Abstract
As its name suggests, tumor necrosis factor (TNF) is known to induce cytotoxicity in a wide variety of tumor cells and cell lines. However, its use as a chemotherapeutic drug has been limited by its deleterious side effects of systemic shock and widespread inflammatory responses. Some nonsteroidal antiinflammatory drugs, such as sodium salicylate, have been shown to have a chemopreventive role in certain forms of cancer. Here, we reveal that sodium salicylate selectively enhances the apoptotic effects of TNF in human erythroleukemia cells but does not affect primary human lymphocytes or monocytes. Sodium salicylate did not affect the intracellular distribution of TNF receptors (TNFRs) but stimulated cell surface TNFR2 shedding. Erythroleukemia cells were shown to possess markedly greater basal NF-κB responses and elevated Fas-associated protein with death domain-like IL-1converting enzyme (FLIP) levels. Sodium salicylate achieved its effects by reducing the elevated NF-κB responsiveness and FLIP levels and restoring the apoptotic response of TNF rather than the proliferative/proinflammatory effects of the cytokine in these cancer cells. Inhibition of NF-κB or FLIP levels in human erythroleukemia cells by pharmacological or molecular-biological means also resulted in switching the character of these cells from a TNF-responsive proliferative phenotype into an apoptotic one. These findings expose that the enhanced proliferative nature of human leukemia cells is caused by elevated NF-κB and FLIP responses and basal levels, reversible by sodium salicylate to allow greater apoptotic responsiveness of cytotoxic stimuli such as TNF. Such findings provide insight into the molecular mechanisms by which human leukemia cells can switch from a proliferative into an apoptotic phenotype.
Keywords: cancer cells, cytokine, receptor, sodium salicylate, leukemia
Tumor necrosis factor-α (TNF) is a 26-kDa transmembrane cytokine protein cleaved and secreted as a 17-kDa form by many inflammatory cells, primarily activated macrophages. Although first recognized for its ability to induce hemorrhagic necrosis of certain tumors (1), TNF also mediates a wide variety of other physiological responses such as differentiation, inflammation, proliferation, and apoptotic and necrotic cell deaths (2). TNF was demonstrated to have cytotoxic, cytostatic, and apoptotic effects when tested with various malignant cell lines, but clinical trials in cancer patients revealed systemic side effects, including toxicity, fever, and cachexia (3). TNF mediates its actions by binding to two distinct cell surface receptor types, TNFR1 and TNFR2, although it is presently unclear which subtype is responsible for each of these TNF effects (4).
It has been shown that the use of nonsteroidal antiinflammatory drugs (NSAIDs) was associated with a reduced risk of developing several malignant diseases (5), and these drugs rank among the most potent and promising agents for cancer chemoprevention. Both colorectal tumorigenesis in man as well as experimental carcinogenesis have been shown to be inhibited by NSAIDs such as indomethacin, sulindac, aspirin, and ibuprofen (6). It was shown that regular intake of aspirin over many years lowered the risk of colorectal, esophageal, stomach, and lung cancers (6). However, the precise mechanism of action of these drugs is presently unknown.
One possible mechanism for the chemopreventive role of NSAIDs is the inhibition of the transcription factor NF-κB activation. In uninduced cells, NF-κB is present in the cytosol where it is bound to a specific inhibitor, IκB, inhibiting DNA binding and cytoplasmic retention of NF-κB (7). The NF-κB family comprises five members: RelA, RelB, c-Rel, p50/NF-κB1, and p52/NF-κB2. Upon induction by a variety of stimuli, including cytokines, mitogens, tumor promoters, and viruses, IκB becomes phosphorylated, polyubiquitinated, and degraded, and the active NF-κB subunits are released (8). It is now known that acceleration of cell death may be achieved by blocking the activity of inhibitors of apoptosis, such as NF-κB (9). When activated, NF-κB limits TNF-induced apoptosis by induction of antiapoptotic genes and protective proteins (10), and inhibiting this activation sensitizes cells to TNF-induced toxicity. Sodium salicylate is one NSAID that has been shown to inhibit NF-κB activation by limiting IκBα phosphorylation and degradation (11).
Fas-associated protein with death domain (FADD)-like IL-1-converting enzyme (FLICE/caspase-8)-inhibitory protein (FLIP) generally acts as an inhibitor of death receptor-mediated apoptosis by blocking caspase pathways initiated by receptor activation (12). FLIP levels are partially under the control of NF-κB-responsive transcriptional elements (13) (as well as multiple AP-1 and nrf2 elements) within its gene sequence that gives rise to multiple splice variants of c-FLIP mRNA generating three distinct protein isoforms, FLIPL, FLIPS, and FLIPR. These isoforms contain two death effector domains (DEDs) involved in recruitment to the DED region of procaspase-8. The short FLIP isoforms, FLIPS and FLIPR, appear to block death receptor-induced apoptosis by inhibiting procaspase-8 activation (14), whereas the role of FLIPL is still controversial (12, 15–17), with both antiapoptotic and proapoptotic functions described. The profile of FLIP isoforms (and associated fragments) and their balance within the cell may determine whether or not a cell will elicit a cytotoxic response upon activation of different death receptors (18). FLIP isoforms have also been reported to play a prominent role in NF-κB signaling processes (19–21), with expression studies showing the ability of FLIP to interact with and influence TNFR machinery involved with TNF-mediated NF-κB responsiveness (19, 22–24).
In this work, we compared levels of TNF-induced NF-κB responsiveness and FLIP in human erythroleukemic cells with noncancerous primary human lymphocytes. We investigated the effects of sodium salicylate and tested the apoptotic effect of this NSAID and how it affected TNF-induced cellular processes. The intracellular and cell surface expression of TNFR subtypes as well as the effect of sodium salicylate and the effect of NF-κB inhibition were also investigated. We show that erythroleukemic cells have enhanced NF-κB responsiveness and display elevated FLIP levels compared with normal, noncancerous lymphocytes. Furthermore, sodium salicylate enhances the apoptotic effects of TNF in cancerous but not primary lymphocytes, and this action was similar to its effects on NF-κB inhibition and reduction of FLIP levels, independent of any alteration of TNFR subtype intracellular distribution but including an effect to enhance TNFR2 shedding. These findings provide important insights into the regulation of cancer cell apoptotic mechanisms and their regulation by death receptor signaling, shedding light on the regulation of life/death switching decisions made in cancerous lymphocytes.
Results and Discussion
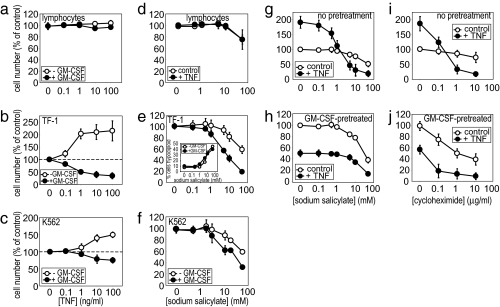
When treated with TNF, noncancerous, primary human lymphocytes displayed no apoptotic response, with a very slight proliferative effect observed (Fig. 1a). This effect was also true of primary monocytes purified from the same healthy volunteer blood samples (data not shown). By contrast, clonal human leukemia cells TF-1 and K562 displayed marked proliferation in response to TNF treatment (Fig. 1 b and c). This TNF-induced proliferation was switched to an alternative apoptotic TNF response by prior treatment of these cells with granulocyte–macrophage colony-stimulating factor (GM-CSF), as has been described previously (25). Therefore, fundamentally, there is a crucial difference between noncancerous blood cells and human leukemia cells, in that (i) TNF life/death “switching” can be observed only in TF-1 and K562 leukemia cells, and (ii) TNF-induced apoptotic responses are only observable in the leukemia cells. By comparison, the NSAID sodium salicylate (which has been suggested as a chemopreventive agent for colon cancer) showed enhanced cytotoxic responses in leukemia cells compared with noncancerous lymphocytes (Fig. 1 d–f). Furthermore, pretreatment of TF-1 and K562 cells with GM-CSF allowed greater cytotoxic effects of sodium salicylate to be observed but had no enhancing effect on primary lymphocytes. The cytotoxic effects of sodium salicylate in TF-1 cells were shown by cell cycle analysis (Fig. 1e Inset) to reveal induction of a hypodiploid state that is indicative of apoptotic DNA fragmentation. These data also show the switching by sodium salicylate of TF-1 cells from a TNF-responsive proliferative state into a TNF-responsive apoptotic state (Fig. 1g) as well as enhancement by sodium salicylate of any TNF-induced apoptosis in GM-CSF-pretreated TF-1 cells (Fig. 1h). Furthermore, cycloheximide has been widely used to allow the cytotoxic effects of apoptotic stimuli to be revealed and (like sodium salicylate) is also capable of enhancing the apoptotic effects of TNF in TF-1 cells (Fig. 1 i and j). Clearly, there is a difference between noncancerous and leukemic cells, and sodium salicylate is able to enhance selectively the apoptotic responses in cancerous blood cells.
Fig. 1.
TNF, sodium salicylate, and cycloheximide responses in normal and leukemic cells. MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] proliferation/cytotoxicity assays were performed on primary lymphocytes (a and d) and TF-1 (b, e, g–j), and K562 (c and f) leukemia cells treated with TNF (a–c), sodium salicylate (d–f, i, and j) or cycloheximide (g and h) stimulation. Except where indicated, cells were pretreated with 1 ng/ml GM-CSF or 50 ng/ml TNF for 24 h before stimulation. Data represent the means ± SEM, n = 4 (n = 3 for i and j).
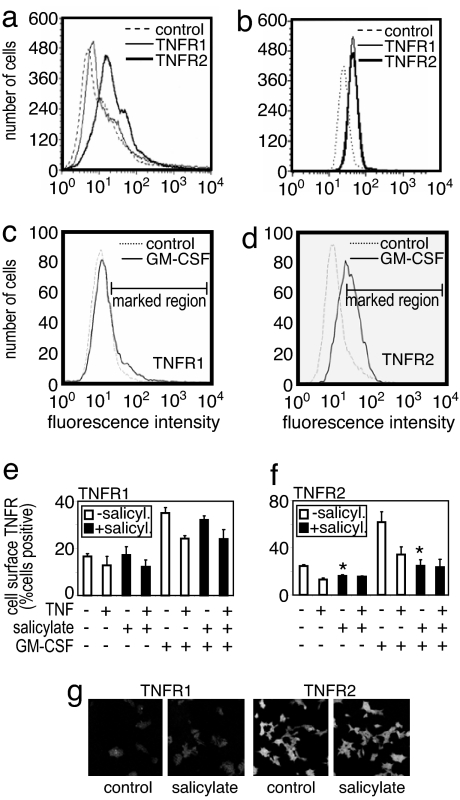
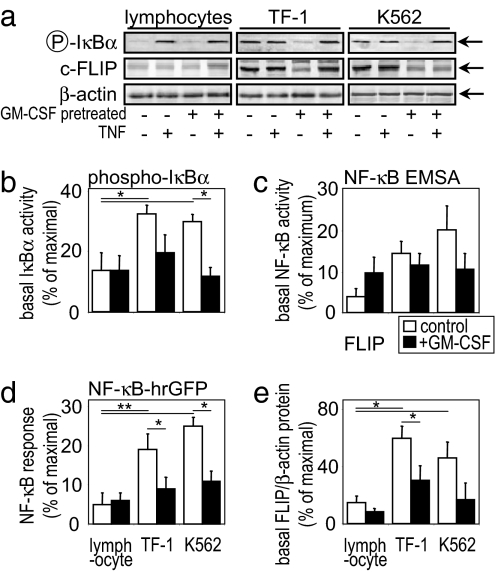
Our work here reveals that the underlying differences between the apoptosis-inducing effects of TNF and sodium salicylate in noncancerous versus leukemic cells are a combination of both the basal NF-κB responsiveness and basal FLIP levels among these different cell types. Interestingly, sodium salicylate did not alter the intracellular distribution of TNFR subtypes found in these cells (or cells overexpressing TNFR2: HeLa-TNFR2; see ref. 26); however, sodium salicylate was found to act like TNF in enhancing the shedding of TNFR2 from the surface of TF-1 or HeLa-TNFR2 cells (Fig. 2) but not normal lymphocytes (data not shown). TNFR2 is known to modulate the apoptotic signals of TNFR1, and its shedding is part of its activation characteristics (4). Fig. 3 clearly shows that activation by TNF of NF-κB responses occurs in noncancerous lymphocytes and leukemic cells. However, the basal levels of activity are significantly elevated in leukemic cell types, as shown by IκBα activation, NF-κB DNA binding via EMSA, and by NF-κB reporter construct transcription measurements. Although definition of the overall NF-κB responsiveness is intrinsically difficult to assess, as far as we can tell, the total TNF induction of NF-κB activity seems to be similar between noncancerous and leukemic cells. It is instead the basal unstimulated levels of NF-κB activity that are significantly greater in the cancer phenotype. That difference between noncancerous and leukemic cells is also true of the basal FLIP levels (Fig. 3 a and e). The elevated FLIP levels in leukemic cells are probably what allow TNF-induced proliferation to be observed in these cells and not in lymphocytes, which have lower basal FLIP levels. Moreover, pretreatment of leukemic cells with GM-CSF reduces the basal NF-κB activity and FLIP levels, thereby switching these cells into their apoptotic phenotype. By contrast, GM-CSF pretreatment of noncancerous lymphocytes does not reduce their basal NF-κB activity and FLIP levels, thereby an apoptotic response to TNF is not revealed, possibly because the basal levels of NF-κB and FLIP are already low/normal. However, we cannot discount other mechanisms that may exist to prevent TNF induction of apoptosis in these normal noncancerous blood cells. Indeed, many cell types will respond to TNF without undergoing cell death, perhaps being proliferative or inflammatory to the cytokine instead. The molecular basis and the differences in TNF-induced signaling underlying this distinct cell type-specific TNF responsiveness are poorly understood.
Fig. 2.
Sodium salicylate alteration of TNFR expression. Primary human lymphocytes (a) and TF-1 cells (b–f) treated as indicated, had their cell surface (a–f) or intracellular (g) TNFR1 and TNFR2 levels measured. TNFR levels of basal (a and b), 24 h, 1 ng/ml GM-CSF-pretreated (c–f), or sodium salicylate-treated (e–g) cells showed that GM-CSF increased TNFR2 expression that could be shed by TNF (50 ng/ml, 30 min) or sodium salicylate (5 mM, 1 h) stimuli. Primary human lymphocytes showed no sodium salicylate-induced shedding of TNFRs (data not shown). (g) TNFR subcellular distribution in HeLa-TNFR2 cells, with TF-1 cells similarly showing no subcellular alteration by sodium salicylate (data not shown). The data are from a single representative experiment repeated at least three other times with similar findings. Histograms represent the means ± SEM., n = 4, of immunofluorescence intensities within the marked regions (c and d).
Fig. 3.
Elevated NF-κB responsiveness in leukemia cells. (a) Western blot analysis of primary lymphocytes and TF-1 and K562 cells pretreated for 24 h with 1 ng/ml GM-CSF and/or 50 ng/ml TNF treatment for a further 24 h where indicated. Quantification of cellular IκBα protein levels (b), p65 NF-κB DNA-binding ELISA (c), NF-κB transcription reporter construct activity (d), and c-FLIP levels (e) is shown. Total β-actin protein levels were used as a loading control. Data represent the means ± SEM of at least four separate experiments.
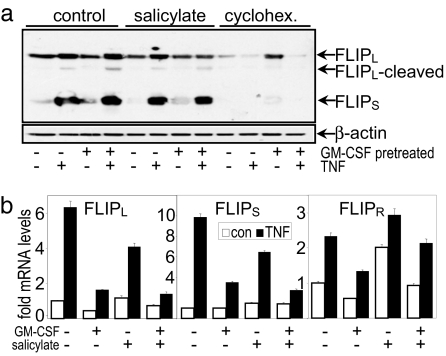
Inhibition of the enhanced NF-κB activity and FLIP levels seen in leukemia cells is achieved by sodium salicylate. Inhibition of basal and TNF-stimulated NF-κB activities by NSAIDs have been observed previously in TF-1 cells (25). As can be seen in Fig. 4, pretreatment of TF-1 cells with sodium salicylate results in a reduction of FLIP levels at the level of both protein and mRNA. Measurement of c-FLIP by the NF6 antibody was able to resolve the FLIPL and FLIPS isoforms of FLIP (as well as a cleaved FLIPL fragment); however, we find that with this antibody we are unable to measure FLIPR levels consistently in these cells. Measurement of FLIP isoforms regulation by sodium salicylate-treated TF-1 cells revealed that salicylate pretreatment reduced basal and TNF-stimulated levels of FLIPL and FLIPS and mRNA. Interestingly, sodium salicylate actually increased FLIPR isoform mRNA levels. This up-regulation of FLIPR may be of significance given that there is some evidence that certain FLIP isoforms (or their relative balance within the cell and at the TNFR complex) may act as proapoptotic signals rather than having the antiapoptotic properties that FLIPL and FLIPS seem to possess here (12). Likewise, cycloheximide also enhances the apoptotic effects of TNF in TF-1 cells (Fig. 1). Because cycloheximide has been used in many studies to permit or enhance TNF-induced cell death (4, 26), its ability to inhibit FLIP levels (Fig. 4a) may provide the mechanism by which this agent contributes to widespread proapoptotic effects in many cell types. Furthermore, its reduction of elevated NF-κB-driven basal FLIP translation, shown here in leukemia cells, is sufficient to switch these cells into an apoptotic rather than proliferative phenotype.
Fig. 4.
Sodium salicylate reduces elevated FLIP levels in leukemia cells. (a) Western blot analysis of TF-1 cells pretreated for 24 h with 1 ng/ml GM-CSF and/or 50 ng/ml TNF for a further 4 h where indicated. Where shown, cells had 5 mM sodium salicylate or 1 μg/ml cycloheximide preincubated for 1 h or 24 h, respectively, before the addition of TNF stimulus. FLIP isoforms were detected by NF-6 antibody. Concurrently run positive controls confirmed the identity of the bands, which are indicated by arrows. (b) Quantification of cellular FLIPL, FLIPS, and FLIPR mRNA levels by real-time PCR. Data represent the means ± SEM of at least six separate experiments.
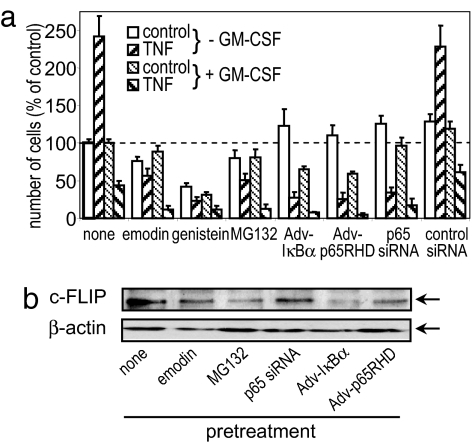
FLIP expression is partially under the control of NF-κB transcription factor, with AP-1 and nrf2 motifs also present in its promoter sequence. Moreover, little is known about the regulation of expression of each of the isoforms, being generated from the same gene. We show here (Fig. 5) that inhibition of NF-κB activity either by selective pharmacological agents or by adenoviral or siRNA knockdown of NF-κB protein reduced the levels of FLIP in TF-1 cells, which is in agreement with the sodium salicylate and cycloheximide data presented above. Selective inhibition or knockdown of NF-κB not only reduced FLIP levels, but also switched TF-1 cells from a proliferative phenotype into a solely apoptotic phenotype. These data demonstrate the crucial importance of NF-κB-driven FLIP expression in the mechanisms that regulate leukemia cell life/death switching. Thus, it is feasible that leukemia cells could be switched from their normal proliferative phenotype into an apoptotic phenotype by decreasing their FLIP activity. It has not escaped the attention of the authors that regulation of NF-κB activation or FLIP would be a useful treatment for the selective deletion of cancerous leukemia cells. Importantly, inhibition of NF-κB activity has serious widespread side effects because of its key role in innate and acquired immunity, leading to immunosuppression; equally molecular biological means of selectively reducing FLIP levels currently preclude this method as a means of therapeutically tackling this disease. Sodium salicylate provides an ideal drug to enhance leukemia cell apoptotic mechanisms because NSAIDs have good tolerance in humans, a wealth of data on their toxicity, and consistently promote safe cellular deletion by modulating cell signaling processes that select for an apoptotic cell phenotype in cancerous but not normal blood cells.
Fig. 5.
Inhibition of NF-κB and FLIP levels in leukemia cells. (a) MTS assay of TF-1 cells pretreated with the indicated pharmacological inhibitor (30 μg/ml emodin, 25 μg/ml genistein, or 5 μM MG132) for 1 h before 50 ng/ml TNF treatment for a further 24 h. Where indicated, cells were pretreated for 24 h with 1 ng/ml GM-CSF. p65 NF-κB siRNA and adenoviral pretreatments were for 24 h before TNF stimulation. Data represent the means ± SEM of at least three separate experiments. (b) c-FLIP and β-actin protein levels in pretreated TF-1 cells. The Western blotting data are from a single experiment representative of at least three independent results.
Unlike acetyl salicylate (aspirin), sodium salicylate does not inhibit platelet aggregation (27). Treatment of leukemia cells and normal cells with sodium salicylate showed NF-κB-dependent toxicity to primitive leukemia cells, which was at least partly a consequence of caspase-mediated events (28). We have also demonstrated that sodium salicylate was toxic and apoptotic when exposed to a human erythroleukemic cell line, in a more sensitive manner than to primary human lymphocytes. Sodium salicylate has previously been shown to cause vascular endothelial cell apoptosis (29) as well as significantly increasing TNF-induced apoptosis of human pancreatic cancer cells (30). Similarly, we have demonstrated that TNF-induced cytotoxicity and apoptosis of TF-1 cells were enhanced by sodium salicylate, regardless of whether TNF alone had a stimulatory or toxic effect. TF-1 erythroleukemic cells depend on GM-CSF or IL-3 for their survival (31). GM-CSF promotes survival and stimulates proliferation of hematopoietic cells (27). We have reported previously that GM-CSF and IL-3 enhanced the ability of TNF to induce apoptosis (31). However, these cytokines also had a proliferative effect that overcame any proapoptotic effect, and previous studies that used these cytokines to recruit leukemia cells into cell cycle, and thus make them more susceptible to chemotherapy, had revealed no benefit (32). Sodium salicylate was able to induce apoptosis in both quiescent and proliferating leukemic cells here, whereas TNF was toxic and apoptotic only when exposed to proliferating TF-1 or K562 cells.
Stimulation of TNFR1 can trigger such opposing cellular responses as NF-κB activation and induction of apoptosis within the same cell (33). The majority of TNF-induced NF-κB activation (34), growth inhibition, and suppression of proliferation also resulted exclusively from activation of TNFR1 (31, 35). In other studies, TNF-induced apoptotic and cytotoxic effects were mediated almost entirely by TNFR1, with TNFR2 having only a minimal effect (34, 36, 37). TNFR1-selective muteins but not TNFR2-selective mutant proteins have also been shown to induce NF-κB activation (38, 39). However, when overexpressed, TNFR2 can mediate apoptosis, NF-κB activation, and JNK activation (26, 40). We have shown here that sodium salicylate enhances the shedding of TNFR2 from TF-1 cells, which is presumably a consequence of its activation. We have shown previously that TNF had a stimulatory effect on quiescent TF-1 cells and an apoptotic effect on proliferating cells, and these effects were mediated predominantly by TNFR1 (31). However, TNFR2 levels are known to be up-regulated in the apoptotic phenotype (Fig. 2 and ref. 41), which may be targeted by sodium salicylate as a proapoptotic mechanism. Whereas both soluble and membrane-bound TNF have comparable bioactivities when acting on TNFR1, membrane-bound TNF is the prime activating ligand for TNFR2 (42). The membrane-bound form of TNF can also trigger effective cytotoxicity by stimulating TNFR2 (43). It is therefore likely that the effect of TNFR2 activation here has been experimentally underestimated. The TNFR1/TNFR2 ratio is likely to be important in determining the cytotoxic response of a cell (43). Sodium salicylate affects the TNFR ratio by causing shedding of TNFR2, and clearly the mode of these proapoptotic receptor mechanisms needs more investigation. TNF is a negative regulator of hematopoiesis and can mediate potent inhibitory signals in hematopoietic cells that induce apoptosis (44, 45), which may be regulated through TNFR2-induced death (46). We have shown that the primary human lymphocytes expressed both subtypes of TNFR, with a particularly high expression level of TNFR2. However, these cells were relatively unaffected by TNF exposure and did not alter the toxic effect of sodium salicylate.
Activation of NF-κB and subsequent induction of protective proteins are involved in TNF-induced cell survival§ and can protect against TNF-induced apoptosis (48). Thus, it has been proposed that the balance between life and death is regulated by NF-κB (9). The pathway that is initially chosen by a stimulus may depend on the presence or absence of low constitutive NF-κB activation, as seen here. Elevated basal NF-κB levels have been implicated in tumorigenicity (49, 50). Our investigations here in leukemia cells suggest that it is the combination of both elevated basal NF-κB/FLIP levels and higher overall NF-κB/FLIP responsiveness that is driving the cancerous phenotype. Recent work (24) has shown that FLIP is able to inhibit the sustained JNK activation by TNF that is found only in death phenotype leukemia cells (25). JNK and NF-κB pathways have many antagonistic features mediated by other intermediary proteins such as MKK7 inhibiting effects of GADD45β (51, 52). Thus, there may be a crucial relationship between the NF-κB, FLIP, and JNK activities of a cell that predetermines its proliferative or apoptotic nature. Our thinking is that whereas TNF is also capable or antiapoptotic signaling (increased NF-κB activation, stimulation of FLIP transcription, etc), sodium salicylate will only stimulate proapoptotic pathways (reduction of NF-κB and FLIP levels) and that these antiapoptotic pathways will be suppressed sufficiently by sodium salicylate to reveal the proapoptotic nature of TNFR signaling. These findings here provide further insight into the mechanisms controlling life/death decisions in leukemia cells and enhance the therapeutic potential of modulating these molecules as prospective treatments of various cancers including leukemia (53, 54).
Experimental Procedures
Materials.
Culture reagents were from Invitrogen (Carlsbad, CA). Recombinant human TNF was from R&D Systems (Minneapolis, MN). Recombinant human GM-CSF was a gift from Meenu Wadhwa (National Institute for Biological Standards and Control, U.K.). c-FLIP polyclonal and NF-6 monoclonal anti-FLIP antibodies were from R&D Systems and Axxora (San Diego, CA), respectively. TNFR-specific monoclonal antisera htr-9 (TNFR1) and utr-1 (TNFR2) were from Bachem, Ltd. (King of Prussia, PA). NF-κB inhibitory retroviruses were a gift from David Kluth (University of Edinburgh, Edinburgh, Scotland, U.K.). MG132 (Z-Leu-Leu-CHO), was from Calbiochem (San Diego, CA).
Cells.
TF-1 human erythroleukemia cells (41) were from ECACC (cell no. 93022307). K562 human chronic myelogenous leukemia cells were donated by Heather Wallace (University of Aberdeen). Cells were cultured as described previously (25).
Mononuclear cells were isolated, with a modification of a procedure described previously (55). Briefly, human blood was freshly drawn from healthy volunteers who gave informed consent. After dextran sedimentation to remove red blood cells, leukocytes were centrifuged on isotonic, discontinuous Percoll (Amersham Pharmacia, Piscataway, NJ) cushions at 700 × g, 20 min, 4°C. The mononuclear layer consisting of lymphocytes and 15–20% monocytes was aspirated, washed, and resuspended in appropriate medium for treatment and analysis. In experiments with isolated monocytes, enrichment was achieved by adherence to culture flasks for 1 h (37°C). Mononuclear cells were resuspended at 4 × 106/ml in Iscove's DMEM and plated out. Nonadherent cells (lymphocytes) were removed by aspiration gentle washing with PBS (Ca/Mg-free). Monocytes were resuspended in appropriate medium for treatment.
Cytotoxicity/Proliferation.
For NF-κB inhibition, TF-1 cells were pretreated with inhibitor for 1 h before TNF exposure. The metabolic activity of the cells (related to viable cell number) was assessed with the MTS assay (25).
Propidium Iodide.
Cells were incubated in 12-well plates with cytokines for 4 or 24 h. Cells were then washed with PBS and fixed with 70% ethanol. Propidium iodide (10 mg/ml) and RNase (1 mg/ml) were added to cells (dark, 25°C) for 30 min. Cells were washed with PBS and then analyzed by FACS.
TNFR Expression.
Cultured cells were washed and resuspended in serum-free medium. A 200-μl aliquot was incubated (1 h, 4°C) in a 1:50 dilution of anti-TNFR1 or anti-TNFR2 antibody (or neither for control). Cells were washed and resuspended in 200 μl of serum-free medium with a 1:50 dilution of FITC-labeled anti-mouse antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 30 min on ice. Before FACS analysis, cells were washed three times in PBS + 2% FBS. FACS analysis was performed on a FACScaliber (Becton Dickinson, San Jose, CA). Intracellular TNFR localization was achieved by the same protocol except cells were incubated with 1% Nonidet P-40 for 5 min (TF-1) or ice-cold methanol (20 s) (HeLa-TNFR2) before antisera incubation. Cells were fixed with 2% paraformaldehyde in PBS and then confocal analyzed on an LSM510 system (Zeiss, Thornwood, NY).
Western Blotting.
SDS/PAGE and Western blot analyses were performed as described previously (26). ECL detection and subsequent quantification were done with a VersaDoc imaging system (Bio-Rad, Hercules, CA).
NF-κB Transcription.
NF-κB DNA binding was measured with TransAM NF-κB p65 96-well colorimetric ELISA plates (Active Motif, Rixensart, Belgium) according to the manufacturer's protocol. NF-κB reporter construct analysis was performed as described (25).
Adenovirus and p65 siRNA.
Cell were infected with 25 pfu/ml Adv-IκB-α or Adv-p65RHD (adenoviral NF-κB inhibitors) as described (25). For siRNA transfection (56), cells were transfected with Lipofectamine (Invitrogen). Lipofectamine micelles were mixed with 200 nM p65 NF-κB-specific siRNA or control nonhomologous siRNA (Ambion, Austin, TX) and then added onto cells in 1 ml of serum-free medium for 4 h. Serum was then added (to 10% final concentration) and cultured for 24 h. More than 75% knockdown of p65 NF-κB compared with control samples was observed by Western blotting.
Real-Time PCR.
Total RNA was extracted from 1 × 106 cells TNF-treated TF-1 cells with the RNeasy kit (Qiagen, Valencia, CA). Reverse transcription and real-time PCR were performed as described (47) with primers for GAPDH and FLIPL, FLIPS, and FLIPR isoforms (Invitrogen). Primer sequences as follows: GAPDH forward, 5′-AACAGCCTCAAGATCATCAGCA-3′; GAPDH reverse, 5′-TGCTAAGCAGTTGGTGGTGC-3′; common FLIP forward, 5′-GTTCAAGGAGCAGGGACAAG-3′; FLIPL reverse, 5′-TCCCATTATGGAGCCTGAAG-3′; FLIPS reverse, 5′-ATCAGGACAATGGGCATAGG-3′; FLIPR reverse, 5′-CTTTCATGCTGGGATTCCATA-3′.
Statistics.
Suitable one-way ANOVA with a Bonferroni post hoc multiple comparison or Student's t test was performed. Significant P values were <0.05 (*) or <0.01 (**) where indicated.
Acknowledgments
This work was supported by the Leukaemia Research Fund Grant 05059 (to D.J.M.).
Abbreviations
- FADD
Fas-associated protein with death domain
- FLICE
FADD-like interleukin 1-converting enzyme (caspase-8)
- FLIP
FLICE-inhibitory protein
- GM-CSF
granulocyte-macrophage colony-stimulating factor
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- NSAID
nonsteroidal antiinflammatory drug
- TNFR
TNF receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Liu, R. Y., Fan, C., Olashaw, N. E., Patel, E., Zisholtz, T. C., and Zuckerman, K. S. (1999) Exp. Hematol. 27:81(abstr.).
References
- 1.Carswell EA, Old LJ, Kassel RL, Green S, Fiore N, Williamson B. Proc Natl Acad Sci USA. 1975;72:3666–3670. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiers W. FEBS Lett. 1991;285:199–212. doi: 10.1016/0014-5793(91)80803-b. [DOI] [PubMed] [Google Scholar]
- 3.Berkova N, Lemay A, Korobko V, Shingarova L, Sagaidak L, Goupil S. Cancer Detection Prevention. 1999;23:1–7. doi: 10.1046/j.1525-1500.1999.00067.x. [DOI] [PubMed] [Google Scholar]
- 4.MacEwan DJ. Cell Signalling. 2002;14:477–492. doi: 10.1016/s0898-6568(01)00262-5. [DOI] [PubMed] [Google Scholar]
- 5.Dannenberg AJ, Altorki NK, Boyle JO, Dang C, Howe LR, Weksler BB, Subbararnaiah K. Lancet Oncol. 2001;2:544–551. doi: 10.1016/S1470-2045(01)00488-0. [DOI] [PubMed] [Google Scholar]
- 6.Marks F, Furstenberger G. Eur J Cancer. 2000;36:314–329. doi: 10.1016/s0959-8049(99)00318-4. [DOI] [PubMed] [Google Scholar]
- 7.Baeuerle PA, Baltimore D. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 8.Beinke S, Ley SC. Biochem J. 2004;382:393–409. doi: 10.1042/BJ20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinon F, Holler N, Richard C, Tschopp J. FEBS Lett. 2000;468:134–136. doi: 10.1016/s0014-5793(00)01212-6. [DOI] [PubMed] [Google Scholar]
- 10.Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 11.Kopp E, Ghosh S. Science. 1994;265:956–959. doi: 10.1126/science.8052854. [DOI] [PubMed] [Google Scholar]
- 12.Peter ME. Biochem J. 2004;382:e1–e3. doi: 10.1042/BJ20041143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J. Mol Cell Biol. 2001;21:5299–5305. doi: 10.1128/MCB.21.16.5299-5305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golks A, Brenner D, Fritsch C, Krammer PH, Lavrik IN. J Biol Chem. 2005;280:14507–14513. doi: 10.1074/jbc.M414425200. [DOI] [PubMed] [Google Scholar]
- 15.Boatright KM, Deis C, Denault CB, Sutherlin DP, Salvesen GS. Biochem J. 2004;382:651–657. doi: 10.1042/BJ20040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Micheau O, Thome M, Schneider P, Holler N, Tschopp J, Nicholson DW, Briand C, Grutter MG. J Biol Chem. 2002;277:45162–45171. doi: 10.1074/jbc.M206882200. [DOI] [PubMed] [Google Scholar]
- 17.Chang DW, Xing Z, Pan Y, Algeciras-Schimnich A, Barnhart BC, Yaish-Ohad S, Peter ME, Yang XL. EMBO J. 2002;21:3704–3714. doi: 10.1093/emboj/cdf356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golks A, Brenner D, Krammer PH, Lavrik IN. J Exp Med. 2006;203:1295–1305. doi: 10.1084/jem.20051556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kataoka T, Tschopp J. Mol Cell Biol. 2004;24:2627–2636. doi: 10.1128/MCB.24.7.2627-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Field N, Low W, Daniels M, Howell S, Daviet L, Boshoff C, Collins M. J Cell Sci. 2003;116:3721–3728. doi: 10.1242/jcs.00691. [DOI] [PubMed] [Google Scholar]
- 21.Kreuz S, Siegmund D, Rumpf JJ, Samel D, Leverkus M, Janssen O, Hacker G, Dittrich-Breiholz O, Kracht M, Scheurich P, et al. J Cell Biol. 2004;166:369–380. doi: 10.1083/jcb.200401036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kataoka T, Budd RC, Holler N, Thome M, Martinon F, Irmler M, Burns K, Hahne M, Kennedy N, Kovacsovics M, et al. Curr Biol. 2000;10:640–648. doi: 10.1016/s0960-9822(00)00512-1. [DOI] [PubMed] [Google Scholar]
- 23.Guiet C, Silvestri E, De Smaele E, Franzoso G, Vito P. Cell Death Differ. 2002;9:138–144. doi: 10.1038/sj.cdd.4400947. [DOI] [PubMed] [Google Scholar]
- 24.Wicovsky A, Muller N, Daryab N, Marienfeld R, Kneitz C, Kavuri S, Leverkus M, Baumann B, Wajant H. J Biol Chem. 2007;282:2174–2183. doi: 10.1074/jbc.M606167200. [DOI] [PubMed] [Google Scholar]
- 25.Tucker SJ, Rae C, Littlejohn AF, Paul A, MacEwan DJ. Proc Natl Acad Sci USA. 2004;101:12940–12945. doi: 10.1073/pnas.0400949101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jupp OJ, McFarlane SM, Anderson HM, Littlejohn AF, Mohamed AAA, MacKay RH, Vandenabeele P, MacEwan DJ. Biochem J. 2001;359:525–535. doi: 10.1042/0264-6021:3590525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klampfer L, Cammenga J, Wisniewski HG, Nimer SD. Blood. 1999;93:2386–2394. [PubMed] [Google Scholar]
- 28.Guzman ML, Neering SJ, Upchurch D, Grimes B, Howard DS, Rizzieri DA, Luger SM, Jordan CT. Blood. 2001;98:2301–2307. doi: 10.1182/blood.v98.8.2301. [DOI] [PubMed] [Google Scholar]
- 29.Madge LA, Sierra-Honigmann MR, Pober JS. J Biol Chem. 1999;274:13643–13649. doi: 10.1074/jbc.274.19.13643. [DOI] [PubMed] [Google Scholar]
- 30.Mcdade TP, Perugini RA, Vittimberga FJ, Carrigan RC, Callery MP. J Surg Res. 1999;83:56–61. doi: 10.1006/jsre.1998.5560. [DOI] [PubMed] [Google Scholar]
- 31.Rae C, MacEwan DJ. Cell Death Differ. 2004;11:S162–S171. doi: 10.1038/sj.cdd.4401494. [DOI] [PubMed] [Google Scholar]
- 32.Hassan HT, Zander AR. Oncol Reports. 1997;4:1141–1149. doi: 10.3892/or.4.6.1141. [DOI] [PubMed] [Google Scholar]
- 33.Wajant H, Henkler F, Scheurich P. Cell Signalling. 2001;13:389–400. doi: 10.1016/s0898-6568(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 34.Hu XT, Tang MQ, Fisher AB, Olashaw N, Zuckerman KS. J Immunol. 1999;163:3106–3115. [PubMed] [Google Scholar]
- 35.Carter A, Haddad N, Draxler I, Israeli E, Raz B, Rowe JM. Br J Haematol. 1996;92:116–126. doi: 10.1046/j.1365-2141.1996.272806.x. [DOI] [PubMed] [Google Scholar]
- 36.Loetscher H, Stueber D, Banner D, Mackay F, Lesslauer W. J Biol Chem. 1993;268:26350–26357. [PubMed] [Google Scholar]
- 37.Horie T, Dobashi K, Iizuka K, Yoshii A, Shimizu Y, Nakazawa T, Mori M. Exp Hematol. 1999;27:512–519. doi: 10.1016/s0301-472x(98)00058-7. [DOI] [PubMed] [Google Scholar]
- 38.Jelkmann W, Hellwig-Buergel T. Exp Hematol. 1999;27:224–228. doi: 10.1016/s0301-472x(98)00054-x. [DOI] [PubMed] [Google Scholar]
- 39.McFarlane SM, Pashmi G, Connell MC, Littlejohn AF, Tucker SJ, Vandenabeele P, MacEwan DJ. FEBS Lett. 2002;515:119–126. doi: 10.1016/s0014-5793(02)02450-x. [DOI] [PubMed] [Google Scholar]
- 40.Haridas V, Darnay BG, Natarajan K, Heller R, Aggarwal BB. J Immunol. 1998;160:3152–3162. [PubMed] [Google Scholar]
- 41.Baxter GT, Kuo RC, Jupp OJ, Vandenabeele P, MacEwan DJ. J Biol Chem. 1999;274:9539–9547. doi: 10.1074/jbc.274.14.9539. [DOI] [PubMed] [Google Scholar]
- 42.Grell M, Douni E, Wajant H, Lohden M, Clauss M, Maxeiner B, Georgopoulos S, Lesslauer W, Kollias G, Pfizenmaier K, et al. Cell. 1995;83:793–802. doi: 10.1016/0092-8674(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 43.Vandenabeele P, Declercq W, Beyaert R, Fiers W. Trends Cell Biol. 1995;5:392–399. doi: 10.1016/s0962-8924(00)89088-1. [DOI] [PubMed] [Google Scholar]
- 44.Sato T, Yamauchi N, Sasaki H, Takahashi M, Okamoto T, Sakamaki S, Watanabe N, Niitsu Y. Cancer Res. 1998;58:1677–1683. [PubMed] [Google Scholar]
- 45.Tsushima H, Imaizumi Y, Imanishi D, Fuchigami K, Tomonaga I. Exp Hematol. 1999;27:433–440. doi: 10.1016/s0301-472x(98)00028-9. [DOI] [PubMed] [Google Scholar]
- 46.Murray J, Barbara JAJ, Dunkley SA, Lopez AF, Vanostade X, Condliffe AM, Dransfield I, Haslett C, Chilvers ER. Blood. 1997;90:2772–2783. [PubMed] [Google Scholar]
- 47.Zhou Z, Connell MC, MacEwan DJ. Cell Signalling. 2007;19:1238–1248. doi: 10.1016/j.cellsig.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 48.Li ZW, Chu WM, Hu YL, Delhase M, Deerinck T, Ellisman M, Johnson R, Karin M. J Exp Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aggarwal BB, Shishodia S, Sandur SK, Pandey MK, Sethi G. Biochem Pharmacol. 2006;72:1605–1621. doi: 10.1016/j.bcp.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 50.Braun T, Carvalho G, Fabre C, Grosjean J, Fenaux P, Kroemer G. Cell Death Differ. 2006;13:748–758. doi: 10.1038/sj.cdd.4401874. [DOI] [PubMed] [Google Scholar]
- 51.Papa S, Zazzeroni F, Bubici C, Jayawardena S, Alvarez K, Matsuda S, Nguyen DU, Pham CG, Nelsbach AH, Melis T, et al. Nat Cell Biol. 2004;6:146–153. doi: 10.1038/ncb1093. [DOI] [PubMed] [Google Scholar]
- 52.De Smaele E, Zazzeroni F, Papa S, Nguyen DU, Jin RG, Jones J, Cong R, Franzoso G. Nature. 2001;414:308–313. doi: 10.1038/35104560. [DOI] [PubMed] [Google Scholar]
- 53.Kataoka T. Crit Rev Immunol. 2005;25:31–58. doi: 10.1615/critrevimmunol.v25.i1.30. [DOI] [PubMed] [Google Scholar]
- 54.Micheau O. Exp Opin Ther Targets. 2003;7:559–573. doi: 10.1517/14728222.7.4.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hart SP, Ross JA, Ross K, Haslett C, Dransfield I. Cell Death Differ. 2000;7:493–503. doi: 10.1038/sj.cdd.4400680. [DOI] [PubMed] [Google Scholar]
- 56.McLaggan D, Adjimatera N, Sepcic K, Jaspars M, MacEwan DJ, Blagbrough IS, Scott RH. BMC Biotechnol. 2006;6:6. doi: 10.1186/1472-6750-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]