signal observed in PS2 originates from pyrrole nitrogen N-IV (247.6 ppm). Two further signals of the Chl donor cofactor are detected, at 211.5 (N-II) and 195.3 (N-III) ppm. No unequivocal signal is obtained from N-I. The intensity ratio between the three light-induced Chl signals shows a strong asymmetry of electron spin density, which appears to be shifted toward the pyrrole ring IV. This is in line with the strong asymmetry of electron spin density detected previously by 13C photo-CIDNP MAS NMR, demonstrating maximum electron spin density at the neighboring C-15 methine carbon (14). Hence, there is a good agreement between photo-CIDNP data obtained from both types of nuclei. Thus, in the donor of PS2 the electron spin density pattern is inverted compared with the donor and acceptor cofactors in PS1 as well as to isolated Chl cofactors (Fig. 4B). On the other hand, there is no indication for a significant disturbance of the electronic ground state. Therefore, the change in the electronic structure seems to be restricted to the photo-oxidized state. Two more signals appear at 295.0 and 138.3 ppm, which can be conveniently assigned to N-IV and N-III of the primary electron acceptor, a Phe cofactor. The donor signals are remarkably narrow [full width at half-height (FWHH) of ≈40 Hz], whereas the acceptor signals are slightly broader (≈70 Hz), indicating a general feature of photosynthetic RCs having a rigid donor site without structural heterogenities (15, 20) and more structural flexibility at the acceptor site.
Matrix Involvement.
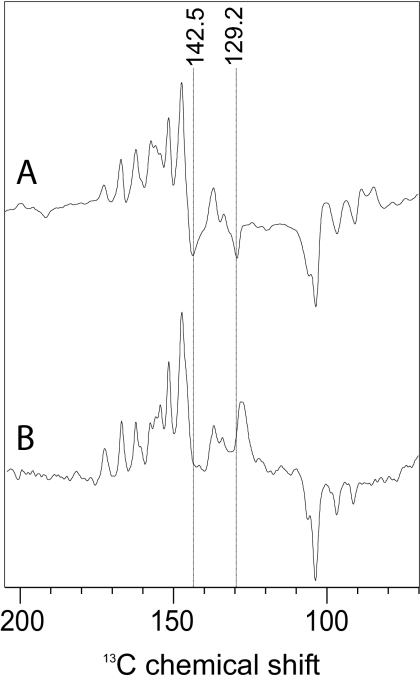
In Fig. 3, spectrum C, a further signal arises at 243.8 ppm. It is possible that this signal arises from a second Chl cofactor having much lower electron spin density. On the other hand, there is no further indication in this spectrum or in the 13C photo-CIDNP MAS NMR data (14, 16) for involvement of a second donor Chl cofactor. Furthermore, the signal is clearly broader (FWHH of 90–100 Hz) than the other signals assigned to the Chl donor. This indicates that another, structurally more flexible unit close to the Chl donor cofactor also carries electron spin density. Hence, an involvement of the protein matrix has to be considered. An assignment to a protonated Schiff base nitrogen (29), as discussed as a chemical modification of the donor Chl (16), is not convincing. The chemical shift value is also difficult to reconcile to any aromatic amino acid other than histidine. In fact, a nitrogen N-π of a Type-1 histidine (i.e., carrying a lone pair at the π-position) resonates at ≈250 ppm (30). With 13C photo-CIDNP MAS NMR at 9.6 T, three emissive signals at 142.5, 139.8, and 129.2 ppm have been detected (16) that also match to a Type-1 histidines (30). As in the 15N data, these three signals have approximately a double FWHH as the Chl signals. The signals at 142.5 and 129.2 ppm are also observed at 4.7 T (Fig. 5, spectrum A), but disappear upon sample orientation (Fig. 5, spectrum B). This observation suggests that these signals originate from a π-system having orientation different as the Chl donor, which has an orientation which does not affect the intensity pattern. The nitrogen N-τ in Type-1 histidines can be either bound to the Mg of the Chl or protonated (30). On basis of the observed chemical shifts, we are not able to distinguish whether the histidine carrying electron spin density is the axial one. On the other hand, an analysis of the x-ray structure (31) does not provide any possible candidate of a nonaxial histidine in the pocket of one of the four central Chl cofactors. Thus, we propose that the electron spin density is distributed over both the donor Chl and its axial histidine. Hence, the reduction of spin density, observed by EPR and originally interpreted in terms of a weakly coupled dimer having ≈82% of the spin density on one Chl cofactor (25), could be explained by the model presented here implying that the rest of the spin density are localized on the axial histidine. Because the two accessory Chls, ChlD1 and ChlD2, are not coordinated to histidines (32), the donor must be an inner Chl, either PD1 or PD2. This conclusion is in line with results of previous pulse EPR studies (33–35).
Fig. 5.
13C photo-CIDNP MAS NMR spectra of oriented and nonoriented PS2. Spectra of PS2 in nonoriented (A) and oriented (B) samples obtained at 4.7 T are shown. Cycle delay was 4 s. Orientation of the membrane normal parallel to rotor axis in the MAS experiment.
Hinge Model of the Donor of PS2.
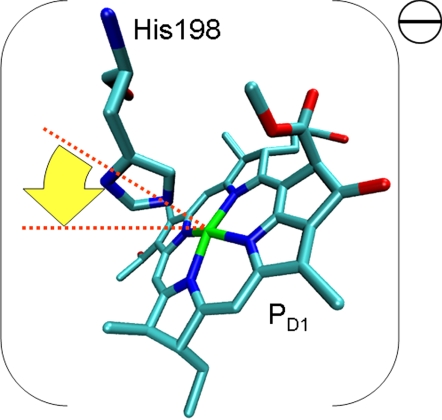
Assuming the signals arise from a Type-1 histidine having a deprotonated π-position, the donor would be a negatively charged [Chl-His]− complex in the ground-state, and a neutral radical in the photo-oxidized state. We propose a hinge-type model for the donor complex unifying those aspects (Fig. 6). Slight bending of the axial histidine toward pyrrole ring IV and the methine bridge C-15 would lead to π-π overlap of both conjugated systems and stabilize the negatively charged electronic ground state of the complex. In the hinge model, the Chl-His distance may be modulated depending on the redox state. Such lowering of the electronic ground state would increase the redox potential. Preliminary density functional computations indicate that such a small tilt indeed causes both a shift of spin density into pyrrole ring IV and some distribution of spin density into the aromatic ring of the histidine itself. We could not yet obtain quantitative agreement of the relative experimental spin densities. Systematic tests are tedious as both the orientation of the histidine ring with respect to the coordinating nitrogens and the tilt angle need to be varied and each structure has to be optimized with respect to deformations of the Chl macrocycle.
Fig. 6.
The hinge model of the electron donor in PS2. Because of the tilt of the axial histidine toward pyrrole ring IV (arrow), the electron spin density pattern is inverted, and electron spin density is partially shifted on the axial histidine.
Nevertheless, the hinge model of the electron donor in PS2 can at least in principle explain the observed inversion of the pattern of electron spin density distribution and the spin density on a histidine. It is well known from metalloporphin macrocycles to conserve its shape during evolution if it is of functional relevance (36). It appears that such functional conservation principle has to be extended to functional cofactor-matrix units.
Materials and Methods
Sample Preparation.
Spinach plants were cultured on half-strength Gamborg's B5 basal media. (15NH4)2SO4 and K15NO3 were used as the source of isotope labeled nitrogen. For preparation of PS1 (PS1–110) and PS2 (D1D2-cytb559), see refs 14 and 15.
PS2 RCs were incorporated into L-α-phosphatidylcholine (egg, chicken; Avanti Polar Lipids, Inc., Alabaster, AL) bilayers. The lipid/protein weight ratio was ≈1:1. Drops of ≈22 μl containing ≈0.38 mg of protein were spread onto round, 0.01-mm thin glass plates with a diameter of 5.4 mm (Marienfeld GmbH, Lauda-Königshofen, Germany) according to a described procedure (37, 38). The disks were mounted into clear 7-mm sapphire MAS rotors and rehydrated.
MAS NMR Measurements.
The NMR experiments were performed on a DMX-200 NMR and a DMX-400 NMR spectrometer (Bruker Biospin GmbH, Karlsruhe, Germany). The illumination setup is described elsewhere (5, 39). The light and dark spectra were collected with a Hahn echo pulse sequence and two pulse phase modulation proton decoupling (40). All NMR spectra were obtained at a temperature of 240 K and at a spinning frequency of 8 kHz. Each spectrum was measured within 2 days. Chemical shifts are given relative to liquid 15NH3, by using the response of solid 15NH3NO4 at δ = 23.5 ppm as reference.
Acknowledgments
We thank F. Lefeber, J. G. Hollander, and K. Erkelens for their help. We also thank A. de Wit and W. van der Meer for their help in sample preparation. This work was supported by Volkswagen-Stiftung Grant I/78010 (to C.G. and J.M.) as well as by Netherlands Organization for Scientific Research (NWO) through Jonge Chemici Award 700.50.521, Open Competition Grant 700.50.004, and Vidi Grant 700.53.423 (to J.M.).
Abbreviations
- Chl
chlorophyll
- Phe
pheophytin
- FWHH
full width at half-height
- MAS
magic-angle spinning
- photo-CIDNP
photochemically induced dynamic nuclear polarization
- PS1
photosystem I
- PS2
photosystem II
- RC
reaction center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.van Gorkom HJ, Schelvis JPM. Photosynth Res. 1993;38:297–301. doi: 10.1007/BF00046753. [DOI] [PubMed] [Google Scholar]
- 2.Webber AN, Lubitz W. Biochim Biophys Acta. 2001;1507:61–79. doi: 10.1016/s0005-2728(01)00198-0. [DOI] [PubMed] [Google Scholar]
- 3.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 4.Jeschke G, Matysik J. Chem Phys. 2003;294:239–255. [Google Scholar]
- 5.Daviso E, Jeschke G, Matysik J. In: Biophysical Techniques in Photosynthesis II. Aartsma TJ, Matysik J, editors. Dordrecht: Springer; 2007. pp. 385–399. Chap 19. [Google Scholar]
- 6.Zysmilich MG, McDermott A. J Am Chem Soc. 1994;116:8362–8363. [Google Scholar]
- 7.Jeschke G. J Chem Phys. 1997;106:10072–10086. [Google Scholar]
- 8.Polenova T, McDermott AE. J Phys Chem B. 1999;103:535–548. [Google Scholar]
- 9.Goldstein RA, Boxer SG. Biophys J. 1987;51:937–946. doi: 10.1016/S0006-3495(87)83421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDermott A, Zysmilich MG, Polenova T. Solid State Nucl Magn Reson. 1998;11:21–47. doi: 10.1016/s0926-2040(97)00094-5. [DOI] [PubMed] [Google Scholar]
- 11.Prakash S, Alia, Gast P, de Groot HJM, Jeschke G, Matysik J. J Am Chem Soc. 2005;127:14290–14298. doi: 10.1021/ja054015e. [DOI] [PubMed] [Google Scholar]
- 12.Prakash S, Alia, Gast P, de Groot HJM, Matysik J, Jeschke G. J Am Chem Soc. 2006;128:12794–12799. doi: 10.1021/ja0623616. [DOI] [PubMed] [Google Scholar]
- 13.Jeschke G. J Am Chem Soc. 1998;120:4425–4429. [Google Scholar]
- 14.Matysik J, Alia, Gast P, van Gorkom HJ, Hoff AJ, de Groot HJM. Proc Natl Acad Sci USA. 2000;97:9865–9870. doi: 10.1073/pnas.170138797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alia, Roy E, Gast P, van Gorkom HJ, de Groot HJM, Jeschke G, Matysik J. J Am Chem Soc. 2004;126:12819–12826. doi: 10.1021/ja048051+. [DOI] [PubMed] [Google Scholar]
- 16.Diller A, Alia, Roy E, Gast P, van Gorkom HJ, Zaanen J, de Groot HJM, Glaubitz C, Matysik J. Photosynth Res. 2005;84:303–308. doi: 10.1007/s11120-005-0411-0. [DOI] [PubMed] [Google Scholar]
- 17.Ishikita H, Loll B, Biesiadka J, Saenger W, Knapp EW. Biochemistry. 2005;44:4118–4124. doi: 10.1021/bi047922p. [DOI] [PubMed] [Google Scholar]
- 18.Prakash S, Tong SH, Alia, Gast P, de Groot HJM, Jeschke G, Matysik J. In: Photosynthesis: Fundamental Aspects to Global Perspectives. van der Est A, Brouce D, editors. Montreal: Allen; 2004. pp. 236–238. [Google Scholar]
- 19.Boxer SG, Closs GL, Katz JJ. J Am Chem Soc. 1974;96:7058–7066. [Google Scholar]
- 20.Fischer MR, de Groot HJM, Raap J, Winkel C, Hoff AJ, Lugtenburg J. Biochemistry. 1992;31:11038–11049. doi: 10.1021/bi00160a013. [DOI] [PubMed] [Google Scholar]
- 21.Witt H, Schlodder E, Teutloff C, Niklas J, Bordignon E, Carbonera D, Kohler S, Labahn A, Lubitz W. Biochemistry. 2002;41:8557–8569. doi: 10.1021/bi025822i. [DOI] [PubMed] [Google Scholar]
- 22.Käβ H, Bittersmannweidlich E, Andreasson LE, Bönigk B, Lubitz W. Chem Phys. 1995;194:419–432. [Google Scholar]
- 23.Käβ H, Lubitz W, Hartwig G, Scheer H, Noy D, Scherz A. Spectrochim Acta. 1998;54:1141–1156. [Google Scholar]
- 24.Käβ H, Lubitz W. Chem Phys Lett. 1996;251:193–203. [Google Scholar]
- 25.Rigby SEJ, Nugent JHA, O'Malley PJ. Biochemistry. 1994;33:10043–10050. doi: 10.1021/bi00199a031. [DOI] [PubMed] [Google Scholar]
- 26.Krabben L, Schlodder E, Jordan R, Carbonera D, Giacometti G, Lee H, Webber AN, Lubitz W. Biochemistry. 2000;39:13012–13025. doi: 10.1021/bi001200q. [DOI] [PubMed] [Google Scholar]
- 27.Käβ H, Fromme P, Witt HT, Lubitz W. J Phys Chem B. 2001;105:1225–1239. [Google Scholar]
- 28.Plato M, Krauss N, Fromme P, Lubitz W. Chem Phys. 2003;294:483–499. [Google Scholar]
- 29.Creemers AFL, Klaassen CHW, Bovee-Geurts PHM, Kelle R, Kragl U, Raap J, de Grip WJ, Lugtenburg J, de Groot HJM. Biochemistry. 1999;38:7195–7199. doi: 10.1021/bi9830157. [DOI] [PubMed] [Google Scholar]
- 30.Alia, Matysik J, Soede-Huijbregts C, Baldus M, Raap J, Lugtenburg J, Gast P, van Gorkom HJ, Hoff AJ, de Groot HJM. J Am Chem Soc. 2001;123:4803–4809. doi: 10.1021/ja002591z. [DOI] [PubMed] [Google Scholar]
- 31.Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Nature. 2005;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- 32.Zouni A, Witt HT, Kern J, Fromme P, Krauss N, Saenger W, Orth P. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]
- 33.Zech SG, Kurreck J, Eckert HJ, Renger G, Lubitz W, Bittl R. FEBS Lett. 1997;414:454–456. doi: 10.1016/s0014-5793(97)01054-5. [DOI] [PubMed] [Google Scholar]
- 34.Lubitz W, Lendzian F, Bittl R. Acc Chem Res. 2002;35:313–320. doi: 10.1021/ar000084g. [DOI] [PubMed] [Google Scholar]
- 35.Kammel M, Kern J, Lubitz W, Bittl R. Biochim Biophys Acta. 2003;1605:47–54. doi: 10.1016/s0005-2728(03)00063-x. [DOI] [PubMed] [Google Scholar]
- 36.Shelnutt JA, Song XZ, Ma JG, Jia SL, Jentzen W, Medforth CJ. Chem Soc Rev. 1998;27:31–41. [Google Scholar]
- 37.Glaubitz C, Watts A. J Magn Reson. 1998;130:305–316. doi: 10.1006/jmre.1997.1344. [DOI] [PubMed] [Google Scholar]
- 38.Lopez JJ, Mason AJ, Kaiser C, Glaubitz C. J Biomol NMR. 2007;37:97–111. doi: 10.1007/s10858-006-9109-7. [DOI] [PubMed] [Google Scholar]
- 39.Matysik J, Alia, Hollander JG, Egorova-Zachernyuk T, Gast P, de Groot HJM. Indian J Biochem Biophys. 2000;37:418–423. [PubMed] [Google Scholar]
- 40.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 41.Diller A, Prakash S, Alia A, Gast P, Matysik J, Jeschke G. J Phys Chem B. 2007 doi: 10.1021/jp072428r. in press. [DOI] [PubMed] [Google Scholar]