Abstract
Production of TNF-α and IL-1 in infectious and autoimmune diseases is associated with fever, fatigue, and sleep disturbances, which are collectively referred to as sickness behavior syndrome. In mice TNF-α and IL-1 increase nonrapid eye movement sleep. Because clock genes regulate the circadian rhythm and thereby locomotor activity and may alter sleep architecture we assessed the influence of TNF-α on the circadian timing system. TNF-α is shown here to suppress the expression of the PAR bZip clock-controlled genes Dbp, Tef, and Hlf and of the period genes Per1, Per2, and Per3 in fibroblasts in vitro and in vivo in the liver of mice infused with the cytokine. The effect of TNF-α on clock genes is shared by IL-1β, but not by IFN-α, and IL-6. Furthermore, TNF-α interferes with the expression of Dbp in the suprachiasmatic nucleus and causes prolonged rest periods in the dark when mice show spontaneous locomotor activity. Using clock reporter genes TNF-α is found here to inhibit CLOCK-BMAL1-induced activation of E-box regulatory elements-dependent clock gene promoters. We suggest that the increase of TNF-α and IL-1β, as seen in infectious and autoimmune diseases, impairs clock gene functions and causes fatigue.
Keywords: behavior, circadian rhythms, cytokines, innate immunity
In microbial infections, host defense mechanisms activate the innate and adaptive arms of the immune response. Microbial recognition by Toll-like receptors (TLR) expressed by macrophages and dendritic cells leads to the activation of signal transduction pathways with induction of various genes including IL-1, TNF-α, IL-6, and IFN-α/β (1–3). These cytokines mediate the acute-phase response, which is a systemic generalized reaction characterized by fever, fatigue, and weight loss, an increase in the number of neutrophils, and the induction of synthesis of acute-phase proteins in the liver with increased haptoglobin, antiproteases, complement components, fibrinogen, ceruloplasmin, and ferritin in the blood (4). An acute phase response with a dose-dependent state of lethargy and severe fatigue has been described in cancer patients treated with TNF-α (5). A link between production of TNF-α and daytime fatigue has also been suggested in rheumatoid arthritis (RA) and in the obstruction sleep apnea syndrome (OSAS). Inhibition of TNF-α by soluble TNF-receptor p75 improves disabling fatigue in patients with RA (6). A TNF-α-308 (A-G) single-nucleotide polymorphism and elevated TNF-α serum levels have been described in OSAS (7, 8). Moreover, neutralization of TNF-α reduces daytime sleepiness in sleep apneics (9). Direct effects of TNF-α on spontaneous sleep are also shown in animal studies. i.v., i.p., or intracerebroventricular injections of TNF-α or IL-1 enhance nonrapid eye-movement (NREM) sleep (for reviews, see refs. 10 and 11). This increase in NREM sleep is independent of the fever-inducing capacity of these cytokines (12). Although there is good evidence for TNF-α and IL-1 as mediators of altered sleep–wake behavior, the underlying mechanisms remain elusive.
The regulation of sleep depends on a circadian control and a homeostatic drive (13, 14). The circadian influence is provided by the suprachiasmatic nuclei (SCN) of the hypothalamus being entrained by light stimuli to the environment. This self-sustaining circadian pacemaker uses a molecular mechanism similar to the one used in subsidiary oscillators present in any type of cell in the organism. The molecular clockwork involves the transcriptional repressor genes Per1, Per2, Cry1, and Cry2, as well as the transcriptional activators Bmal1 and Clock. The heterodimerized transcription factor BMAL1:CLOCK activates Per and Cry gene transcription by binding to E-box motives in their promoters. PER and CRY proteins inhibit BMAL1:CLOCK complexes, thereby inhibiting their own gene expression. This feedback-loop mechanism generates circadian oscillations of Per and Cry expression. The same positive and negative regulatory components also govern the rhythmic expression of the nuclear orphan receptor Rev-Erbα, which in turn represses the transcription of Bmal1 through direct binding to a REV-ERBα response element in the Bmal1 promoter. Thereby, REV-ERBα interconnects the cyclic expression of positive- and negative-loop members (for reviews, see refs. 15 and 16). The targeted inactivation of Bmal1 showed that this gene is indispensable for the maintenance of circadian functions (17). Clock-deficient mice show robust circadian patterns of locomotor activity with period lengths shortened by only 20 min; Dbp mRNA rhythm, however, was severely blunted in both the SCN and the liver (18). The deletion of the clock-controlled genes (CCG) PAR bZip transcription factors Dbp, Tef, and Hlf only moderately affects the circadian clock but leads to pronounced disturbances of locomotor activity. In the present study, our objective was to examine (i) whether and how TNF-α influences the circadian timing system and modulates the expression of clock genes and CCGs, and (ii) whether TNF-α affects locomotor activity, rest time, and periodicity in mice.
Results
Suppressed Expression of Clock Genes in TNF-α-Treated Synchronized Fibroblasts.
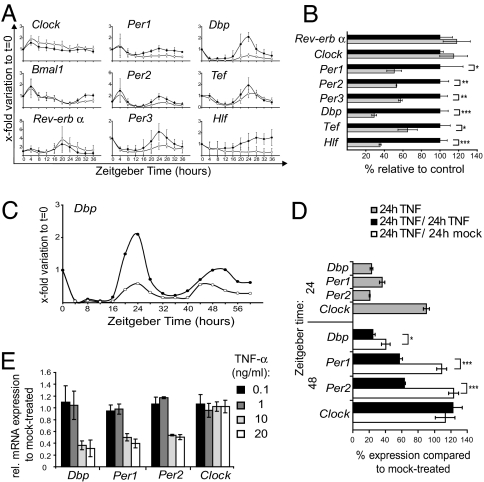
To determine whether TNF-α interferes with circadian gene expression, we used NIH 3T3 fibroblast cultures synchronized by a 2-h serum shock; this system elicits a well described circadian expression of central clock genes and CCGs for at least three cycles (19). We analyzed serum shocked fibroblasts challenged over time with or without TNF-α (10 ng/ml). RNA was extracted every 4 h thereafter up to Zeitgeber time (ZT) 36 and analyzed by using quantitative real-time RT-PCR methods. Within the first 12 h, the course of the examined clock gene expression was not altered by TNF-α treatment. However, around the peak (ZT 20–28), TNF-α strikingly suppressed the expression of the central clock genes Per1, Per2, and Per3 and of the CCGs Dbp, Tef, and Hlf. Peak expression of Rev-erbα mRNA at ZT 20 was slightly increased, whereas Bmal1 expression was not affected by TNF-α. Clock expression was slightly increased. For all genes, TNF-α did not affect the phases but rather attenuated the amplitude of expression (Fig. 1A). Therefore, we assessed the extent of clock gene expression at their main peak at ZT 24. TNF-α induced a significant reduction of gene expression for Per1, Per2, Per3, Dbp, Tef, and Hlf genes. Rev-Erbα expression, as assessed at its peak of expression at ZT 20, remained unchanged by TNF-α, as did Clock (Fig. 1B). We extended the time of observation over a second circadian cycle and analyzed rhythmic expression up to 60 h. In this, emphasis was placed on Dbp expression, because its amplitude was the most affected by TNF-α. Rhythmicity and period length of Dbp expression were not changed by TNF-α, but its amplitude was severely suppressed at both peaks (Fig. 1C). To determine whether the TNF-α-mediated suppression is reversible, we washed out TNF-α after the first cycle (at ZT 24) and analyzed the second peak at ZT 48. By washing out TNF-α, the second peak of expression of Per1 and Per2 was restored, and Dbp expression increased compared with the control where TNF-α still led to reduced peak expression. The expression of Clock was not affected when extending the time of TNF-α treatment to 48 h (Fig. 1D). Moreover, the suppression of Dbp, Per1, and Per2 gene expression in synchronized NIH 3T3 fibroblasts is clearly dose-dependent, being most significantly affected with concentrations >1 ng/ml TNF-α. The expression of Clock remained unchanged irrespective of the dose of TNF-α (Fig. 1E).
Fig. 1.
TNF-α impairs expression of clock genes in synchronized NIH 3T3 fibroblasts. (A) TNF-α attenuates circadian Per1/2/3 and PAR bZIP family (Dbp, Tef, and Hlf) gene expression. After serum shock (ZT 0–2), cells were kept in serum-free medium with TNF-α (10 ng/ml; open circles) or without the cytokine (filled circles) and analyzed every 4 h with quantitative real-time RT-PCR. Results are shown as x-fold variations to nonsynchronized fibroblast cultures at ZT 0; three independent experiments; mean values ± SD. (B) Per and PAR bZIP family genes are significantly down-regulated at the 24-h peak (TNF-α: gray bars; controls: black bars), whereas Clock and RevErbα are not affected. Expression of Rev-Erbα was assessed at its peak at ZT 20. Data show one representative experiment done in triplicate (mean ± SD) of four experiments. (C) The amplitude of Dbp expression in serum-shocked NIH 3T3 fibroblasts was attenuated during 60 h in the presence of TNF-α (open circles) compared with controls (filled circles). (D) Withdrawal of TNF-α after the first peak at ZT 24 shows that the suppression of Dbp, Per1, and Per2 at the second peak at ZT 48 is reversible. After serum shock, cells were treated with TNF-α for 24 h (gray bars), or for 48 h with or without a withdrawal of TNF-α after 24 h (white and black bars, respectively). Gene expression was compared with mock-treated cells at the respective ZT (100% expression). Data show one representative experiment done in triplicates (mean ± SD) of three experiments. (E) The suppression of the expression of Dbp, Per1, and Per2 in synchronized fibroblasts at ZT 24 is dose-dependent being significant (P < 0.005) at doses higher than 1 ng/ml TNF-α. Data show the mean ± SD of three independent experiments performed in triplicates. For BDE, we used the independent-sample t test; *, P ≤ 0,05; **, P ≤ 0,005; ***, P ≤ 0.0005.
IL-1β, but Neither IFN Nor IL-6, Shares with TNF-α the Effect to Down-Regulate Clock Genes.
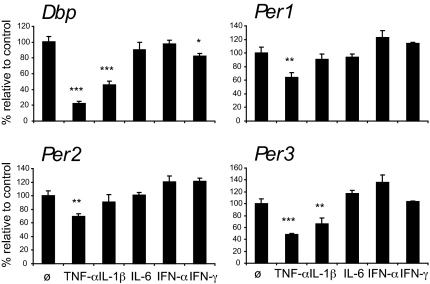
Besides TNF-α, other cytokines are produced by TLR-activated macrophages and dendritic cells that have been implicated in the sickness behavior syndrome, namely the proinflammatory cytokines IL-1, IL-6, and IFN-γ and type I IFN cytokines IFN-α and -β (20). Therefore, we examined whether the effect of TNF-α on clock genes is a unique property of TNF-α or is shared by other cytokines. Besides TNF-α, also IL-1β suppressed the expression of both, Dbp and Per3 (Fig. 2). The expression of Dbp, Per1, Per2, and Per3 was not inhibited by treatment of synchronized fibroblasts with IL-6 and IFN-α. IFN-γ induced a significant, albeit minor, suppression of Dbp expression.
Fig. 2.
Cytokine effects on Dbp, Per1, Per2, and Per3 in NIH 3T3 fibroblasts. Confluent cells were synchronized with 50% horse serum for 2 h (ZT 0–2). After serum shock, cells were kept in serum-free medium with TNF-α (10 ng/ml), IL-1β (10 ng/ml), IL-6 (10 ng/ml), IFN-α (10 ng/ml), IFN-γ (20 ng/ml) or without cytokines and analyzed at ZT 24 with quantitative real-time RT-PCR. Results are shown as percent of expression to noncytokine-treated fibroblast cultures; one representative experiment of three; mean values of triplicates ± SD; independent sample t test; *, P ≤ 0,05; **, P ≤ 0,005; ***, P ≤ 0.0005.
TNF-α Interferes with E-Box-Dependent Transcription of Clock Genes.
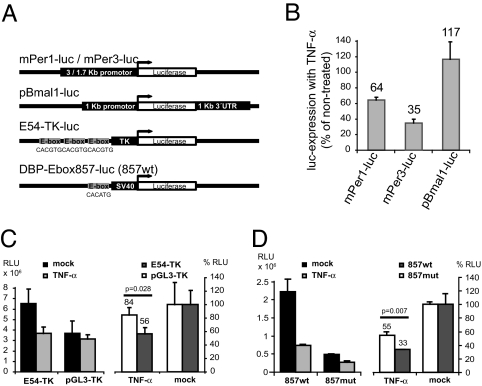
Circadian transcriptional activation of Per genes and of the PAR bZip transcription factor genes Dbp, Tef, and Hlf is thought to depend on the binding of the heterodimer BMAL1:CLOCK to the canonical or noncanonical E-box, a basic helix–loop–helix transcription factor-binding site ideally harboring the sequence CACGTG (21). In our study, TNF-α affected suppression only of the transcription of those clock genes harboring an E-box upstream of the transcription initiation site (Per genes and Dbp, Tef, and Hlf). No suppression was observed on Bmal1 and Clock expression, their transcription not being E-box-dependent. Thus, TNF-α may interfere with E-box-mediated transcriptional activation. To test this hypothesis, we performed transient transfections of NIH 3T3 cells with luciferase reporter genes under the control of the native 3- or 1.7-kB promoter sequences of mouse Per1 and Per3, respectively, and of the 1-kB promoter of Bmal1 (Fig. 3A). Consistent with the gene expression studies, Per1 and Per3 promoter activity was suppressed after TNF-α administration but not that of the Bmal1 promoter, which is devoid of E-box elements (Fig. 3B). To specify the possible effect on E-boxes, we performed assays using NIH 3T3 cells stably transfected with a luciferase reporter plasmid consisting of three E-boxes within 2.0 kb of the 5′ flanking region of the mouse Per1 gene with their immediate flanking sequence linked together and joined to the thymidine kinase promoter (22) (Fig. 3A). Again, treatment with TNF-α reduced luciferase activity by 45%, corresponding to the basal activity of the basic pGL3-TK vector without E-boxes (Fig. 3C). An E-box reporter construct of the Dbp gene (Dbp-E-box857; Fig. 3A) that was cotransfected with plasmids expressing CLOCK and BMAL1 proteins showed expression reduced by 67%, indicating that overexpressed CLOCK and BMAL1 are still efficiently blocked. In contrast, with the mutated E-box (ACCAGT instead of CACATG) reporter construct (Dbp-857 mut), the activity was significantly derepressed (Fig. 3D). We suggest therefore that TNF-α suppresses E-box-mediated transcription of clock genes. Thus, selective down-regulation of clock genes or CCGs is likely to depend on the presence of E-box elements in the respective genes.
Fig. 3.
TNF-α suppresses E-box-mediated transcription of clock genes. (A) Schematic representation of the luciferase reporter genes used. mPer1-luc (3-kb promoter fragment) and mPer3-luc (1.7-kb promoter fragment) contain promoter sequences upstream of their genes in the pGL3basic vector. pBmal1-luc is regulated by a 1-kb promoter fragment and a 1-kb sequence of the 3′UTR from mBmal1 gene. E54-TK consists of the three E-boxes of the mouse Per1 gene and their immediate flanking sequences in front of a TK promoter. DBP-Ebox857-luc contains one of four E-boxes of the Dbp promoter regulating the activity of a SV40 promoter. (B) Native clock gene promoters bearing an E-box are affected by TNF-α. Percent luciferase expression of mPer1-luc, mPer3-luc, and pBmal1-luc transfected NIH 3T3 cells after treatment with 10 ng/ml TNF-α overnight compared with untreated controls (100% luciferase expression) is shown. (C) E-boxes of mPer1 are affected by TNF-α treatment. TNF-α significantly reduces E54-TK-dependent luciferase expression but not pGL3-TK lacking E-boxes and flanking sequences as shown by raw data (RLU, relative light units; Left) and percent inhibition (Right). (D) The E-box of Dbp gene at position +857 is repressed by TNF-α even when CLOCK and BMAL1 are overexpressed. Cotransfection with CLOCK and BMAL1 leads to four to five times higher relative luciferase activity of 857wt compared with the mutated E-box vector (857mut) indicating that the interaction of CLOCK:BMAL1 with the E-box is functional. Luciferase activity is efficiently suppressed by overnight treatment with TNF-α as shown by raw data (Left) and percent inhibition (Right). TNF-α leads to a higher repression in 857wt than in 857mut. For all assays, mean ± SD of triplicates from one representative experiment of three; independent sample t test.
Reduction of Clock Genes and CCGs in TNF-α-Treated Mice.
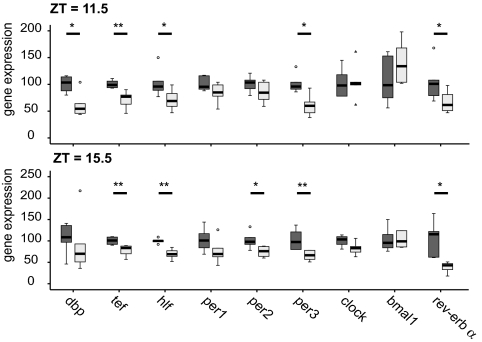
To assess the effect of TNF-α on the expression of clock genes in vivo, C57BL/6 mice were constantly infused with TNF-α (1.5 μg/day or 0.075 mg/kg per day) via an osmotic minipump inserted s.c. on the back of the mice over a period of 7 days (see Materials and Methods). The dose chosen is ≈10-fold lower than the dose used to induce a septic shock-like disease in rats (0.7 mg/kg, administered intravenously) (23). Histopathology revealed no evidence for TNF-α-induced vascular damage, hemorrhages, or inflammation in the liver, lung, and kidney based on morphological criteria and on the expression of heme oxygenase (HO-1), a marker for oxidative stress. Macrophages of mice infused with TNF-α showed signs of being activated in the liver and kidney [supporting information (SI) Fig. 6]. We found TNF-α serum concentrations at 43 (±10.9) pg/ml at day three in TNF-α-treated mice compared with 10 (±0.8) pg/ml in controls (SI Fig. 7). In mice with experimental septic shock the respective value for TNF-α exceeds 10 ng/ml (24). In controls, osmotic pumps were filled with diluent (PBS, 0.1% BSA). Total RNA was extracted from the liver of mice treated with TNF-α or control; expression of clock genes was tested at day three at ZT 11.5 and ZT 15.5 (ZT 0 = 6 a.m. lights on, ZT 12 = 6 p.m. lights off), when we encountered maximal inhibition of locomotion (Fig. 5A). As shown for fibroblasts in vitro, TNF-α also suppressed the expression of Dbp, Tef and Hlf, and Per3, but not of Bmal1 and Clock (Fig. 4). Although Per1 and Per2 were down-regulated, this was not significant. However, at a later time point (ZT 15.5), when Per2 normally reaches its peak of expression, significant suppression of Per2 was observed (Fig. 4 Lower). In contrast to the fibroblast data, we found that TNF-α suppressed the expression of Rev-Erbα. TNF-α induced reduction of expression of the Per genes and of PAR bZip transcription factor genes is likely to influence the expression of clock output genes. Among the genes that are positively regulated by DBP is the liver-specific albumin gene (25). The capacity of TNF-α to interfere with Dbp mRNA expression is congruent with the observation of a decrease in albumin serum concentration by 41% in TNF-α-treated mice compared with controls (SI Fig. 8). TNF-α has already been described as inhibiting albumin synthesis in liver cells, but the mechanisms remain elusive (26). Taken collectively, these data provide evidence that the suppressive role of TNF-α on Dbp is associated with impaired activation of their target gene. TNF-α-treated mice also show higher endogenous expressions of IL-1β and TNF-α in the liver [both genes are well known to be induced by TNF-α (28)]; the level of expression of these cytokines correlates with each other (SI Fig. 9). Furthermore, the increased expression of the cytokines in the liver is associated with a decrease of the CCG Dbp, Hlf, and Tef; the extent of the decrease of these PAR bZip transcription factors fits to the TNF-α expression in individual mice (SI Fig. 9).
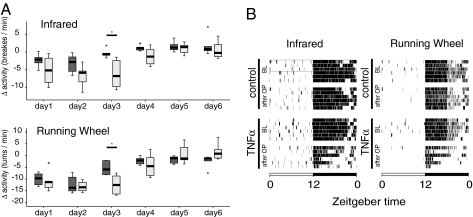
Fig. 5.
TNF-α reduces locomotor activity of mice but does not alter their circadian rest-activity cycles in free running conditions. (A) Reduced locomotor activities in mice with constant TNF-α infusions are detected from days 1–4. Boxplot showing the changes of locomotor activity for each day after minipump implantation (TNF-α, 1.5 μg/day, for 6 days) or saline as control, compared with the baseline (mean of 3 days immediately before the operation). The TNF-α-treated group is shown as light gray bars and the control group as dark gray bars; circles are outliers, and triangles are extreme values. Six mice per group were analyzed. Data show one experiment (ANOVA for repeated measures, followed by independent-sample t tests; *, P ≤ 0.05). The effect of TNF-α to impair locomotor activity was confirmed at day 3 in two independent experiments. (B) TNF-α reduces locomotor activity in the second half of the dark phase. Actograms (as measured by infrared and running wheel) show data from six individual mice before and after the minipump insertion. Each line represents one individual mouse per treatment, 1 day before the implantation of minipumps (baseline, BL) and 3 days after the operation (OP).
Fig. 4.
Impaired expression of clock genes in livers of TNF-α-infused mice. Animals were entrained to a LD cycle during 2 weeks, and their locomotor activity was constantly monitored via a passive infrared sensor and a running wheel. Mice were then implanted with osmotic minipumps delivering TNF-α (light gray bars) or saline as control (dark gray bars). On the third day after the operation, mice were killed at ZT = 11.5 or = 15.5 (ZT = 0, light on; ZT = 12, light off). Livers were extracted and gene expression assessed by real-time PCR. (A) Per1/2/3, RevErbα, and PAR bZip family genes, Dbp, Hlf, and Tef, are down-regulated at both time points, the suppression of PAR bZip family genes is highly significant. In contrast to fibroblast data, RevErbα is strongly down-regulated. Results are shown as percentages relative to the mean of the control group. One representative experiment of three is shown (n = 6 per group; Mann–Whitney test. *, P ≤ 0,05; **, P ≤ 0,005.)
TNF-α Reduces Dbp Expression in the SCN in the Hypothalamic Region.
To verify whether the effect of TNF-α is sustained across the blood–brain barrier and reaches the SCN, the expression of Dbp was quantified. As described above, mice were implanted with minipumps releasing TNF-α or diluent; mice were killed after 3 days at ZT 6, when peak expression of Dbp was expected. A slight (−15%) but statistically significant reduction of Dbp expression (P = 0.003) can be seen by in situ hybridization in the SCN of TNF-α-treated mice (SI Fig. 10). To verify the time point of the rhythm, we used Bmal1, which, at peak expression of Dbp, is not detectable. Indeed, unlike Dbp mRNA-positive cells, Bmal1 transcripts were not identified by in situ hybridization at ZT 6.
TNF-α Reduces Locomotor Activity and Promotes Increased Rest Time in Mice.
The function of TNF-α to interfere with the transcription of distinct clock genes or CCGs prompted us to evaluate whether TNF-α alters the circadian rhythm and/or the amount of locomotor activity in vivo. The timing and extent of locomotor activity were assessed with infrared sensors as well as by monitoring running-wheel activity in mice recorded under a 12-h light/12-h dark (LD) schedule. The surgical procedure led to a decrease in locomotor activity that lasted for 48 h (infrared; Fig. 5A Upper). However, beside the more pronounced impairment of locomotor activity on days 1 and 2, TNF-α led also to a severe suppression of locomotor activity on day 3; on this day, control mice had attained baseline levels of activity. When running-wheel activity was assessed, an analogous picture emerged with maximum inhibition at day 3 (Fig. 5 A Lower and B). At later time points, especially on days 5 and 6, TNF-α no longer exerted any effect on locomotor activity. This is most likely due to a failed release of TNF-α from the pump, because starting at day 4, hemorrhagic necrosis developed at the site of the pump. This local side effect was noticed when pumps were filled with TNF-α but not with control solution and has been described in areas of s.c. injection of TNF-α in mice (3 μg for 5 days) (29). Our results show that TNF-α leads to a decrease in the total amount of locomotor activity as measured by a running-wheel and infrared sensors until day 4 (Fig. 5A). The activity in both groups was still restricted to the dark period, and highest loss of activity in TNF-α treated animals can be observed during the second half of the active period (Fig. 5B). Further, based on these observations, we chose day 3 to investigate the frequency and duration of rest episodes during the LD period (SI Fig. 11A). During the light period, no change was observed. But in the dark period, when mice are usually active, a significant increase in rest episodes lasting 6–60 min was detected. As a readout of the endogenous circadian clock, we assessed locomotor activity in constant darkness (“free-running condition”) after implantation of the minipumps and calculated the period length (τ). We observed no changes in the periodicity of τ (SI Fig. 11B). Taken together, mice treated continuously with TNF-α show reduced motor activity and more consolidated rest time but no change in period length under free-running conditions. These findings are consistent with experiments with fibroblasts showing that the amplitude of the expression of clock genes and CCGs are attenuated by TNF-α, rather than their circadian rhythm itself.
Discussion
Whereas the molecular mechanisms provided by clock genes to maintain the circadian rhythm are becoming increasingly clear, the potential influences of the immune system on the molecular clockwork remain to be explored. Here, we provide evidence that TNF-α interferes with the expression of clock genes, namely the Per genes and the PAR bZip genes Dbp, Tef, and Hlf. TNF-α suppresses the expression of these genes in fibroblasts (and attenuates their amplitudes) in vitro and in vivo in the liver of mice infused with this cytokine. That the same genes (with the exception of RevErbα) are prone to TNF-α stimulation in vitro compared with in vivo speaks for a rather direct effect of TNF-α, although we cannot exclude other intermediates involved leading to the same response. In situ hybridization shows that the s.c. administration of TNF-α also leads to reduction of Dbp expression in the central circadian pacemaker, the SCN. Thus, it is tempting to speculate that TNF-α is likely to reach the SCN via the blood and to bind to TNF receptor (TNF-R) on neurons. At least in the hippocampus, both TNF-RI and -RII are expressed (30). In the present study E-box regulatory elements of clock genes are found here to play a pivotal role in the effect of TNF-α to inhibit clock gene expression. First, only clock genes with E-boxes in their promoter, the PAR bZip genes Dbp, Tef, and Hlf and the Per genes, are affected by TNF-α, whereas clock genes devoid of E-boxes such as Clock and Bmal1 are not affected by TNF-α. Second, mutated E-boxes provide protection of TNF-α-induced suppression of clock reporter genes. E-boxes are functionally important components of DNA promoters that guide the expression of clock genes and thereby influence the circadian rhythm, including the sleep–wake cycle. Rhythmic binding of CLOCK and BMAL1 depends on E-boxes and is a prerequisite for robust waves of gene expression characteristic of circadian transcription (31, 32).
The effects of TNF-α on clock gene expression also become apparent when studying clock-dependent genes. Dbp has been described to mediate transcription of the albumin gene in hepatocytes (25). Albumin serum concentrations are found here to be lowered by 41% in TNF-α-treated mice compared with controls.
Recording of locomotor activity of TNF-α-treated mice shows more rest episodes during spontaneous activity. In line with the finding that TNF-α does not alter circadian rhythm in cultured fibroblasts but rather lowers the extent of expression of distinct clock genes, TNF-α did not influence period length of the circadian rhythm under “free running” conditions. Of interest for the findings presented here are data showing that the deletion of the Dbp gene in mice results in only a slight reduction of period length but in a striking impairment of spontaneous locomotor activity and running-wheel activity (33, 34). Whereas Tef and Hlf single-knockout mice show an increased period length, the inactivation of all three genes, Dbp, Tef, and Hlf, resulted in an unchanged circadian period length (27). Thus, a normal period length may be due to opposite effects leading to mutual neutralization of indirect clock gene dysfunction. Besides lowering the expression of PAR bZip transcription factors, TNF-α impaired Per1, Per2, and Per3 mRNA. Target disruption of these genes results in slightly (Per3−/− mice) or more dramatically (Per1−/− mice) shortened period lengths or eventually gives rise to arrhythmic behavior (Per2−/− mice). Taken collectively, TNF-α interferes with the expression of E-box-dependent clock genes and leads to prolonged rest episodes during spontaneous activity of mice.
Sleep architecture in humans is affected by the endogenous circadian pacemaker, which regulates the timing of the sleep–wake cycle, presumably by circadian expression of clock genes (13, 35). Enhanced NREM sleep has been shown in mice treated with muramyl dipeptides, which activate TLR2 and TLR4 on macrophages (36, 37). TNF-α, as well as IL-1, is produced by TLR2- and TLR4-activated macrophages and is well described to enhance NREM sleep (11, 38, 39). In light of the overlapping properties of TNF-α and IL-1 on NREM sleep, it is interesting that our studies show IL-1 to share with TNF-α the inhibitory effect on expression of the Dbp and Per3 genes in fibroblasts. As recently outlined, the effects of IL-6 on sleep differs from IL-1 and TNF-α, in that it may also act on systems involved in NREM sleep but not in REM sleep, which is suppressed by TNF-α and IL-1 but not by IL-6 (40). In this context, it may be of relevance that the expression of Dbp is found here not to be affected by IL-6. Although the expression of Dbp and Per1 is also not inhibited by IFN-α, IFN-γ only slightly interfered with Dbp expression. This is remarkable, because in mice, the daily injection of IFN-α or -γ has been reported to lower Per1, Per2, Per3, and Clock after 6 days of treatment (41). The absence of effects of IFN-α and -γ on expression of the Per genes by synchronized fibroblasts in vitro indicates that IFNs down-regulate the Per and Clock genes by E-box-independent mechanisms or induce production of other clock gene-regulating factors when mice are treated over a long time period with the cytokines.
In infectious diseases, TNF-α serves to successfully eliminate the infectious agent. The function of TNF-α to interfere with the expression of clock genes, to impair locomotor activity, and to enhance rest may provide the link between the activation of the innate immunity and fatigue associated with infectious and autoimmune diseases, such as multiple sclerosis, RA, or Crohn's disease. In these disorders, both fatigue and elevated TNF-α concentrations have been described (6, 42, 43). It is still debated whether sleep changes in infections are beneficial to the host defense. Rabbits infected with E. coli, Staphylococcus aureus, or Candida albicans showed an improved prognosis when their sleep duration was prolonged (44). In this context, it is to be noted that Per2−/− mice are partially protected from LPS-induced shock (45). During the TNF-α-induced “inflammatory clock gene response,” the expression of Per1, Per2, and Per3 genes and of the PAR bZip transcription factors Dbp, Tef, and Hlf is down-regulated, the locomotor activity reduced, and rest episodes prolonged. Although this pathway may induce an adaptive state in infectious diseases, the “inflammatory clock gene response” may, by inducing fatigue, diminish the quality of life in autoimmune diseases. Our study will serve to lay an important foundation for further exploration of the connection between the TNF-α-induced “inflammatory clock gene response” and the TNF-α-triggered reduction of locomotor activity.
Materials and Methods
Cytokines.
Recombinant murine (rm) TNF-α, rm IL-1β, and rm IL-6 were purchased either from Sigma (St. Louis, MO) (in vitro time course assays) or from Peprotech (London, U.K.) (in vivo assays); rm IFN-α from Immunotools (Friesoythe, Germany), and rm IFN-γ from Roche (Rotkreuz, Switzerland).
Synchronization of Fibroblasts by Serum Shock.
NIH 3T3 fibroblasts were grown in DMEM (Gibco, Basel, Switzerland) supplemented with 10% FBS (PAA Laboratories, Pasching, Austria) and Glutamax (Gibco). For serum shock, cells were grown to confluency in 6-cm tissue culture dishes. At time t = 0, the medium was exchanged by 50% horse serum (Gibco) in DMEM/Glutamax; after 2 h, the medium was replaced with serum-free DMEM/Glutamax, with or without TNF-α. At the indicated time points, tissue culture dishes were washed once with Hanks; solution, frozen on a layer of liquid nitrogen, and kept at −70°C until the extraction of whole-cell RNA.
Transfection and Luciferase Assays.
Unsynchronized NIH 3T3 cells were transfected with the following constructs by using Lipofectamine Plus (Life Technologies, Basel, Switzerland) or TransFectin (Bio-Rad, Hercules, CA) according to the manufacturer's protocol: mPer1-luc (46), kindly provided by David Earnest [Texas A&M, College Station, TX; E54-TK (22)], kindly provided by Sato Honma (Hokkaido University, Sapporo, Japan); Bmal1-luc (19), DBP-Ebox-luc and mut Ebox, pCDNA3.1-Clock and pCDNA3.1-Bmal1 (47), kindly provided by U. Schibler (University of Geneva, Geneva, Switzerland); mPer3-luc (48), kindly provided by P. Sassone-Corsi (IGBMC, Illkirch, France). As an internal control for transfection efficiency, a GFP construct (pMax-GFP; Amaxa, Cologne, Germany) was cotransfected 1:10.
Twenty-four hours after transfection, the medium was replaced with serum-free DMEM/glutamax with or without TNF-α (10 ng/ml). After ≈15 h, cells were lysed by using Passive Lysis Buffer (Promega, Wallisellen, Switzerland) and enzyme activity was measured by the Luciferase Assay System (Promega). Bioluminescence was measured with a Luminometer (Berthold Technologies, Regensdorf, Switzerland) and normalized to transfection efficiency or protein concentration.
Animal Groups and Locomotor Activity Recording.
Seven-week-old C57BL/6 male mice (Harlan Breeding Laboratory, AD Horst, The Netherlands) were housed in individual cages, equipped with a running-wheel and a passive infrared sensor in a temperature-controlled sound-proof light-tight room. Food and water were available ad libitum. We allowed mice 10–15 days of acclimatization to a LD cycle (light on at 0600, i.e., ZT 0; light off at 1800, i.e., ZT 12). Mice were operated under deep isoflurane anesthesia, and 30 μg of Temgesic anesthetic (buprenorphine; Essex Chemie, Lucerne, Switzerland) was applied. TNF-α (1.5 μg/day, diluted in 0.1% BSA/PBS) or 0.1% BSA/PBS as a control was administered s.c. by using osmotic minipumps (Model 1007D; Alzet, Cupertino, CA) implanted on the back, for 6 days. Locomotor activity was continuously measured via running-wheel and infrared sensors based on 1-min episodes by using the Chronobiology Kit software (Stanford Software Systems, Santa Cruz, CA), as described (49, 50). Rest episodes were defined as 1-min units with activity = zero. The free-running period of locomotion was calculated by periodogram analysis for days 2–6 after minipump implantation, when the mice were kept in constant darkness. In gene expression studies, livers were extracted 3 days after minipump insertion at two different ZTs known to approximately represent the peak of expression of Dbp (ZT = 11.5) or Per2 (ZT = 15.5). Livers were frozen in TRIzol (Invitrogen, Basel, Switzerland) for subsequent RNA extraction. All experimental procedures were approved by the local committee of the veterinary office and in strict accordance with Swiss regulations on animal welfare.
RNA Isolation and Gene Expression Analysis.
The method for RNA extraction, RT-PCR, and quantification of gene expression is described in SI Text.
In Situ Hybridization.
The method for in situ hybridization was published (51) and is described in SI Text.
Supplementary Material
Acknowledgments
We thank Dr. U. Schibler (University of Geneva) for helpful discussions and critical feedback on this manuscript and Dr. Michael Kurrer (Department of Pathology, University Hospital Zurich) for analysis of histopathological findings in TNF-α-treated mice. This study was supported by the Swiss National Science Foundation (Project no. 310000-109469/1 and NCCR Neural Plasticity and Repair, project 6), the Swiss Multiple Sclerosis Society, and the Gianni Rubatto Foundation.
Abbreviations
- TLR
Toll-like receptor
- RA
rheumatoid arthritis
- NREM
nonrapid eye movement
- SCN
suprachiasmatic nuclei
- CCG
clock-controlled gene
- ZT
Zeitgeber time
- LD
12-h light/12-h dark.
Footnotes
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701466104/DC1.
References
- 1.Akira S, Uematsu S, Takeuchi O. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Kishimoto T. Annu Rev Immunol. 2005;23:1–21. doi: 10.1146/annurev.immunol.23.021704.115806. [DOI] [PubMed] [Google Scholar]
- 3.Beutler B, Cerami A. N Engl J Med. 1987;316:379–385. doi: 10.1056/NEJM198702123160705. [DOI] [PubMed] [Google Scholar]
- 4.Dinarello CA. J Endotoxin Res. 2004;10:201–222. doi: 10.1179/096805104225006129. [DOI] [PubMed] [Google Scholar]
- 5.Spriggs DR, Sherman ML, Michie H, Arthur KA, Imamura K, Wilmore D, Frei E, 3rd, Kufe DW. J Natl Cancer Inst. 1988;80:1039–1044. doi: 10.1093/jnci/80.13.1039. [DOI] [PubMed] [Google Scholar]
- 6.Pollard LC, Choy EH, Gonzalez J, Khoshaba B, Scott DL. Rheumatology (Oxford) 2006;45:885–889. doi: 10.1093/rheumatology/kel021. [DOI] [PubMed] [Google Scholar]
- 7.Vgontzas AN, Papanicolaou DA, Bixler EO, Kales A, Tyson K, Chrousos GP. J Clin Endocrinol Metab. 1997;82:1313–1316. doi: 10.1210/jcem.82.5.3950. [DOI] [PubMed] [Google Scholar]
- 8.Riha RL, Brander P, Vennelle M, McArdle N, Kerr SM, Anderson NH, Douglas NJ. Eur Respir J. 2005;26:673–678. doi: 10.1183/09031936.05.00130804. [DOI] [PubMed] [Google Scholar]
- 9.Vgontzas AN, Zoumakis E, Lin HM, Bixler EO, Trakada G, Chrousos GP. J Clin Endocrinol Metab. 2004;89:4409–4413. doi: 10.1210/jc.2003-031929. [DOI] [PubMed] [Google Scholar]
- 10.Opp MR. Sleep Med Rev. 2005;9:355–364. doi: 10.1016/j.smrv.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Krueger JM, Majde JA. Ann NY Acad Sci. 2003;992:9–20. doi: 10.1111/j.1749-6632.2003.tb03133.x. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi S, Kapas L, Fang J, Krueger JM. Am J Physiol. 1999;276:R1132–R1140. doi: 10.1152/ajpregu.1999.276.4.R1132. [DOI] [PubMed] [Google Scholar]
- 13.Daan S, Beersma DG, Borbely AA. Am J Physiol. 1984;246:R161–R183. doi: 10.1152/ajpregu.1984.246.2.R161. [DOI] [PubMed] [Google Scholar]
- 14.Dijk DJ, Czeisler CA. J Neurosci. 1995;15:3526–3538. doi: 10.1523/JNEUROSCI.15-05-03526.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schibler U, Sassone-Corsi P. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 16.Reppert SM, Weaver DR. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 17.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 19.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Iwasaki A, Medzhitov R. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 21.Cermakian N, Sassone-Corsi P. Nat Rev Mol Cell Biol. 2000;1:59–67. doi: 10.1038/35036078. [DOI] [PubMed] [Google Scholar]
- 22.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 23.Tracey KJ, Beutler B, Lowry SF, Merryweather J, Wolpe S, Milsark IW, Hariri RJ, Fahey TJ, 3rd, Zentella A, Albert JD, et al. Science. 1986;234:470–474. doi: 10.1126/science.3764421. [DOI] [PubMed] [Google Scholar]
- 24.Evans TJ, Moyes D, Carpenter A, Martin R, Loetscher H, Lesslauer W, Cohen J. J Exp Med. 1994;180:2173–2179. doi: 10.1084/jem.180.6.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cereghini S. FASEB J. 1996;10:267–282. [PubMed] [Google Scholar]
- 26.Perlmutter DH, Dinarello CA, Punsal PI, Colten HR. J Clin Invest. 1986;78:1349–1354. doi: 10.1172/JCI112721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gachon F, Fonjallaz P, Damiola F, Gos P, Kodama T, Zakany J, Duboule D, Petit B, Tafti M, Schibler U. Genes Dev. 2004;18:1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian B, Nowak DE, Brasier AR. BMC Genomics. 2005;6:137. doi: 10.1186/1471-2164-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erickson SL, de Sauvage FJ, Kikly K, Carver-Moore K, Pitts-Meek S, Gillett N, Sheehan KC, Schreiber RD, Goeddel DV, Moore MW. Nature. 1994;372:560–563. doi: 10.1038/372560a0. [DOI] [PubMed] [Google Scholar]
- 30.Neumann H, Schweigreiter R, Yamashita T, Rosenkranz K, Wekerle H, Barde YA. J Neurosci. 2002;22:854–862. doi: 10.1523/JNEUROSCI.22-03-00854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munoz E, Brewer M, Baler R. J Biol Chem. 2002;277:36009–36017. doi: 10.1074/jbc.M203909200. [DOI] [PubMed] [Google Scholar]
- 32.Ripperger JA, Schibler U. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 33.Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franken P, Lopez-Molina L, Marcacci L, Schibler U, Tafti M. J Neurosci. 2000;20:617–625. doi: 10.1523/JNEUROSCI.20-02-00617.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borbely AA, Achermann P. In: Principles and Practice of Sleep Medicine. Kryger MH, Roth T, Dement WC, editors. Philadelphia: Elsevier Saunders; 2005. pp. 405–417. [Google Scholar]
- 36.Krueger JM, Pappenheimer JR, Karnovsky ML. Proc Natl Acad Sci USA. 1982;79:6102–6106. doi: 10.1073/pnas.79.19.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uehori J, Fukase K, Akazawa T, Uematsu S, Akira S, Funami K, Shingai M, Matsumoto M, Azuma I, Toyoshima K, Kusumoto S, Seya T. J Immunol. 2005;174:7096–7103. doi: 10.4049/jimmunol.174.11.7096. [DOI] [PubMed] [Google Scholar]
- 38.Krueger JM, Walter J, Dinarello CA, Wolff SM, Chedid L. Am J Physiol. 1984;246:R994–R999. doi: 10.1152/ajpregu.1984.246.6.R994. [DOI] [PubMed] [Google Scholar]
- 39.Shoham S, Davenne D, Cady AB, Dinarello CA, Krueger JM. Am J Physiol. 1987;253:R142–R149. doi: 10.1152/ajpregu.1987.253.1.R142. [DOI] [PubMed] [Google Scholar]
- 40.Hogan D, Morrow JD, Smith EM, Opp MR. J Neuroimmunol. 2003;137:59–66. doi: 10.1016/s0165-5728(03)00038-9. [DOI] [PubMed] [Google Scholar]
- 41.Ohdo S, Koyanagi S, Suyama H, Higuchi S, Aramaki H. Nat Med. 2001;7:356–360. doi: 10.1038/85507. [DOI] [PubMed] [Google Scholar]
- 42.Heesen C, Nawrath L, Reich C, Bauer N, Schulz KH, Gold SM. J Neurol Neurosurg Psychiatry. 2006;77:34–39. doi: 10.1136/jnnp.2005.065805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pimentel M, Chang M, Chow EJ, Tabibzadeh S, Kirit-Kiriak V, Targan SR, Lin HC. Am J Gastroenterol. 2000;95:3458–3462. doi: 10.1111/j.1572-0241.2000.03361.x. [DOI] [PubMed] [Google Scholar]
- 44.Toth LA, Tolley EA, Broady R, Blakely B, Krueger JM. Proc Soc Exp Biol Med. 1994;205:174–181. doi: 10.3181/00379727-205-43694. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Mankani G, Shi X, Meyer M, Cunningham-Runddles S, Ma X, Sun ZS. Infect Immun. 2006;74:4750–4756. doi: 10.1128/IAI.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen G, Rappe J, Earnest DJ, Cassone VM. J Neurosci. 2001;21:7937–7943. doi: 10.1523/JNEUROSCI.21-20-07937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ripperger JA, Shearman LP, Reppert SM, Schibler U. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- 48.Doi M, Hirayama J, Sassone-Corsi P. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 49.Deboer T, Tobler I. J Comp Physiol [A] 2000;186:969–973. doi: 10.1007/s003590000150. [DOI] [PubMed] [Google Scholar]
- 50.Tobler I, Gaus SE, Deboer T, Achermann P, Fischer M, Rulicke T, Moser M, Oesch B, McBride PA, Manson JC. Nature. 1996;380:639–642. doi: 10.1038/380639a0. [DOI] [PubMed] [Google Scholar]
- 51.Albrecht U, Lu HC, Revelli JP, Xu XC, Lotan RGE. In: Human Genome Methods. Adolph KW, editor. New York: CRC; 1998. pp. 93–119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.