Abstract
The mechanisms by which DNA-damaging agents trigger the induction of the stress response protein p53 are poorly understood but may involve alterations of chromatin structure or blockage of either transcription or replication. Here we show that transcription-blocking agents can induce phosphorylation of the Ser-15 site of p53 in a replication-independent manner. Furthermore, microinjection of anti-RNA polymerase II antibodies into the nuclei of cells showed that blockage of transcription is sufficient for p53 accumulation even in the absence of DNA damage. This induction of p53 occurs by two independent mechanisms. First, accumulation of p53 is linked to diminished nuclear export of mRNA; and second, inhibition specifically of elongating RNA polymerase II complexes results in the phosphorylation of the Ser-15 site of p53 in a replication protein A (RPA)- and ATM and Rad3-related (ATR)-dependent manner. We propose that this transcription-based stress response involving RPA, ATR, and p53 has evolved as a DNA damage-sensing mechanism to safeguard cells against DNA damage-induced mutagenesis.
Keywords: antibody microinjection, DNA damage response, RNA polymerase II, nuclear export, phosphorylation
The mechanisms by which DNA lesions are detected and how damage-response pathways are activated in cells are not well understood (1, 2). The stress-response kinases ataxia telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) link DNA damage to p53 activation by phosphorylating the Ser-15 site of p53 (3–6). Although the ATM kinase is activated after exposure to ionizing radiation (7), the ATR kinase responds to agents that interfere with replication such as UV light and hydroxyurea (6). The tumor suppressor p53 is induced in response to a variety of different agents in all phases of the cell cycle (8, 9). By acting as a transcription factor, p53 can induce the expression of gene products involved in DNA repair, cell cycle arrest, or apoptosis (10, 11). It can also regulate DNA repair and apoptosis by transcription-independent mechanisms (12). Under normal conditions, the p53 protein is rapidly targeted for nuclear export and degradation in a process regulated by the MDM2 protein (13). After cellular stress, the MDM2-mediated negative regulation of p53 is abrogated, and p53 proteins accumulate in the cell nucleus. This nuclear accumulation can be accomplished by (i) DNA damage-induced phosphorylation of p53 and MDM2, leading to the interference of the interaction between MDM2 and p53 (14); (ii) inactivation of proteasomes (15); or (iii) inhibition of the nuclear export machinery (13, 16).
DNA lesions that block RNA polymerase II induce the recruitment of transcription-coupled repair (TCR) and chromatin remodeling factors to recover RNA synthesis (17–19). Cells defective in TCR induce p53 and apoptosis at much lower doses than cells with proficient TCR, suggesting that lesions in the transcribed strand of active genes trigger these responses (9, 20–22). Although these studies have shown a correlation between blockage of transcription and the accumulation and phosphorylation of p53, the agents used have multiple targets in cells, and therefore it has not been possible to link blockage of transcription conclusively to the activation of the p53 stress response. Furthermore, agents that block transcription are likely also to affect replication. To investigate whether blockage of transcription is sufficient to induce p53 even in the absence of initial DNA damage, we microinjected anti-RNA polymerase antibodies into the nucleus of human fibroblasts. We show that this approach efficiently inhibits transcription and leads to the accumulation of p53 in the nucleus of injected cells. Phosphorylation of p53 at Ser-15 occurred specifically when elongating RNA polymerases were targeted and occurred in a replication protein A (RPA)- and ATR-dependent manner in all injected cells, whereas targeting of preelongation RNA polymerases resulted in nuclear accumulation of p53 without concomitant phosphorylation of the Ser-15 site. We show that the export of p53 is normally linked to the export of mRNA so that when mRNA synthesis is inhibited, p53 accumulates in the nucleus by default. Taken together, our studies suggest that cells monitor RNA synthesis and transcription elongation and launch a transcription stress response involving RPA, ATR, and p53 when transcription elongation is stalled.
Results
Phosphorylation of the Ser-15 Site of p53 Can Occur Independently of Replication.
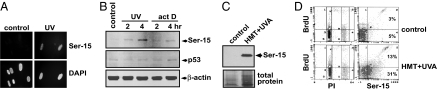
Many agents that block RNA polymerase II elongation also block replication elongation. To investigate whether phosphorylation of p53 at the Ser-15 site can occur in a replication-independent manner, we serum-starved confluent human fibroblasts for 7 days and challenged them with the transcription-blocking agents UVC light (254 nm), actinomycin D, or photoactivated 4′-hydroxymethyl-4,5′,8-trimethylpsoralen (HMT+UVA). UV light induces pyrimidine dimers and (–4)photoproducts, actinomycin D intercalates into DNA, and HMT+UVA forms interstrand DNA cross-links (23). All of these agents were capable of inducing Ser-15 phosphorylation of p53 in confluent, serum-starved cells (Fig. 1 A–C). To verify that phosphorylation occurred in a replication-independent manner, we treated asynchronously growing human fibroblasts with HMT+UVA in the presence of BrdU and then analyzed Ser-15 phosphorylation of p53 as a function of BrdU incorporation. It was found that Ser-15 phosphorylation occurred in both BrdU-negative as well as BrdU-positive cells (Fig. 1D). These results show that Ser-15 phosphorylation can occur in a replication-independent manner after exposure to UV light, actinomycin D, or photoactivated psoralen.
Fig. 1.
Replication-independent phosphorylation of the Ser-15 site of p53 after DNA damage. (A) Diploid human fibroblasts were grown to confluence and then serum-starved for 48 h (0.1% FBS) before being treated with UV light (10 J/m2) and analyzed 6 h later for Ser-15 phosphorylation of p53. (B and C) Cells grown as described above were treated with UV light (10 J/m2), actinomycin D (B), or HMT+UVA (C) and collected 2, 4, or 6 h later, and Ser-15 phosphorylation of p53 was analyzed by using Western blotting. We used β-actin or Coomassie blue staining of total proteins as loading controls. (D) Asynchronously growing diploid fibroblasts were pretreated for 15 min with BrdU to label replicating cells at the time of treatment. The cells were then mock-treated (control) or treated with HMT+UVA. Cells were collected 2 h later, and the induction of Ser-15 phosphorylation of p53 was analyzed in S phase cells (BrdU+, above the line) and in non-S phase cells (BrdU−, under the line). The percentages of Ser-15 phosphorylation-positive cells are indicated for each group.
Specific Blockage of RNA Polymerase II-Mediated Transcription Induces p53.
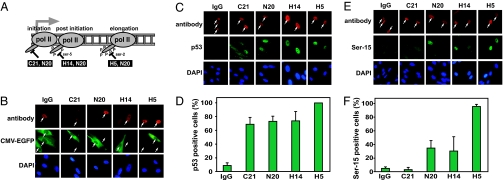
We next sought to answer a question left ambiguous by previous studies, namely, is blockage of RNA polymerase II in the absence of DNA damage sufficient to induce p53 accumulation? To specifically inhibit RNA polymerase II transcription without initially inducing DNA damage, we microinjected into the nuclei of human fibroblasts anti-RNA polymerase II antibodies, which has been shown to specifically inhibit transcription within 3 h of microinjection (24). Four different antibodies recognizing RNA polymerase II at different stages of transcription were used (Fig. 2A). The C21 antibody recognizes the preelongating form of RNA polymerase II with an unphosphorylated C-terminal domain (CTD), the H14 antibody recognizes a promoter-proximal postinitiation form of RNA polymerase II with a Ser-5-phosphorylated CTD, and the H5 antibody recognizes the elongating form that is phosphorylated at Ser-2 of the CTD (25). Finally, the N20 antibody recognizes all forms of RNA polymerase II. We used injection of IgG as a control for effects induced by the microinjection procedure itself and as an antibody control that is not expected to interact with any specific antigens in the cells. To confirm that microinjection of these antibodies inhibits RNA polymerase II-mediated transcription, we microinjected the CMV-EGFP-Luc vector into cells premicroinjected with the different anti-RNA polymerase II antibodies. This vector was used to monitor RNA polymerase II activity because it expresses GFP from a RNA polymerase II-dependent promoter. We found that microinjection of IgG did not interfere with the expression of EGFP from the CMV-EGFP-Luc vector (Fig. 2B). However, microinjection of any of the four different anti-RNA polymerase II antibodies resulted in little or no expression of EGFP from the CMV-EGFP-Luc vector compared with neighboring cells injected with the expression vector alone. Thus, our microinjection approach using anti-RNA polymerase II antibodies efficiently inhibits RNA polymerase II-mediated transcription in human fibroblasts.
Fig. 2.
Microinjection of anti-RNA polymerase II antibodies in the nucleus inhibits transcription in human fibroblasts and induces p53 accumulation and phosphorylation. (A) Drawing illustrating what stage in the transcription cycle the different antibodies are expected to abrogate. (B) Human fibroblasts were microinjected in the nucleus with IgG or one of the anti-RNA polymerase II (pol II) antibodies, incubated for 3 h at 37°C, and remicroinjected with the CMV-EGFP-Luc expression vector. After a 1-h incubation at 37°C, the cells were fixed, and the expression of GFP was monitored by immunocytochemistry. White arrows indicate microinjected cells. (C) Human fibroblasts were microinjected in the nucleus with IgG or one of the anti-RNA polymerase II antibodies and incubated for 4 h at 37°C. After fixation, p53 levels were determined by immunocytochemistry. White arrows indicate microinjected cells. (D) Analysis of the percentage of cells in C expressing elevated levels of nuclear p53. (E) Cells were microinjected as in C, but levels of Ser-15-phosphorylated p53 were determined with phospho-specific antibodies. White arrows indicate microinjected cells. (F) Analysis of the percentage of cells in E expressing elevated nuclear levels of Ser-15-phosphorylated p53. The bars in D and F represent the average number of positive cells from five different experiments with 20 microinjected cells per experiment, with error bars representing the SD.
We next microinjected human fibroblasts with the different anti-RNA polymerase II-specific antibodies, and 4 h later the cells were fixed, and the presence of p53 was determined by immunocytochemistry. It was found that microinjection of any of the four RNA polymerase II antibodies resulted in a substantial increase of the number of cells with nuclear accumulation of p53 (Fig. 2 C and D). In contrast, microinjection of IgG did not result in the induction of p53, indicating that the microinjection procedure itself or the presence of high concentration of antibody in the nucleus does not induce a stress response leading to p53 induction. We next explored whether the different antibodies could induce Ser-15 phosphorylation of p53 and found that microinjection of the C21 antibody did not induce elevated levels of Ser-15 phosphorylation of p53 (Fig. 2 E and F) despite causing significant nuclear accumulation of p53 (see Fig. 2 C and D). However, microinjection of the N20 and H14 antibodies resulted in nuclear accumulation of Ser-15-phosphorylated p53 in 30–40% of the microinjected cells, and microinjection of the H5 antibody resulted in accumulation of Ser-15-phosphorylated p53 in >90% of the cells. Thus, only the antibodies able to recognize the elongating form of RNA polymerase II were able to trigger Ser-15 phosphorylation of p53. The reason that the H5 antibody caused a more robust induction of Ser-15 phosphorylation than the N20 and H14 antibodies may be that it specifically targets the elongating form of RNA polymerase II. The N20 antibody, on the other hand, targets all forms of RNA polymerase II, even initiating polymerases, leading to less new initiation and therefore fewer elongating polymerases that can be forced to stall. Finally, the H14 antibody targets promoter-proximal elongation that may generate a weaker stress signal because normally, RNA polymerases frequently pause in these promoter-proximal regions.
Nuclear Accumulation of p53 After Transcription Blockage Is Linked to Diminished Nuclear Export of mRNA.
Phosphorylation of the Ser-15 site of p53 is thought to be one of the initiating phosphorylation events after cellular stress that stimulates subsequent modifications and stabilization of the p53 protein (14). Our findings that p53 accumulates in the nucleus without concomitant Ser-15 phosphorylation after inhibition of the preelongating phase of transcription are thus intriguing. We have previously demonstrated that generic nuclear export of proteins containing nuclear export signals is reduced after inhibition of transcription (26). This reduction was found to be specifically linked to diminished nuclear export of mRNA. Because the p53 protein contains two nuclear export signals and p53 proteins accumulate in the nucleus after the inhibition of transcription, we next explored whether p53 nuclear export is linked to mRNA export.
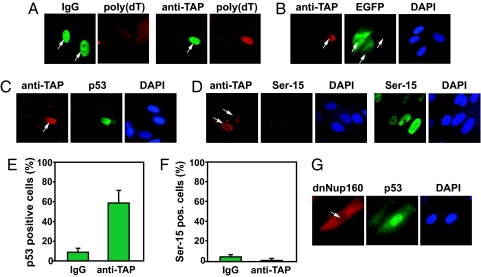
To specifically block mRNA export, we microinjected antibodies raised against TAP, the main nuclear export factor for mRNA (27). As expected, microinjection of anti-TAP induced the nuclear accumulation of poly(A) mRNA (Fig. 3A) and reduced the expression of the RNA polymerase II-driven CMV-EGFP-Luc construct (Fig. 3B). Importantly, microinjection of anti-TAP antibodies resulted in nuclear accumulation of p53 (Fig. 3 C and E). The nuclear accumulation of p53 after microinjection of anti-TAP antibodies was not accompanied by Ser-15 phosphorylation, suggesting that the accumulation of nuclear p53 was the result of diminished nuclear export rather than induction of a stress response (Fig. 3 D and F). Similar results were obtained when expressing a dominant negative form of NUP160, a nucleoporin protein required for efficient nuclear export of mRNA (Fig. 3G) (28). These results strongly suggest that nuclear export of p53 is normally linked to the nuclear export of mRNA and that p53 accumulates in the nucleus by default when mRNA synthesis or export is inhibited.
Fig. 3.
Inhibition of mRNA export induces nuclear accumulation of p53 without concomitant phosphorylation of the Ser-15 site. (A) Cells were microinjected with rabbit IgG or anti-TAP antibodies and incubated at 37°C for 4 h followed by fixation and immunofluorescence staining of poly(A) RNA by using biotinylated poly(dT) and Texas red-conjugated streptavidin. White arrows indicate microinjected cells. (B) Human fibroblasts were microinjected in the nucleus with anti-TAP antibodies, incubated for 3 h at 37°C, remicroinjected with the CMV-EGFP-Luc expression vector, and then incubated for 1 h at 37°C. White arrows indicate the microinjected cells. Cells were microinjected with anti-TAP and incubated at 37°C for 4 h followed by fixation and immunocytochemistry for p53 (C) and Ser-15-phosphorylated p53 (D). White arrows indicate the microinjected cells, and 20 J/m2 UV light was used as a positive control of Ser-15 phosphorylation in D. (E and F) Analysis of the level of nuclear p53 (E) and Ser-15-phosphorylated p53 (F) in cells microinjected as in C and D, respectively. The bars represent the average number of positive cells from five different experiments with 20 microinjected cells per experiment. (G) Cells were microinjected with a DNA construct expressing a dominant negative Nup160 protein, and 6 h later the cells were fixed and stained. The white arrow indicates the microinjected cell.
Phosphorylation of the Ser-15 Site of p53 After Inhibition of Transcription Elongation Depends on RPA and ATR.
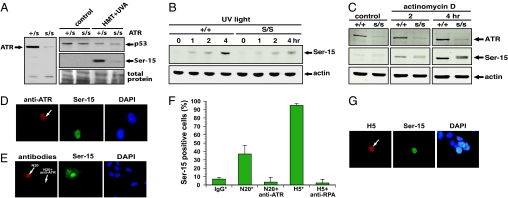
We first investigated whether Ser-15 phosphorylation of p53 is defective in ATR-deficient Seckel syndrome cells after blockage of transcription elongation. Our results show that Ser-15 phosphorylation was attenuated in the ATR-deficient cells after treatment with photoactivated psoralen (Fig. 4A), UV light (Fig. 4B), or actinomycin D (Fig. 4C). We next asked whether the Ser-15 phosphorylation induced by specific blockage of transcription elongation by microinjection of the anti-RNA polymerase II antibodies H5 and N20 is also mediated by the ATR kinase. To inhibit the ATR kinase in cells specifically and rapidly, we microinjected anti-ATR antibodies into the nuclei of human fibroblasts. Microinjection of anti-ATR antibodies fully abrogated the ATR-dependent phosphorylation of the Ser-15 site of p53 after UV irradiation (6), validating this approach (Fig. 4D). When the anti-ATR antibodies were microinjected together with the N20 antibodies, we found that the anti-ATR antibodies abolished the ability of the N20 antibody to induce Ser-15 phosphorylation of p53 (Fig. 4 E and F). This finding strongly suggests that Ser-15 phosphorylation of p53 after blockage of transcription elongation is mediated by ATR. Because the RPA protein is thought to activate the ATR kinase, we next explored whether sequestering of the RPA protein in cells by microinjection of anti-RPA antibodies also would abolish induction of Ser-15 phosphorylation of p53 after blockage of transcription elongation. It was found that comicroinjection of anti-RPA antibodies with the anti-RNA polymerase II antibody H5 abolished Ser-15 phosphorylation (Fig. 4F). We also microinjected the H5 antibody into AT fibroblasts and found that these cells induced Ser-15 phosphorylation similarly to wild-type cells, indicating that the ATM kinase is not involved in the induction of Ser-15 phosphorylation after blockage of transcription elongation (Fig. 4G). Taken together, these results suggest that RPA and the ATR link blocked transcription elongation complexes with the p53 response pathway by the phosphorylation of the Ser-15 site of p53.
Fig. 4.
Induction of the Ser-15 site of p53 after blockage of transcription elongation is mediated by RPA and ATR. (A) (Left) Lymphocytes from an individual affected by the Seckel syndrome (s/s) express very little ATR protein compared with heterozygote cells from a parent (+/s). (Right) (+/s) and (s/s) cells were mock-treated or treated with HMT+UVA. Cells were harvested 6 h later, and the levels of total p53 (Top) and Ser-15-phosphorylated p53 (Middle) were determined by Western blotting. Coomassie blue staining was used to evaluate protein transfer to the membrane. (B) (+/+) and (s/s) cells were irradiated with 10 J/m2 and incubated for different periods of time before analysis of Ser-15 phosphorylation of p53. We used β-actin as a loading control. (C) (+/+) and (s/s) cells were treated with actinomycin D for 2 or 4 h before analysis of Ser-15 phosphorylation of p53 was performed with Western blotting. We used β-actin as a loading control. (D) Human fibroblasts were microinjected in the nucleus with anti-ATR antibodies and incubated for 1 h before being irradiated with 20 J/m2 UV light (254 nm). After 2 h of incubation at 37°C, the cells were fixed and stained. (E) Human fibroblasts were microinjected with either N20 antibodies alone or with both N20 and anti-ATR antibodies. After incubation for 4 h at 37°C, the cells were fixed and stained. (F) Analysis of the percentage of cells with elevated Ser-15 phosphorylation after comicroinjection with N20 and anti-ATR or H5 and anti-RPA. Because the anti-ATR antibodies are rabbit, they had to be comicroinjected with the rabbit N20 antibodies so that Ser-15 phosphorylation could be detected with mouse antibodies. Conversely, mouse anti-RPA had to be microinjected with mouse H5 antibodies so that Ser-15 phosphorylation could be detected with rabbit antibodies. Bars represent the average of at least five different experiments with 20 injected cells per experiment. The error bars represent SD. The asterisk indicates that the data were taken from Fig. 2F. (G) Ataxia telangiectasia fibroblasts were microinjected with the H5 antibody, and 4 h later the cells were fixed and stained for the presence of H5 antibody, Ser-15 phosphorylation, and DNA. This image is representative of at least 10 injected cells that all stained positive for p53.
Discussion
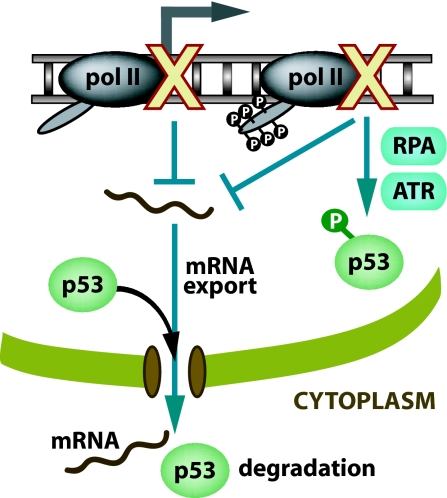
In this work, we show that specific inhibition of transcription by microinjection of anti-RNA polymerase II antibodies into the nucleus of human fibroblasts induced p53 and that this induction occurred by two different mechanisms. First, our work suggests that p53 is normally exported out of the nucleus in a process dependent on the export of mRNA so when synthesis or export of mRNA is blocked, p53 accumulates in the nucleus by default. Second, inhibition specifically of transcription elongation leads to the phosphorylation of the Ser-15 site of p53 in an RPA- and ATR-dependent manner (Fig. 5). These results show unambiguously that blockage of transcription is sufficient for the induction of nuclear accumulation of p53. Furthermore, our work suggests a role for RPA and ATR in stress response signaling by monitoring the elongation phase of RNA polymerase II-mediated transcription.
Fig. 5.
A model of the transcription stress response leading to the induction of p53 by two different mechanisms. First, inhibition of transcription elongation induces a transcription stress response resulting in phosphorylation of the Ser-15 site of p53 in an RPA- and ATR-mediated manner. Second, loss of mRNA synthesis results in diminished amounts of mRNA available to be exported out of the nucleus (mRNA export also inhibited by anti-TAP antibody microinjection) leading to attenuated nuclear export of p53.
It has been shown that the ATR kinase is activated in an RPA-dependent manner after interference with DNA replication leading to Ser-15 phosphorylation of p53 (29). Our work suggests that RPA and ATR may also be involved in monitoring transcription elongation. Our finding that p53 is phosphorylated in an RPA- and ATR-dependent fashion after inhibition of transcription elongation even in the absence of initial DNA damage suggests that generically stalled RNA polymerase II complexes or some altered DNA structure generated when transcription stalls triggers the stress signal (Fig. 5). We hypothesize that a region of single-stranded DNA is formed after blockage of transcription elongation that attracts RPA, leading to the recruitment of ATR and the activation of p53 by phosphorylation of the Ser-15 site. Consistent with this hypothesis is a recent study showing that RPA and ATR preferentially accumulate on transcribed DNA sequences after UV irradiation, presumably at sites of blocked RNA polymerases (30). This accumulation occurs even in XP-A cells, suggesting that the recruitment of RPA to active genes is not related to its role in nucleotide excision repair. Furthermore, because we did not find any role for ATM in the phosphorylation of Ser-15 after blockage of transcription elongation, it is unlikely that transcription stalling results in DNA double-stranded breaks that in turn would attract RPA during repair processing.
What is the physiological role of p53 accumulation at times when transcription is inhibited? One possibility is that the induced p53 can mediate transcription-independent apoptosis by translocating to mitochondria (31). However, we did not observe any accumulation of p53 at mitochondria or in the cytoplasm after blockage of transcription in our work. It has been shown that p53 protects both human fibroblasts (32) and keratinocytes (33) against UV light, which is the most physiological transcription inhibitor to which skin cells are exposed. It is possible that nuclear accumulation of p53 after UV-induced transcription blockage ensures a rapid and strong induction of target genes as transcription resumes after removal of transcription-blocking lesions by transcription-coupled repair. Only at high doses, UV light may stimulate p53-dependent apoptosis by preferentially inducing small apoptosis-promoting genes (34). Alternatively, the protective effect of nuclear accumulation of p53 after moderate doses of UV irradiation may be the result of the ability of p53 to enhance nucleotide excision repair by increasing DNA damage accessibility in chromatin (35).
The safeguarding of the genome requires a substantial effort because of the large number of lesions that are formed in the DNA on a daily basis from both endogenous and exogenous sources (23). Some of these lesions may interfere with transcription, directly or indirectly (36) leading to the induction of stress responses and apoptosis that protect the organism against carcinogenesis but may also contribute to aging (20, 37–39). Although protein-coding genes only constitute ≈2% of the genome, recent studies suggest that RNA polymerase II may transcribe as much as 60–70% of the genome (40). The purpose of this vast engagement in transcription is not well understood, but it has been suggested that the noncoding RNAs generated may participate in gene regulation (40). Based on the presumptive role of the elongating RNA polymerase II complex as a sensor of DNA damage (39), we propose that this vast transcription in the genome may serve as an effective damage-scanning mechanism. By triggering transcription-coupled repair and induction of p53 after transcription blockage, RNA polymerase II may act as both a sensor and dosimeter for DNA damage and as such may play an important role in tumor suppression.
Materials and Methods
Cell Culture, Treatments, and Antibodies.
Diploid human fibroblasts and AT fibroblasts (GM01588) were grown as monolayers in culture dishes or on coverslips in MEM supplemented with 10% FBS and antibiotic/antimycotic (GIBCO, Grand Island, NY). Human lymphoblast cells (Seckel-ATR, 536 wild-type, 1526 AT) were grown in RPMI supplemented with 15% FBS (GIBCO). Cells were treated with 20 nM actinomycin D (Sigma, St. Louis, MO) in medium for the indicated time periods at 37°C, 1 μg/ml HMT (HRI Associates, Concord, CA) in PBS for 10 min in the dark at 4°C followed by a 10-min irradiation from a UVA source or UVC irradiation at room temperature with a germicidal UVC light (254 nm). The dose rate of the UVC light source (Philips, New York, NY) was measured with a UVX radiometer (UVP, Inc., Upland, CA). Antibodies used for microinjection were rabbit IgG (Sigma), rabbit polyclonal anti-RNA polymerase II (N20; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal anti-RNA polymerase II (C21; Santa Cruz Biotechnology), mouse monoclonal anti-Ser-2-phosphorylated RNA polymerase II (H5; Covance, Berkeley, CA), mouse monoclonal anti-Ser-5-phosphorylated RNA polymerase II (H14; Covance), goat polyclonal anti-TAP (R20; Santa Cruz Biotechnology), rabbit polyclonal anti-ATR (Abcam, Cambridge, MA), and mouse monoclonal anti-RPA, 70 kDa (ab-1; Oncogene, Cambridge, MA).
Western Blotting.
After treatment, cells were rinsed with ice-cold PBS, detached by scraping, and collected by centrifugation. Cell lysates were extracted in 1% Nonidet P-40 lysis buffer by rotation at 4°C for 20 min followed by a 10-min centrifugation at 16,060 × g at 4°C. The cell samples were boiled for 5 min in loading buffer [1.6% SDS/5% (vol/vol) glycerol/5% 2-mercaptoethanol/0.05% bromophenol blue/62.5 mM Tris, pH 6.8] before loading. Protein concentrations were quantified by using a protein assay (Bio-Rad, Hercules, CA), and ≈40 μg of protein was loaded per lane. After SDS/PAGE, proteins were electrophoretically transferred to Immobilon-P or Immobilon-FL transfer membranes (Millipore, Billerica, MA). Antibodies used for the Western blotting were anti-Ser-15 phospho-specific p53 antibody (Ser-15; Cell Signaling Technology, Danvers, MA), anti-p53 antibody (Ab-2; Oncogene Research Products, Boston, MA), and anti-ATR (Abcam). X-ray film (blue sensitive autoradiographic film; Marsh Bio Products, Rochester, NY) and chemiluminescence (Super Signal CL-HRP substrate system, Pierce, Rockford, IL) were used to visualize the p53 proteins. Western blotting was also carried out by using the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE).
Two-Parameter Flow Cytometry.
Immediately after psoralen and UVA treatment, 30 μM BrdU (Sigma) was added to the growth medium and was maintained in the medium for the entire postincubation period. For experiments measuring phosphorylation of the Ser-15 site of p53 and BrdU incorporation, cells were harvested 6 h after the psoralen treatment and fixed by adding ice-cold 70% ethanol dropwise under vortexing. The staining procedure for BrdU was performed essentially as described previously (41). We incubated the fixed cells with mouse anti-human BrdU antibodies conjugated to Alexa Fluor 647 (Molecular Probes, Eugene, OR) and rabbit phospho-specific Ser-15 anti-p53 antibodies for 30 min at room temperature. The cell samples were analyzed for BrdU content (Alexa Fluor 647) and Ser-15 phosphorylation of p53 (FITC) by using flow cytometry (Coulter Elite ESP cell sorter; Beckman Coulter, Fullerton, CA).
Antibody Microinjections.
Antibodies used to inhibit RNA polymerase II were microinjected into the nuclei of human fibroblasts at a concentration of 2 mg/ml. Microinjection was performed as described previously (26). Cells were then fixed by using 3.7% paraformaldehyde followed by permeabilization with permeabilization buffer (PBS/0.2% Triton X-100/0.5% BSA).
Immunocytochemistry.
Immunocytochemistry was performed as described previously (26). The antibodies used were mouse monoclonal anti-p53 antibody 1801 (a gift from Jiayuh Lin, Ohio State University, Columbus, OH), rabbit monoclonal anti-p53 antibody (FL; Santa Cruz Biotechnology), mouse monoclonal phospho-specific anti-Ser-15-phosphorylated p53 (Cell Signaling Technology), and rabbit monoclonal phospho-specific anti-Ser-15-phosphorylated p53 (Cell Signaling Technology). The cells microinjected with the rabbit and mouse antibodies were visualized by incubation with anti-rabbit or anti-mouse secondary antibodies conjugated with Alexa Fluor 555 (red), Alexa Fluor 488 (green) (Molecular Probes), or FITC (Sigma). Images were captured by using a Akioskop fluorescent microscope (Zeiss, Thornwood, NY) with Plan-NEUFLUAR ×63/1.25 oil lens and a CoolSnapPro digital camera (MediaCybernetics, Bethesda, MD) with associated software.
Expression of Dominant Negative Nup160 and Localization of Poly(A)+ RNA.
These experiments were performed as described previously (26, 28).
Acknowledgments
We thank all of the members of the M.L. laboratory for valuable input and critique. We also thank Dr. Gabriel Nuñez (University of Michigan) for technical support and Dr. Douglass Forbes (University of California at San Diego, La Jolla, CA) for the generous gift of the dnNup160 expression vector. This work was supported by National Institutes of Health Grant CA-82376 (to M.L.), University of Michigan Comprehensive Cancer Center Core Grant NCI 5 P30 CA46592 (to M.L.), University of Michigan Cancer Biology Training Program (to H.M.O.), Rackham Graduate School, and the Department of Radiation Oncology (to S.H.).
Abbreviations
- HMT
4′-hydroxymethyl-4,5′,8-trimethylpsoralen
- HMT+UVA
photoactivated HMT
- RPA
replication protein A
- ATM
ataxia telangiectasia mutated
- ATR
ATM and Rad3-related
- TCR
transcription-coupled repair
- CTD
carboxyl-terminal domain.
Footnotes
The authors declare no conflict of interest.
References
- 1.Sancar A, Lindsey-Boltz LA, Unsal-Kaccmaz K, Linn S. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 2.Ljungman M. Mutat Res. 2005;577:203–217. doi: 10.1016/j.mrfmmm.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 4.Banin S, Moyal L, Shieh SY, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 5.Lakin N, Hann B, Jackson S. Oncogene. 1999;18:3989–3995. doi: 10.1038/sj.onc.1202973. [DOI] [PubMed] [Google Scholar]
- 6.Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakkenist CJ, Kastan MB. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 8.Fritsche M, Haessler C, Brandner G. Oncogene. 1993;8:307–318. [PubMed] [Google Scholar]
- 9.Yamaizumi M, Sugano T. Oncogene. 1994;9:2775–2784. [PubMed] [Google Scholar]
- 10.Levine AJ. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 11.Ford JM, Hanawalt PC. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 12.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 13.Freedman DA, Levine AJ. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shieh SY, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 15.Maki CG, Huibregtse JM, Howley PM. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 16.Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellon I, Bohr VA, Smith CA, Hanawalt PC. Proc Natl Acad Sci USA. 1986;83:8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanawalt PC. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 19.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Mol Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 20.Ljungman M, Zhang F. Oncogene. 1996;13:823–831. [PubMed] [Google Scholar]
- 21.Ljungman M, Zhang FF, Chen F, Rainbow AJ, McKay BC. Oncogene. 1999;18:583–592. doi: 10.1038/sj.onc.1202356. [DOI] [PubMed] [Google Scholar]
- 22.Ljungman M, O'Hagan HM, Paulsen MT. Oncogene. 2001;20:5964–5971. doi: 10.1038/sj.onc.1204734. [DOI] [PubMed] [Google Scholar]
- 23.Friedberg E, Walker G, Siede W, Wood R, Schultz R, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: Am Soc Microbiol; 2006. [Google Scholar]
- 24.Mercer WE, Avignolo C, Galanti N, Rose KM, Hyland JK, Jacob ST, Baserga R. Exp Cell Res. 1984;150:118–130. doi: 10.1016/0014-4827(84)90707-9. [DOI] [PubMed] [Google Scholar]
- 25.Bregman DB, Du L, van der Zee S, Warren SL. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Hagan HM, Ljungman M. Exp Cell Res. 2004;297:548–559. doi: 10.1016/j.yexcr.2004.03.051. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 28.Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ. J Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou L, Elledge SJ. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 30.Jiang G, Sancar A. Mol Cell Biol. 2006;26:39–49. doi: 10.1128/MCB.26.1.39-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arima Y, Nitta M, Kuninaka S, Zhang D, Fujiwara T, Taya Y, Nakao M, Saya H. J Biol Chem. 2005;280:19166–19176. doi: 10.1074/jbc.M410691200. [DOI] [PubMed] [Google Scholar]
- 32.McKay B, Ljungman M. Neoplasia. 1999;1:276–284. doi: 10.1038/sj.neo.7900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaturvedi V, Sitailo LA, Qin JZ, Bodner B, Denning MF, Curry J, Zhang W, Brash D, Nickoloff BJ. Oncogene. 2005;24:5299–5312. doi: 10.1038/sj.onc.1208650. [DOI] [PubMed] [Google Scholar]
- 34.McKay BC, Stubbert LJ, Fowler CC, Smith JM, Cardamore RA, Spronck JC. Proc Natl Acad Sci USA. 2004;101:6582–6586. doi: 10.1073/pnas.0308181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubbi CP, Milner J. EMBO J. 2003;22:975–986. doi: 10.1093/emboj/cdg082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanamadala S, Ljungman M. Mol Cancer Res. 2003;1:747–754. [PubMed] [Google Scholar]
- 37.Gorgels TGM, van der Pluijm I, Brandt RMC, Garinis GA, van Steeg H, van den Aardweg G, Jansen GH, Ruijter JM, Bergen AA, van Norren D, et al. Mol Cell Biol. 2007;27:1433–1441. doi: 10.1128/MCB.01037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andressoo JO, Hoeijmakers JH. Mutat Res. 2005;577:179–194. doi: 10.1016/j.mrfmmm.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Ljungman M, Lane DP. Nat Rev Cancer. 2004;4:727–737. doi: 10.1038/nrc1435. [DOI] [PubMed] [Google Scholar]
- 40.Mattick JS, Makunin IV. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 41.Hoy C, Seamer L, Schimke R. Cytometry. 1989;10:718–725. doi: 10.1002/cyto.990100608. [DOI] [PubMed] [Google Scholar]