Abstract
Duplication of opsin genes has a crucial role in the evolution of visual system. Zebrafish have four green-sensitive (RH2) opsin genes (RH2–1, RH2–2, RH2–3, and RH2–4) arrayed in tandem. They are expressed in the short member of the double cones (SDC) but differ in expression areas in the retina and absorption spectra of their encoding photopigments. The shortest and the second shortest wavelength subtypes, RH2–1 and RH2–2, are expressed in the central-to-dorsal retina. The longer wavelength subtype, RH2–3, is expressed circumscribing the RH2–1/RH2–2 area, and the longest subtype, RH2–4, is expressed further circumscribing the RH2–3 area and mainly occupying the ventral retina. The present report shows that a 0.5-kb region located 15 kb upstream of the RH2 gene array is an essential regulator for their expression. When the 0.5-kb region was deleted from a P1-artificial chromosome (PAC) clone encompassing the four RH2 genes and when one of these genes was replaced with a reporter GFP gene, the GFP expression in SDCs was abolished in the zebrafish to which a series of the modified PAC clones were introduced. Transgenic studies also showed that the 0.5-kb region conferred the SDC-specific expression for promoters of a non-SDC (UV opsin) and a nonretinal (keratin 8) gene. Changing the location of the 0.5-kb region in the PAC clone conferred the highest expression for its proximal gene. The 0.5-kb region was thus designated as RH2-LCR analogous to the locus control region of the L-M opsin genes of primates.
Keywords: gene duplication, GFP reporter, transgenesis
Functional differentiation of duplicated genes is important for organismal evolution and is achieved through modifications of not only coding regions but also regulatory mechanisms of the gene expression in the duplicates. The regulatory mechanism for the spatial and the temporal coordination of gene expression among duplicated genes has been a subject of intense interest (1, 2). Gene duplications have an essential role in the evolution of the visual system. The visual opsins in vertebrates are classified into five phylogenetic types that originated before the vertebrate radiation: rod opsin or rhodopsin (RH1); RH1-like, or green cone opsin (RH2); short wavelength-sensitive type 1, or UV-blue cone opsin (SWS1); short wavelength-sensitive type 2, or blue cone opsin (SWS2); and middle- to long-wavelength-sensitive, or red-green cone opsin (M/LWS) (3). However, the understanding of the regulatory mechanisms for the differential expression among the five types is still fragmentary. Subtypes of the visual opsins, created by more recent gene duplications, provide an excellent model to study the regulatory evolution of duplicated genes. A well documented example is the locus control region (LCR) of the L-M opsin genes (subtypes of M/LWS) arrayed on the X chromosome in the catarrhine primates (Old World monkeys, apes, and humans) (4).
The primate L-M opsin LCR is located at ≈3.5 kb upstream of the gene array and interacts with only the most proximal or the second proximal gene of the array, often an L and an M opsin gene, respectively, through their proximal promoters (4–6). The choice of the promoters by the LCR is largely a stochastic process (7, 8). There is only a single active X chromosome in a photoreceptor cell becaue of the random inactivation of one X chromosome in females and the hemizygosity in males. As a result, the distribution of L and M cone cells is nearly random in the retina, although there is a slight tendency for clumping of L and M cones in humans (9). Together with the scattered distribution of S cones, this system enables the trichromatic color vision of the primates.
It is not known whether the randomness of the gene choice of the LCR is controlled, and if so, how. There is a tendency for the L:M mRNA ratio to increase toward the periphery of the retina in humans and baboons, but there is no well defined central-to-peripheral gradient in macaques (10). The overall L:M mRNA ratio in the retina differs among species; L opsin expression is dominant in humans, but M opsin is dominant in baboons (10). A comparative study of gene regulation of opsin duplication is necessary for more a comprehensive understanding of visual system evolution.
Zebrafish (Danio rerio) have nine visual opsin genes, including two spectrally distinct M/LWS and four separate RH2 opsin subtypes in addition to single-copy genes of the SWS1, SWS2, and RH1 types (11). The four RH2 genes, RH2–1, RH2–2, RH2–3, and RH2–4, which are most closely related to the goldfish RH2 opsin genes (12), are positionally arrayed in this sequential order and are functionally differentiated with shorter to longer wavelength absorption spectra in this order in their encoding photopigments (11). All these opsin genes are expressed in a single type of cone cell, the short member of double cones (SDC), but differ in the expression areas in the retina. The shortest and the second shortest wavelength subtypes, RH2–1 and RH2–2, are expressed in the central-to-dorsal retina, the longer wavelength subtype, RH2–3, circumscribing the RH2–1/RH2–2 area, and the longest wavelength subtype, RH2–4, further circumscribing the RH2–3 area and mainly occupying the ventral retina (13). In the vertebrate retina, peripheral cells are developmentally younger and have a longer history of cell divisions than the central cells. Therefore, the ringed expression pattern of the zebrafish RH2 opsin genes could be partly ascribed to their temporally ordered expression through the gene order (13).
The zebrafish RH2 system is more developmentally restricted than the primate L-M opsin system. These systems result in different visual functions, different spectral sensitivity, and possibly different color vision, which are caused by visual angles in zebrafish and the improvement of color dimension from dichromacy to trichromacy in primates. Despite these differences, the present study shows that the zebrafish RH2 system is controlled by an analogous regulatory component found in the primate L-M system, a single upstream-located region regulating the duplicated genes, designated as RH2-LCR.
Results
A Large Insert P1-Artificial Chromosome (PAC) DNA Clone Contains a Sufficient Regulator for Expression of the RH2 Opsin Genes.
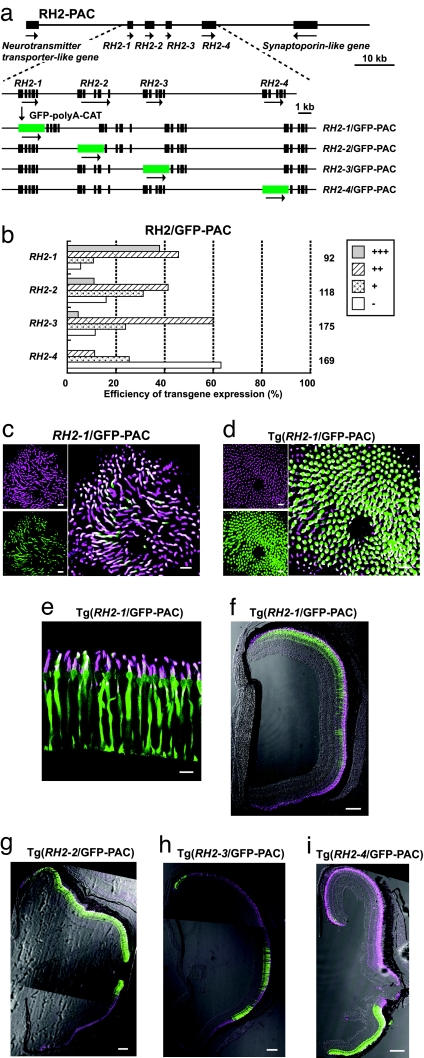
It was shown that the adjacent upstream regions of RH2–1 and RH2–3 of 10.5 and 8 kb, respectively, failed to drive the GFP reporter expression (14). These negative results were confirmed for the four RH2 opsin genes by injecting the GFP reporters conjugated to their adjacent upstream regions of 7.3, 2.8, 2.6, and 6.7 kb in RH2–1, RH2–2, RH2–3, and RH2–4, respectively, into zebrafish embryos. There was no GFP expression in the SDCs (data not shown). A PAC clone with an ≈85-kb insert encompassing the four RH2 opsin genes (RH2-PAC) was used to learn whether it contains a distant regulatory region(s) sufficient for their proper gene expression. A series of modified RH2-PAC (RH2/GFP-PAC) clones, RH2–1/GFP-PAC, RH2–2/GFP-PAC, RH2–3/GFP-PAC and RH2–4/GFP-PAC were constructed in which the first exon of one of the four RH2 genes after the initiation codon was replaced with a GFP-encoding DNA segment (Fig. 1a).
Fig. 1.
GFP expression in the zebrafish retina with the RH2/GFP-PAC transgenes. (a) Construction of the RH2/GFP-PAC clones. The RH2-PAC clone contains the four RH2 opsin genes as well as synaptoporin-like and neurotransmitter transporter-like genes. In the expanded view, boxes indicate exons of the four RH2 genes. The orientation of transcription is given by the arrow for each gene. In the RH2/GFP-PAC clones, the first exon of one of the RH2 opsin genes was replaced with the GFP-polyA-CAT cassette by site-specific homologous recombination. (b) Expression levels at 5 dpf of the GFP reporter in zebrafish injected with the RH2/GFP-PAC clones. The histogram shows the percentage of eyes graded into four levels (+++, ++, +, and −) according to the number of GFP-expressing cells in the retina. The names to the left of the histogram indicate the gene replaced with GFP. The numbers to the right of the histogram show the total number of eyes examined for each of the RH2/GFP-PAC transgenes. (c–i) Images of immunostained SDCs with the anti-RH2 antibody (magenta) and GFP-expressing cells in the retina (green). Overlap of the two signals appears as white. Note that the antibody recognizes the four RH2 subtype opsins (13, 15). (c) A whole-mount retina of a 7-dpf zebrafish embryo injected with the RH2–1/GFP-PAC. (d) A whole-mount retina of a 7-dpf Tg(RH2–1/GFP-PAC) fish. (c and d) Right is the overlay of Left Upper and Lower. (e) A tangential section of the photoreceptor layer in an adult Tg(RH2–1/GFP-PAC) fish. Note that the opsin (magenta) localizes in the outer segment, whereras the GFP (green) is found mostly in the cell body of a SDC. (f–i) Transverse sections of the adult transgenic zebrafish retinas: Tg(RH2–1/GFP-PAC) (f); Tg(RH2–2/GFP-PAC) (g); Tg(RH2–3/GFP-PAC) (h); and Tg(RH2–4/GFP-PAC) (i). The dorsal side is at the top and the ventral side is at the bottom. (Scale bars: c–e, 10 μm; f–i, 100 μm.)
At 5 days postfertilization (dpf) the zebrafish larvae injected with one of the RH2/GFP-PAC clones were graded for GFP expression into four levels, +++, ++, +, −, according to Luo et al. (14). GFP expression was observed for the four RH2/GFP-PAC clones in ≈40–90% of eyes examined (Fig. 1b). Immunostaining of the 7 dpf embryos using the antibody against the zebrafish green (RH2) opsin (15) showed that the GFP signals overlapped with the RH2 immunostaining signals of the SDCs [Fig. 1c and supporting information (SI) Fig. 6 a–c].
Separate transgenic lines of zebrafish were produced for each of the four RH2/GFP-PAC transgenes, Tg(RH2–1/GFP-PAC), Tg(RH2–2/GFP-PAC), Tg(RH2–3/GFP-PAC), and Tg(RH2–4/GFP-PAC). In these four lines, GFP and immunostaining signals appeared perfectly overlapped. GFP was specifically expressed in the SDCs of the retina and not in other cells/tissues from embryos examined at 7 dpf (Fig. 1d) and in adults (Fig. 1e and SI Fig. 6 d–f). Retinal areas of GFP expression in the four transgenic lines were also consistent with the expression patterns of the corresponding RH2 genes in adults (13). GFP signals were present in the central-to-dorsal area of the retina in Tg(RH2–1/GFP-PAC) (Fig. 1f), in a larger central to dorsal area in Tg(RH2–2/GFP-PAC) (Fig. 1g), in a narrow dorsal and a part of ventral areas in Tg(RH2–3/GFP-PAC) (Fig. 1h), and in the most dorsal area and a large ventral area in Tg(RH2–4/GFP-PAC) (Fig. 1i). These results indicate that the RH2-PAC clone contains sufficient regulatory region(s) for the proper expression of the four RH2 opsin genes.
A Distal Regulatory Region, RH2-LCR, Was Found in the RH2-PAC Clone.
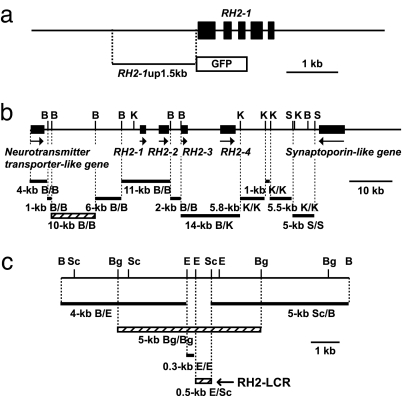
The existence of distal regulatory region(s) in the RH2-PAC clone was investigated by employing a coinjection protocol using mixed concatamers of separate fragments formed upon integration (16). A restriction fragment of the clone DNA was injected together with a GFP-reporter construct conjugated to the 1.5-kb adjacent upstream region of RH2–1 (Fig. 2a). Among the restriction fragments analyzed (see Fig. 2b), only the 10-kb BamHI fragment drove GFP expression in the eyes of 35 of 62 (56%) surviving embryos examined at 5 dpf; the other fragments did not result in a GFP signal in 53–159 surviving embryos examined. Further examination of the BamHI 10-kb region (Fig. 2c) revealed that GFP signals appeared only when the injected segments contained the 0.5-kb EcoRI-SacI region (Fig. 2c). GFP signals appeared in the eyes examined at 5 dpf in 46 of 106 (43%) and in 46 of 135 (34%) of the surviving embryos when the 4.8-kb BglII and the 0.5-kb EcoRI-SacI fragments were used, respectively. There were no embryos having a GFP signal among the 96–142 surviving embryos examined at 5 dpf that received DNA segments without the 0.5-kb EcoRI-SacI region. The 0.5-kb region was designated “RH2-LCR” (Fig. 2c and SI Fig. 7).
Fig. 2.
Coinjection strategy to search for a distal regulatory region(s). (a) The 1.5-kb upstream region of RH2–1 conjugated to the GFP reporter. (b) Restriction fragments used for the coinjections. The 10-kb BamHI (B/B) fragment capable of inducing GFP expression is depicted as a hatched bar. (c) Restriction fragments from the 10-kb BamHI region used for the coinjection. The fragments capable of inducing GFP expression are depicted as hatched bars. B, BamHI; Bg, BglII; E, EcoRI; K, KpnI; Sc, SacI; S, SalI.
The RH2-LCR Is Sufficient for Gene Expression in SDCs.
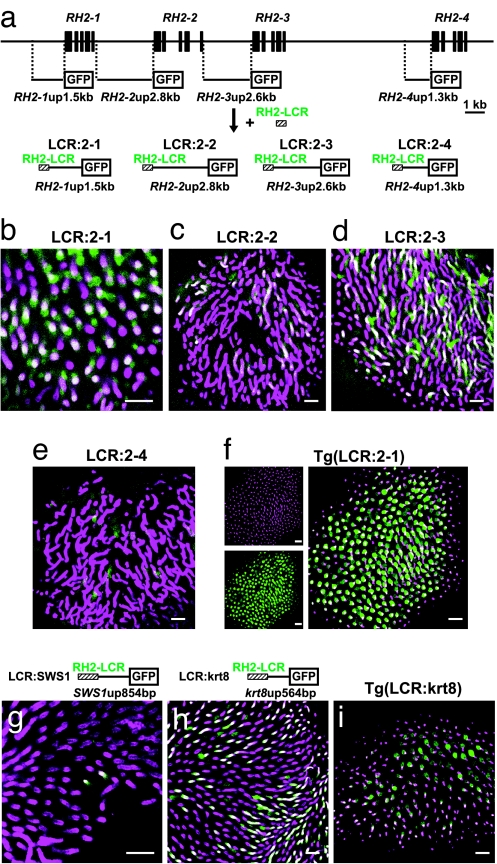
The RH2-LCR was attached to the GFP-reporter constructs in which the reporter promoters were from the adjacent upstream regions of RH2–1 (1.5 kb; designated RH2–1up1.5kb), RH2–2 (2.8 kb; designated RH2–2up2.8kb), RH2–3 (2.6 kb; designated RH2–3up2.6kb) and RH2–4 (1.3 kb; designated RH2–4up1.3kb), the resulting constructs were designated as LCR:2–1, LCR:2–2, LCR:2–3 and LCR:2–4, respectively (see Fig. 3a). Zebrafish embryos injected with these construct showed GFP signals in the SDCs immunostained with the anti-RH2 antibody (Fig. 3 b–e) at 7 dpf, although the overlap of these signals was relatively obscure for LCR-2–4 (Fig. 3e). Using the LCR:2–1, a transgenic line of zebrafish was established and was designated Tg(LCR:2–1). In the Tg(LCR:2–1) retina, most GFP signals overlapped with the immunostaining for the anti-RH2 antibody in the SDCs at 7 dpf (Fig. 3f). A few GFP signals were found overlapping in the long single cones immunostained with the antibody against zebrafish blue (SWS2) opsin. There were no overlapping signals found in the other cone cell types examined using antibodies against zebrafish red (M/LWS) and UV (SWS1) opsins at 7 dpf (SI Fig. 8).
Fig. 3.
GFP expression in SDCs by RH2-LCR-coupled promoters. (a) Construction of RH2-LCR-coupled GFP reporters, LCR:2–1, LCR:2–2, LCR:2–3, and LCR:2–4, with the upstream region of RH2–1, RH2–2, RH2–3, and RH2–4, respectively. (b–i) Images of the immunostained SDCs with the anti-RH2 antibody (magenta) and GFP expressing cells in the retina (green). Overlap of the two signals appears as white. (b–e) Whole-mount retinas of 7-dpf zebrafish embryos injected with LCR:2–1 (b), LCR:2–2 (c), LCR:2–3 (d), and LCR:2–4 (e). (f) A whole-mount retina of a 7-dpf Tg(LCR:2–1) fish. Right is the overlay of Left Upper and Lower. (g) A whole-mount retina of a 7-dpf zebrafish embryo injected with LCR:SWS1. (h) A whole-mount retina of a 7-dpf zebrafish embryo injected with the LCR:krt8. (i) A whole-mount retina of a 7-dpf Tg(LCR:krt8) fish. (Scale bars: 10 μm.)
The RH2-LCR was further tested for its ability to induce gene expression in the SDCs by using the promoter regions of a non-SDC, UV (SWS1) opsin (17), and a nonretinal, keratin 8 (18) gene. The 854- and 564-bp adjacent upstream regions of the SWS1 and keratin 8 genes, designated SWS1up854bp and krt8up564bp, respectively, were sandwiched between the RH2-LCR and the GFP reporter and these resulting constructs were referred to as LCR:SWS1 and LCR:krt8, respectively (Fig. 3 g and h). The SWS1up854bp is incapable of inducing gene expression in the retina (14). The krt8up564bp induces gene expression specifically in the epithelial tissues, but not in the retina, and has been used for enhancer trapping as a basal promoter (19). Zebrafish embryos injected with these constructs showed GFP signals in the SDCs immunostained with the anti-RH2 antibody at 7 dpf (Fig. 3 g and h). In the LCR:SWS1-injected fish, more than half of the GFP-expressing cells were the SDCs, 87 of 162 in eight eyes examined, and in the LCR:krt8 injected fish, nearly all GFP-expressing cells in eyes were the SDCs in addition to the expression found in the epithelial tissues. Using the LCR:krt8, a transgenic line of zebrafish was established and was designated Tg(LCR:krt8). In the Tg(LCR:krt8) retina, GFP-expressing cells in the eyes overlapped with SDCs (Fig. 3i), as well as in the epithelial tissues.
The RH2-LCR Is Necessary for Gene Expression in SDCs.
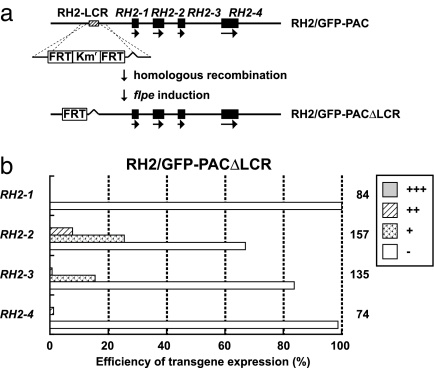
The RH2-LCR region was removed from the series of RH2/GFP-PAC clones, and these constructs were collectively referred to as RH2/GFP-PACΔLCR clones (Fig. 4). Microinjection of the RH2–1/GFP-PACΔLCR and RH2–4/GFP-PACΔLCR clones into zebrafish embryos resulted in the absence of GFP expression at 5 dpf (Fig. 4b). Microinjection of the RH2–2/GFP-PACΔLCR and RH2–3/GFP-PACΔLCR clones resulted in GFP signals in ≈20–40% of the eyes examined at 5 dpf (Fig. 4b). These signals did not overlap with SDCs immunostained by the anti-RH2 antibody at 7 dpf. The majority of signals were located in the inner retinal layer, and a few were located in the photoreceptor layer, but not in SDCs (SI Fig. 9a). Transgenic lines of zebrafish with each of the RH2/GFP-PACΔLCR clones did not show GFP expression in the SDCs of adults (SI Fig. 9 b–d). These results indicate that the RH2-LCR is necessary for each of the four RH2 opsin genes to be expressed in the SDCs.
Fig. 4.
Effect of RH2-LCR elimination from the RH2/GFP-PAC clones on GFP expression in SDCs. (a) Construction of the RH2/GFP-PACΔLCR clones from the RH2/GFP-PAC clones. The RH2-LCR was replaced with the Kmr gene flanked on both sides by the FRT sequences through homologous recombination; the Kmr and one FRT were removed by flpe induction. (b) Expression levels of the GFP reporter in zebrafish injected with the RH2/GFP-PACΔLCR clones. The histogram is shown as in Fig. 1b.
The RH2-LCR Location Affects the Expression Level of the Four RH2 Opsin Genes.
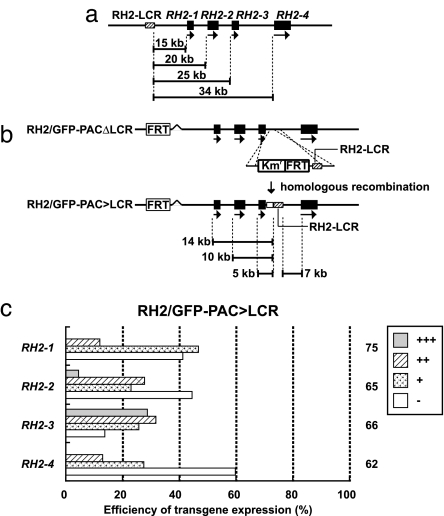
The possibility that the distance from the RH2-LCR to the RH2 genes has relevance to their expression differences was investigated because, as shown in Fig. 1b, the expression level of a RH2 opsin gene, evaluated by its GFP-reporter expression, seems to decrease as its distance from the RH2-LCR increases (see also Fig. 5a). The location of the RH2-LCR was changed to the position immediately downstream of RH2–3 in the four RH2/GFP-PAC clones by homologous recombination using the RH2/GFP-PACΔLCR clones as templates; the resulting constructs were collectively referred to as RH2/GFP-PAC>LCR clones (Fig. 5b). Microinjection of the series of the RH2/GFP-PAC>LCR clones caused dramatic changes of the expression level of the GFP reporter compared with their original RH2/GFP-PAC clones (compare Fig. 1b with Fig. 5c). RH2–3, the most proximal to the RH2-LCR, showed the greatest GFP expression, followed by RH2–2 and RH2–1/RH2–4 at 5 dpf. A retinal image of the GFP expression recovered in the RH2–3/GFP-PAC>LCR injection is shown in SI Fig. 9e.
Fig. 5.
Positional effect of the RH2-LCR in the RH2/GFP-PAC clones on GFP expression in SDCs. (a) Distances from the RH2-LCR to the four RH2 opsin genes in the RH2-PAC clone. (b) Construction of RH2/GFP-PAC>LCR clones from the RH2/GFP-PACΔLCR clones. The RH2-LCR, coupled with Kmr and one FRT sequence, was inserted into the position immediately downstream of RH2–3 in RH2/GFP-PACΔLCR clones by homologous recombination. The Kmr-FRT region is depicted as an open box after the recombination. Distances from the RH2-LCR to the four RH2 opsin genes in the RH2/GFP-PAC>LCR clones are shown. (c) Expression levels of the GFP reporter in zebrafish injected with the RH2/GFP-PAC>LCR clones. The histogram is shown as in Fig. 1b.
Discussion
The present study showed that a 0.5-kb region located at 15 kb upstream of the RH2 opsin gene array in the zebrafish genome is necessary and sufficient for expression of the four RH2 opsin genes in the SDCs of the retina. This 0.5-kb region is capable of inducing gene expression in the SDCs of non-SDC and nonretinal genes. Deletion of this region abolishes gene expression in SDCs. Differences of the expression level among the RH2 genes appear to be correlated with their distance from the 0.5-kb region, with the most proximal gene being expressed at the highest level. This is analogous to human L-M opsin in which a regulatory region, called LCR, is located upstream of the L-M opsin gene array (4). LCR is necessary for the expression of both L and M opsin genes (20) and preferentially interacts with the closer L-M opsin genes in the array (5, 6). The zebrafish 0.5-kb region was designated as RH2-LCR, being the first LCR-type regulator reported for non-primate opsin genes and the first RH2 opsin regulator identified in vertebrates.
In the RH2-LCR region, one OTX sequence (TAATCC/T) (21, 22) and two octamer sequences similar to the Oct-1 recognition site (23, 24) were found (SI Fig. 7). The OTX sequence is present in the primate L-M opsin LCR (22) and is a recognition site of CRX, which is a transcription factor regulating photoreceptor-specific genes (21, 22). However, no other similarity was detected in the primate L-M opsin LCR, in other regulatory regions identified to date, or in other zebrafish opsin genes. To clarify whether these and possibly other unidentified motifs are involved in the RH2-LCR function, and if so, how, transgenic studies using the RH2-LCR with a series of deletions and mutations are required.
It was apparent that not all of the GFP signals originated from the SDCs of the transgenic fish carrying the GFP reporter coupled to the RH2-LCR (SI Fig. 8). It remains to be determined whether these are merely accidental ectopic expressions often observed in transgenic experiments. Additional transgenic lines would be necessary to draw any conclusion, but it is likely that the signals would reflect some RH2-LCR function because they were all observed in photoreceptor cells including SDCs, which are ontogenically/developmentally closely related.
It should be noted that the GFP expression levels in the embryos injected with the RH2–1/GFP-PAC>LCR and RH2–2/GFP-PAC>LCR clones were lower than those injected with the original RH2–1/GFP-PAC and RH2–2/GFP-PAC clones (compare Fig. 1b with Fig. 5c) even though they were closer to the RH2-LCR region in the modified clones than in the original clones (Fig. 5a and 5b). These results suggest that the relative distance, but not the absolute distance, from the RH2-LCR to the genes affects their expression levels. This implies that the genes compete for their interaction with the RH2-LCR and that those closer genes had a greater chance to interact with the RH2-LCR. This distance dependency could further be tested, for example, by keeping the RH2-LCR at its original position and moving the proximal promoter region of a RH2 gene with a GFP conjugated to the promoter to the location of another RH2 gene.
It should also be noted that the expression level of RH2–4, as evaluated by its GFP reporter expression, is relatively insensitive to the distance effect and remains at a low level (see Figs. 1b and 5 a–c). This suggests that the proximal region of RH2–4 might contain some repressive component, being consistent with the low expression level of the GFP reporter when the RH2-LCR is directly connected to the proximal region of the RH2–4 (Fig. 3e).
The four RH2 opsin genes of zebrafish differ in their expression areas in the retina, with RH2–1 and RH2–2 expressed in the central-to-dorsal retina, RH2–3 circumscribing the central area, and RH2–4 further circumscribing the RH2–3 area and mainly occupying the ventral retina (13). The expression starts with RH2–1, followed by RH2–2 and RH2–3/RH2–4 in the retina (13). In vertebrate retina, eyes grow by adding new cells to the peripheral zones, and the peripheral cells are developmentally younger than central cells (25). In fish, the eyes continue to grow through the lifetime by adding new cells to the peripheral zones (26), resulting in a larger gradient of developmental timing in the fish retina than in the primate retina. The peripheral cells have a longer history of cell divisions and have gone through a longer history in chromatin remodeling for transcriptional regulation than central cells in the retina. Assuming that gene switching is affected by change of chromatin configuration through development, the ringed expression pattern of RH2 opsin genes in zebrafish might be related to the larger gradient of chromatin-remodeling history throughout the retina than that in primate retina, in which L and M cones are intermingled with a less conspicuous gradient of L/M cone ratio (10). It is the focus of our ongoing study to clarify whether and how the RH2-LCR is involved in the gene switching process through the development and the formation of the ringed-expression pattern.
Materials and Methods
Construction of GFP-Expression Plasmids.
The pEGFP-1 plasmid (BD Biosciences Clontech, Tokyo, Japan) was used as the source of the GFP gene to assemble GFP-expression constructs. The promoter of keratin 8 was provided by V. Korzh (Institute of Molecular and Cell Biology, Proteos, Singapore) (19). The GFP-expression plasmid constructs were linearized with a restriction enzyme at one site in the vector backbone for microinjection.
RH2-PAC Clone.
Through the screening service of the Resource Center Primary Database (RZPD, Berlin, Germany) of a zebrafish PAC library [no. 706, originally created by C. Amemiya (Benaroya Research Institute, Seattle WA)], a clone DNA (BUSMP706P12115Q2), designated RH2-PAC here, was obtained using the RH2–1 cDNA as a probe (11), which encompasses the four RH2 opsin genes in its ≈85-kb insert. The nucleotide sequence of the RH2-PAC corresponds to the nucleotide position 32275795–32361831 of the chromosome 6 in the Ensembl zebrafish assembly version 6 (www.ensembl.org/Danio_rerio/index.html). Employing this sequence information, restriction sites of BamHI, SalI, and KpnI were mapped on the clone. The GFP-expression constructs and the restriction fragments derived from the RH2-PAC were purified by agarose gel electrophoresis.
Construction of RH2/GFP-PAC Clones.
The RH2-PAC clone was modified by the site-specific recombination system coupled with drug selection by using either the Escherichia coli strain DY380 or EL250, which possesses the heat-inductive gam, beta, and exo for facilitating homologous recombination, and by the flpe-FRT recombination system for excision of a DNA region sandwiched by FRT sequences using E. coli EL250 which possesses the arabinose-inductive flpe as described by Lee et al. (2001) (27). The I-SceI meganuclease system (28) was used for efficient transgenesis of the PAC-derived constructs. A kanamycin-resistance (Kmr) gene in the vector backbone of the RH2-PAC clone was replaced by using site-specific recombination with an ampicillin-resistance (Ampr) cassette having two I-SceI recognition sites sandwiching the Ampr gene. The first exon after the initiation codon of one of the RH2 opsin genes was replaced with a GFP-polyA-CAT cassette having a GFP coding sequence connected to the SV40 polyadenylation signal (polyA) and the chloramphenicol acetyl transferase (CAT) gene (Fig. 1a).
Construction of RH2/GFP-PACΔLCR and RH2/GFP-PAC>LCR Clones.
The RH2-LCR was PCR amplified from the RH2-PAC clone with its 5′ and 3′ flanking sequences of ≈200 bp each, designated 5FL and 3FL, respectively, and was cloned into the pBluescript II (SK-) plasmid (Stratagene, Tokyo, Japan). A Kmr gene flanked by the FRT sequences at both sides, FRT-Kmr-FRT, was inserted into the EcoRI site of the RH2-LCR in the plasmid; the resulting cassette was designated 5FL-FRT-Kmr-FRT-LCR-3FL.
For construction of the RH2/GFP-PACΔLCR clones, the RH2-LCR was removed from the 5FL-FRT-Kmr-FRT-LCR-3FL plasmid by PCR amplification outside the RH2-LCR and reconnection of the resulted FRT and 3FL ends. The assembled cassette, 5FL-FRT-Kmr-FRT-3FL, was isolated from the plasmid by PCR and subjected to site-specific recombination to replace the RH2-LCR in the RH2-PAC clone with the FRT-Kmr-FRT cassette (see Fig. 4a). In EL250, induction of flpe resulted in the excision of the FRT-Kmr-FRT cassette, leaving one FRT sequence (27).
For convenience, to construct the RH2/GFP-PAC>LCR clones, the 5FL-FRT-Kmr-FRT-LCR-3FL plasmid was used as a source of Kmr, with the RH2-LCR for a drug-selection purpose. The Kmr-FRT-LCR cassette was PCR-amplified from the plasmid, although the FRT in the cassette was irrelevant for this construction. To insert the cassette to the 3′ UTR of RH2–3 by the homologous recombination, the forward PCR primer contained the 3′ UTR sequence after the stop codon of RH2–3, and the reverse primer contained its adjacent downstream sequence. The RH2/GFP-PAC>LCR clones were produced by the homologous recombination of the amplified cassette with the RH2/GFP-PACΔLCR clones, with the Kmr-FRT sequence remaining at the recombination site.
Microinjection.
Zebrafish were maintained at 28.5°C in a 14-h light/10-h dark cycle as described by Westerfield (29). The linearized GFP-expression constructs, at a final concentration of 30–50 ng/μl, were resuspended in 0.1 M KCl with tetramethyl-rhodamin dextran added as a tracer and were microinjected into the cytoplasm of embryos at the one-cell stage. The GFP-expression constructs of the 1.5-kb proximal promoter of RH2–1 was coinjected with each of the restriction fragments derived from the RH2-PAC at a final concentration of ≈5 and 10–15 ng/μl, respectively, in 0.1 M KCl and tetramethyl-rhodamin dextran tracer. The RH2-PAC derived constructs (20 ng/μl) were injected with I-SceI meganuclease (0.5 units/μl) (New England Biolabs, Beverly, MA) in a solution of 0.5 × commercial meganuclease buffer with tetramethyl-rhodamin dextran tracer (28). Embryos were grown in 0.003% 1-phenyl-2-thiourea after 12–24 h postfertilization (hpf) to disrupt pigment formation.
The eyes of injected embryos were examined at 5 dpf for GFP fluorescence under a dissecting fluorescent microscope, and the number of eyes expressing GFP was determined as described by Luo et al. (14). The eyes were scored as +++, ++, +, and − when GFP was expressed in more than 50 cells, in 5–50 cells, in 1–4 cells, and in no cells per eye, respectively.
For the generation of transgenic lines, the injected embryos were grown to sexual maturity and crossed with noninjected fish in a pair-wise fashion. The genomic DNA extracted from a pool of the resulting embryos was examined for the presence of the transgene by PCR amplification of the GFP DNA segment as described in Hamaoka et al. (30). Fish of the subsequent generations were screened for the presence of the transgene by PCR amplification of the GFP from genomic DNA extracted from a fin clip or GFP fluorescence in the eyes.
Immunohistochemistry.
Immunostaining was carried out using whole-mount retinas of 7 dpf larvae or with adult retinal sections following the procedure of Luo et al. (14). Antibodies against zebrafish opsins raised in rabbits were provided by T. Vihtelic (University of Notre Dame, South Bend, IN) (15) and were used to stain cone photoreceptors. The Cy3-conjugated anti-rabbit IgG was used as a secondary antibody. Images of GFP and Cy3 fluorescence were captured using a Zeiss 510 laser-scanning confocal microscope (Zeiss, Thornwood, NY).
Supplementary Material
Acknowledgments
We are grateful to Dr. N. G. Copeland (National Cancer Institute, Frederick, MD), for the E. coli strains DY380 and EL250, to Drs. T. Vihtelic and D. Hyde (University of Notre Dame), for the antibodies against the zebrafish opsins, and to Dr. V. Korzh for the GFP expression construct of keratin 8. This study was supported by Japan Society for the Promotion of Science Scientific Research Grant-in-Aid B 16405015 (to S.K.) and Fellowship 14-08073 (to A.C.)
Abbreviations
- dpf
days postfertilization
- Kmr
kanamycin-resistance
- LCR
locus control region
- PAC
P1 artificial chromosome
- RH2
green-sensitive cone opsin
- SDC
short member of double cones
- SWS1
ultraviolet-sensitive cone opsin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704061104/DC1.
References
- 1.Orkin SH. Cell. 1990;63:665–672. doi: 10.1016/0092-8674(90)90133-y. [DOI] [PubMed] [Google Scholar]
- 2.Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- 3.Yokoyama S. Prog Retin Eye Res. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Macke JP, Merbs SL, Zack DJ, Klaunberg B, Bennett J, Gearhart J, Nathans J. Neuron. 1992;9:429–440. doi: 10.1016/0896-6273(92)90181-c. [DOI] [PubMed] [Google Scholar]
- 5.Winderickx J, Battisti L, Motulsky AG, Deeb SS. Proc Natl Acad Sci USA. 1992;89:9710–9714. doi: 10.1073/pnas.89.20.9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi T, Motulsky AG, Deeb SS. Nat Genet. 1999;22:90–93. doi: 10.1038/8798. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Smallwood PM, Cowan M, Blesh D, Lawler A, Nathans J. Proc Natl Acad Sci USA. 1999;96:5251–5256. doi: 10.1073/pnas.96.9.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smallwood PM, Wang Y, Nathans J. Proc Natl Acad Sci USA. 2002;99:1008–1011. doi: 10.1073/pnas.022629799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofer H, Carroll J, Neitz J, Neitz M, Williams DR. J Neurosci. 2005;25:9669–9679. doi: 10.1523/JNEUROSCI.2414-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neitz M, Balding SD, McMahon C, Sjoberg SA, Neitz J. Vis Neurosci. 2006;23:379–385. doi: 10.1017/S095252380623325X. [DOI] [PubMed] [Google Scholar]
- 11.Chinen A, Hamaoka T, Yamada Y, Kawamura S. Genetics. 2003;163:663–675. doi: 10.1093/genetics/163.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinen A, Matsumoto Y, Kawamura S. Mol Biol Evol. 2005;22:1001–1010. doi: 10.1093/molbev/msi086. [DOI] [PubMed] [Google Scholar]
- 13.Takechi M, Kawamura S. J Exp Biol. 2005;208:1337–1345. doi: 10.1242/jeb.01532. [DOI] [PubMed] [Google Scholar]
- 14.Luo W, Williams J, Smallwood PM, Touchman JW, Roman LM, Nathans J. J Biol Chem. 2004;279:19286–19293. doi: 10.1074/jbc.M400161200. [DOI] [PubMed] [Google Scholar]
- 15.Vihtelic TS, Doro CJ, Hyde DR. Vis Neurosci. 1999;16:571–585. doi: 10.1017/s0952523899163168. [DOI] [PubMed] [Google Scholar]
- 16.Muller F, Williams DW, Kobolak J, Gauvry L, Goldspink G, Orban L, Maclean N. Mol Reprod Dev. 1997;47:404–412. doi: 10.1002/(SICI)1098-2795(199708)47:4<404::AID-MRD6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 17.Takechi M, Hamaoka T, Kawamura S. FEBS Lett. 2003;553:90–94. doi: 10.1016/s0014-5793(03)00977-3. [DOI] [PubMed] [Google Scholar]
- 18.Gong Z, Ju B, Wang X, He J, Wan H, Sudha PM, Yan T. Dev Dyn. 2002;223:204–215. doi: 10.1002/dvdy.10051. [DOI] [PubMed] [Google Scholar]
- 19.Parinov S, Kondrichin I, Korzh V, Emelyanov A. Dev Dyn. 2004;231:449–459. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]
- 20.Nathans J, Davenport CM, Maumenee IH, Lewis RA, Hejtmancik JF, Litt M, Lovrien E, Weleber R, Bachynski B, Zwas F, Klingaman R, Fishman G. Science. 1989;245:831–838. doi: 10.1126/science.2788922. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 22.Furukawa T, Morrow EM, Cepko CL. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 23.Bendall AJ, Sturm RA, Danoy PA, Molloy PL. Eur J Biochem. 1993;217:799–811. doi: 10.1111/j.1432-1033.1993.tb18308.x. [DOI] [PubMed] [Google Scholar]
- 24.Pankratova EV, Polanovsky OL, Polanovasky OL. FEBS Lett. 1998;426:81–85. doi: 10.1016/s0014-5793(98)00316-0. [DOI] [PubMed] [Google Scholar]
- 25.La Vail MM, Rapaport DH, Rakic P. J Comp Neurol. 1991;309:86–114. doi: 10.1002/cne.903090107. [DOI] [PubMed] [Google Scholar]
- 26.Moshiri A, Close J, Reh TA. Int. J. Dev Biol. 2004;48:1003–1014. doi: 10.1387/ijdb.041870am. [DOI] [PubMed] [Google Scholar]
- 27.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 28.Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. Mech Dev. 2002;118:91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- 29.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) Eugene: Univ Oregon Press; 1995. [Google Scholar]
- 30.Hamaoka T, Takechi M, Chinen A, Nishiwaki Y, Kawamura S. Genesis. 2002;34:215–220. doi: 10.1002/gene.10155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.