Abstract
In the fasted state, induction of hepatic glucose output and fatty acid oxidation is essential to sustain energetic balance. Production and oxidation of glucose and fatty acids by the liver are controlled through a complex network of transcriptional regulators. Among them, the transcriptional coactivator PGC-1α plays an important role in hepatic and systemic glucose and lipid metabolism. We have previously demonstrated that sirtuin 1 (SIRT1) regulates genes involved in gluconeogenesis through interaction and deacetylation of PGC-1α. Here, we show in vivo that hepatic SIRT1 is a factor in systemic and hepatic glucose, lipid, and cholesterol homeostasis. Knockdown of SIRT1 in liver caused mild hypoglycemia, increased systemic glucose and insulin sensitivity, and decreased glucose production. SIRT1 knockdown also decreased serum cholesterol and increased hepatic free fatty acid and cholesterol content. These metabolic phenotypes caused by SIRT1 knockdown tightly correlated with decreased expression of gluconeogenic, fatty acid oxidation and cholesterol degradation as well as efflux genes. Additionally, overexpression of SIRT1 reversed many of the changes caused by SIRT1 knockdown and depended on the presence of PGC-1α. Interestingly, most of the effects of SIRT1 were only apparent in the fasted state. Our results indicate that hepatic SIRT1 is an important factor in the regulation of glucose and lipid metabolism in response to nutrient deprivation. As these pathways are dysregulated in metabolic diseases, SIRT1 may be a potential therapeutic target to control hyperglycemia and hypercholesterolemia.
Keywords: fasting response, glucose metabolism, lipid metabolism, deacetylase, transcriptional coactivator
In response to nutrient deprivation mammals trigger many tissue-specific metabolic pathways to maintain organismal survival. In particular, the liver functions as a major metabolic buffering system that controls macro- and micronutrient homeostasis, allowing other tissues to function normally under physiological stresses (1–3). Among its many functions, production of glucose by the liver is an essential process that contributes to normalization of systemic glucose levels (4, 5), ensuring that glucose-dependent tissues such as brain and red blood cells will have access to an energy supply during periods of nutrient deprivation. However, chronic elevation of hepatic glucose production is also a key contributor in diabetes that exacerbates hyperglycemia in both the fed and fasted states (6, 7). The importance of hepatic glucose production is underscored by the fact that current antidiabetic drugs such as metformin decrease blood glucose levels through inhibition of gluconeogenesis in liver (8, 9).
The liver also plays an important role in lipid homeostasis. In the fasted state, oxidation of hepatic free fatty acids supplies energy for glucose production (10). Dysregulation of hepatic fatty acid β-oxidation and/or fatty acid synthesis enzymes leads to hepatic steatosis or fatty liver (11). Moreover, cholesterol is synthesized and degraded in the liver accordingly to the needs by other tissues (12). Cyp7A1 is the key rate-limiting enzyme controlling cholesterol degradation through hepatic synthesis of bile acids (13, 14). Efflux and influx through scavenge receptor B1 (SR-B1), low-density lipoprotein (LDL) receptor, and ABC transporters also plays an important role in regulating cholesterol levels (15–17).
The majority of rate-limiting enzymes in key pathways involved in glucose and lipid homeostasis are controlled at the transcriptional level (17). In the last several years, the peroxisome proliferator activated receptor coactivator (PGC)-1α/β and liver X receptor (LXR)/sterol response element-binding protein (SREBP) have been identified as key transcriptional regulators of many metabolic enzymes and pathways (18–20). The PGC-1α transcriptional network is the target of hormonal and nutrient signals. Positively regulated by glucagon through TORC2 and CREB activation, (21) and glucocorticoids. PGC-1α function is negatively regulated by insulin, directly (22) and through FOXO1 (23, 24). This regulation ultimately leads to coordinated changes in the expression of glucose and lipid metabolic genes (25). In the case of lipid metabolism, main regulatory transcription factors include the SREBP family and hormone nuclear receptors LXRs and peroxisome proliferator-activated receptors (PPARs). These factors are also subject to hormonal and nutrient regulation via insulin and cholesterol (12, 26, 27).
We have previously identified a nutrient regulation of glucose homeostasis through the NAD+-dependent deacetylase sirtuin 1(SIRT1). Under low nutrient conditions, up-regulation of SIRT1 promotes hepatic glucose production through interaction and deacetylation of PGC-1α (28). Other groups have shown that SIRT1 represses peroxisome proliferator-activated receptor γ (PPARγ) function, increasing lipolysis in white adipose tissue and insulin secretion in pancreatic β-cells (29–31). Interestingly, a nutrient connection to SIRT1 has been previously established in lower eukaryotes, including yeast (32), worms (33), and flies (34). In these species, SIRT1 homologs were required to extend life span in response to calorie restriction, although its impact on metabolic pathways is unknown. A crucial unsolved question from our previous studies was to what extent SIRT1 was required to control hepatic glucose and lipid metabolic pathways and how this affects systemic nutrient homeostasis in vivo. Here, we report that hepatic knockdown of SIRT1 results in mild hypoglycemia, increased glucose tolerance, insulin sensitivity, and decreased hepatic glucose production. Furthermore, we found accumulation of free fatty acids and cholesterol in hepatic tissue as well as decreased serum cholesterol in SIRT1-deficient mice. These effects correlate with changes in gene expression of enzymes involved in gluconeogenesis, glycolysis, fatty acid oxidation/synthesis, and cholesterol degradation and efflux pathways. Importantly, many of these changes were reversed by hepatic overexpression of SIRT1 and were dependent on PGC-1α. Taken together our results implicate SIRT1 as an important regulator of hepatic glucose and lipid homeostasis in response to fasting.
Results
Impaired Glucose Homeostasis in Hepatic SIRT1 Knockdown Mice.
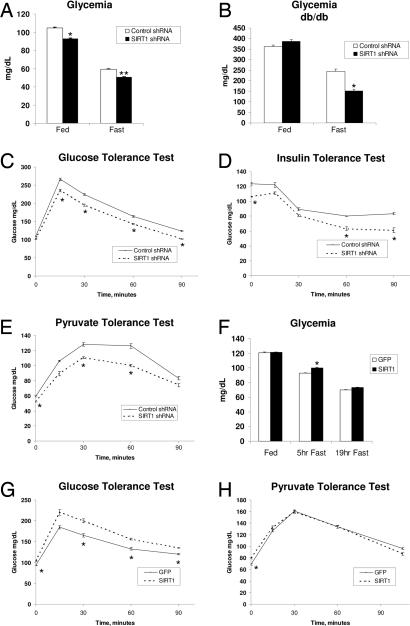
Our previous work demonstrated that SIRT1 was required to induce gluconeogenic genes in response to a nutrient fasting signal in cultured liver cells (28). We therefore initiated studies in mice to further investigate the role of hepatic SIRT1 in systemic glucose homeostasis. Tail vein injection of adenoviruses-expressing SIRT1 small hairpin RNA (shRNA) resulted in a significant knockdown of SIRT1 protein levels. Expression levels of other proteins that physically and functionally interact with SIRT1 such as PGC-1α and FOXO1 were largely not affected [supporting information (SI) Fig. 5A]. Acetylation of PGC-1α was decreased in the fasted state, and knockdown of SIRT1 was sufficient to increase acetylation of endogenous PGC-1α in both fed and fasted mice (SI Fig. 5B). Acetylation of FOXO1, another SIRT1 target, also increased in response to SIRT1 knockdown (SI Fig. 5E). Conversely, overexpression of SIRT1 decreased PGC-1α acetylation (SI Fig. 5 C and D). Blood glucose levels were modestly but consistently lower in SIRT1 shRNA-infected mice compared with control shRNA mice, both in fed and fasted states (Fig. 1A). Hepatic knockdown of SIRT1 also lowered glycemia in fasted diabetic db/db mice (Fig. 1B). To further analyze whether hepatic SIRT1 plays a role in systemic glucose and insulin sensitivity, we performed glucose (GTT) and insulin (ITT) tolerance tests. Fig. 1C shows that in a GTT, mice infected with SIRT1 shRNA adenoviruses displayed significantly lower blood glucose concentrations. Consistent with these results, insulin had a greater effect reducing blood glucose levels in an ITT (Fig. 1D). To determine whether the reductions in blood glucose levels observed in mice with hepatic SIRT1 knockdown were due to a deficit in hepatic glucose production, we performed a pyruvate tolerance test (PTT). As shown in Fig. 1E, hepatic SIRT1 knockdown mice displayed lower blood glucose concentrations at every time point after pyruvate administration. We next analyzed the effects of hepatic SIRT1 overexpression on glucose metabolism. SIRT1 overexpression caused moderate hyperglycemia in short-term fasting (5 h) and a slight hyperglycemic trend upon longer fasting (Fig. 1F). Mice with hepatic SIRT1 overexpression were less glucose tolerant in a GTT (Fig. 1G). However, in a PTT, SIRT1 overexpression did not increase blood glucose levels (Fig. 1H). Taken together, these data indicate that modulation of SIRT1 protein causes changes in blood glucose levels that correlate with effects on glucose and insulin sensitivity. Moreover, our data suggests that these effects are likely due, at least in part, to effects on hepatic glucose production.
Fig. 1.
Hepatic SIRT1 controls glucose metabolism. (A) Fed and fasted blood glucose levels of control and SIRT1 shRNA-infected mice. Data are presented as the average ± SEM of two independent experiments. Shown are control shRNA-infected mice [fed (n = 12), 20-h fasted (n = 12)] and SIRT1 shRNA-infected mice [fed (n = 13), 20-h fasted (n = 12)]. (B) Blood glucose levels from db/db mice infected with control shRNA (n = 5) or SIRT1 shRNA (n = 5) during feeding and during a short fast. (C) GTT. Control shRNA-infected (n = 5) and SIRT1 shRNA-infected (n = 6) mice were fasted 5 h before i.p. injection of 2 g/kg dextrose. (D) ITT. Control shRNA-infected (n = 5) and SIRT1 shRNA-infected (n = 5) mice were fasted 5 h before i.p. injection of 0.6 unit/kg insulin. (E) PTT. Control shRNA-infected (n = 7) and SIRT1 shRNA-infected (n = 7) mice were fasted 18 h before i.p. injection of 2 g/kg sodium pyruvate. (F) Blood glucose levels from mice infected with GFP or SIRT1 overexpression adenovirus. Feeding GFP and SIRT1 (n = 12), following a short 5-h fast (n = 6) or following a 19-h fast (n = 12). (G) GTT from GFP-infected (n = 6) or SIRT1-infected (n = 6) mice fasted for 5 h before injection of 2 g/kg dextrose. (H) PTT from GFP-infected (n = 6) and SIRT1-infected (n = 6) mice fasted for 18 h before injection with 2 g/kg pyruvate. All tolerance tests were performed in at least two independent experiments with similar results. Data are presented as the average ± SEM. Significance was determined by Student's t test. *, P < 0.05; **, P < 0.01.
Hepatic SIRT1 Controls Expression of Gluconeogenic and Glycolytic Genes.
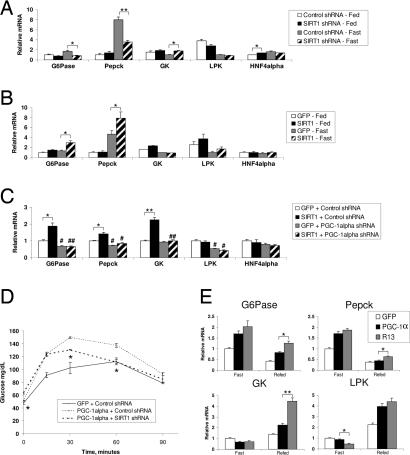
Our previous work in cultured liver cells demonstrated that nutrient signaling through pyruvate increases SIRT1 levels. Furthermore, pyruvate treatment induced gluconeogenic genes [phosphoenolpyruvate carboxylase kinase (Pepck) and glucose 6-phosphatase (G6Pase)] but repressed glycolytic genes [glucokinase (GK) and liver pyruvate kinase (LPK)] in a SIRT1-dependent manner (28). As shown in Fig. 2A, infection with SIRT1 shRNA resulted in reduced expression of the gluconeogenic G6Pase and Pepck under fasting conditions. Interestingly, no changes in the expression of these genes were observed in the fed state. In contrast, SIRT1 knockdown increased glycolytic GK gene expression upon fasting, but no changes were detected in LPK gene expression (Fig. 2A). Under fasting conditions, hepatic overexpression of SIRT1 further caused an increase in G6Pase and Pepck mRNA levels but did not significantly change glycolytic gene expression (Fig. 2B). These results indicate that hepatic SIRT1 functions in the regulation of gluconeogenic gene expression. Notably, in both knockdown and overexpression studies, it is the regulation in response to fasting that is affected by SIRT1.
Fig. 2.
Hepatic SIRT1 regulates genes involved in gluconeogenesis. (A) Quantitative RT-PCR analysis of expression of genes involved in liver glucose metabolism from mice in the fed state infected with control shRNA (n = 5) or SIRT1 shRNA (n = 6) or mice infected with control shRNA (n = 5) and SIRT1 shRNA (n = 5) and fasted for 20 h. (B) Fed, GFP (n = 6) or SIRT1 (n = 6); fasted for 19 h, GFP (n = 6) or SIRT1 (n = 6). (C) Double infection of GFP or SIRT1 overexpression and Control or PGC-1α shRNA (each bar, n = 9) following a 20-h fast. (D) PTT, PGC-1α overexpression, and SIRT1 knockdown. GFP + control shRNA-infected mice (n = 4), PGC-1α + control shRNA-infected mice (n = 4), and PGC-1α + SIRT1 shRNA-infected mice (n = 4) were fasted 18 h before i.p. injection of 2 g/kg sodium pyruvate. Significance indicated is between PGC-1α + control shRNA and PGC-1α + SIRT1 shRNA. (E) Quantitative RT-PCR analysis of gene expression of mice infected with GFP, PGC-1α overexpression, or R13 adenovirus fasted for 22 h (each bar, n = 4) or fasted for 22 h and refed overnight (16 h) (each bar, n = 5). All data are presented as the average ± SEM. Gene expression was normalized to 36b4 expression. Significance was determined by Student's t test. GFP/control shRNA vs. SIRT1/SIRT1 shRNA: *, P < 0.05; **, P < 0.01. Control shRNA vs. PGC-1α shRNA: #, P < 0.05; ##, P < 0.01.
We next investigated whether PGC-1α was required for the effects of SIRT1 on glucose metabolism. We used a combination of adenovirus to simultaneously overexpress SIRT1 and knockdown PGC-1α (PGC-1α shRNA) (SI Fig. 6A). Consistent with previous reports (35, 36), knockdown of PGC-1α resulted in decreased expression of gluconeogenic genes (Fig. 2C) and reduced glycemia (SI Fig. 6B). Interestingly, knockdown of PGC-1α also blocked the induction of gluconeogenic genes (Fig. 2C), and the reduced glucose tolerance caused by SIRT1 overexpression (SI Fig. 6C), suggesting that SIRT1 requires PGC-1α for its regulation of hepatic glucose metabolism.
SIRT1 Controls PGC-1α-Dependent Increase of Hepatic Glucose Production.
Because ectopic PGC-1α expression is sufficient to induce hepatic glucose production, we next tested whether these effects require SIRT1. To do this, we infected mice with a combination of adenoviruses, resulting in PGC-1α overexpression and SIRT1 knockdown (SI Fig. 7A). As expected, overexpression of PGC-1α in the liver resulted in increased blood glucose levels in a PTT. Hepatic knockdown of SIRT1 significantly reduced this increase by PGC-1α (Fig. 2D). Moreover, PGC-1α-induced Pepck gene expression was also reduced by SIRT1 shRNA (SI Fig. 7B) despite a large induction of PGC-1α protein (SI Fig. 7A). Together, these results suggest that SIRT1 is required, at least in part, for PGC-1α-dependent hepatic glucose production.
To determine whether acetylation of PGC-1α regulates its activity in vivo, we used a mutant PGC-1α construct in which 13 lysines have been mutated to arginine-R13 (SI Fig. 7D). This mutant is no longer acetylated when cells are treated with nicotinamide (28). As expected, PGC-1α overexpression caused mild hyperglycemia. Interestingly, the R13 mutant caused a larger increase in blood glucose levels compared with control and wild-type PGC-1α. This effect was particularly pronounced in the short-term fasting condition (SI Fig. 7E). We also observed that R13 was more transcriptionally active, inducing gluconeogenic genes, particularly in the refed situation (Fig. 2E). These data provide evidence that PGC-1α activity is, in part, regulated by SIRT1 and acetylation in vivo.
Defects in Fatty Acid and Cholesterol Metabolism in Mice with Altered Hepatic SIRT1 Expression.
In addition to glucose metabolism, the liver has key roles in lipid homeostasis. We therefore investigated whether SIRT1 plays a role in hepatic lipid metabolism. SIRT1 knockdown or overexpression did not significantly alter serum triglyceride and free fatty acid levels (SI Fig. 8). However, SIRT1 knockdown very strongly increased intracellular hepatic free fatty acids (Fig. 3A) while having no effect on liver triglycerides (SI Fig. 8A). We also observed that SIRT1 had very pronounced effects on systemic and hepatic cholesterol. Hepatic knockdown of SIRT1 resulted in reduced systemic levels of total cholesterol in the fed and fasted state (Fig. 3A). Interestingly, SIRT1 overexpression reversed this effect, increasing systemic cholesterol most significantly in the fasted state (Fig. 3 B and C). In the fasted state, knockdown of SIRT1 also caused a significant accumulation of hepatic cholesterol (Fig. 3A) and overexpression modestly but significantly depleted liver cholesterol (Fig. 3 B and C). Knockdown of SIRT1 in db/db mice had a similar effect on systemic and hepatic lipid levels (SI Fig. 9). Interestingly, we note that unlike the effects on glucose metabolism, SIRT1 regulated cholesterol levels independently of PGC-1α. SIRT1 increased systemic cholesterol and decreased hepatic cholesterol just as potently when PGC-1α levels were reduced by shRNA (Fig. 3C).
Fig. 3.
Hepatic SIRT1 controls systemic and hepatic cholesterol and fatty acid homeostasis. (A) Liver free fatty acids (nonesterified fatty acids), serum total cholesterol, and liver cholesterol from control shRNA-infected fed (n = 5) and 20-h fasted (n = 5) mice and SIRT1 shRNA-infected fed (n = 6) and fasted (n = 5) mice. (B) Fed, GFP (n = 6) and SIRT1 (n = 6); 19-h fasted, GFP (n = 10) and SIRT1 (n = 10). (C) GFP or SIRT1 overexpression and control or PGC-1α shRNA double-infected mice (each bar, n = 5) following a 20-h fast. Liver measurements were normalized to protein content. Similar results for all measurements have been observed in at least two independent experiments. All data are presented as the average ± SEM. Significance was determined by Student's t test. GFP/control shRNA vs. SIRT1/SIRT1 shRNA: *, P < 0.05; **, P < 0.01. Control shRNA vs. PGC-1α shRNA: #, P < 0.05.
Hepatic SIRT1 Controls Gene Expression of Enzymes Involved in Triglyceride, Fatty Acid, and Cholesterol Metabolic Pathways.
The accumulation of liver free fatty acids by SIRT1 knockdown prompted us to analyze the gene expression of enzymes involved in fatty acids and triglyceride metabolism. As expected, fasting induced gene expression of fatty acid oxidation enzymes medium chain acyl-CoA deyhydrogenase (MCAD) and carnitine palmitoyltransferase-1a (CPT-1a) (Fig. 4A). Notably, mice with hepatic SIRT1 knockdown displayed lower expression of these enzymes compared with control mice. In both cases, the fasting induction was largely reduced by SIRT1 knockdown. Other genes involved in fatty acid and mitochondrial oxidation such as cytochrome c (Cyto-C) and L-FABP (data not shown) remained unchanged. In contrast, SIRT1 knockdown induced the expression of FAS, a key lipogenic gene in the fasted state (Fig. 4A). Expression of the transcription factor SREBP-1c/ADD1, a controller of lipogenic gene expression (12), followed a similar pattern to that of its target gene, FAS. Again, the fasting response, repression of these lipogenic genes, was impaired by SIRT1 knockdown. Because SIRT1 knockdown caused an accumulation of hepatic free fatty acids but no change in triglycerides, we analyzed expression of enzymes involved in triglyceride synthesis. Expression of DGAT2, an enzyme involved in fatty acid esterification to glycerol, was markedly reduced in SIRT1 knockdown mice (Fig. 4A). These results suggest that the accumulation of hepatic free fatty acids caused by SIRT1 knockdown could be due to changes in gene expression of enzymes involved in fatty acid oxidation and fatty acid and triglyceride synthesis. Although SIRT1 overexpression did not alter levels of liver free fatty acids (Fig. 3 B and C), it partially reversed the effect of SIRT1 knockdown, inducing MCAD, CPT-1a, and DGAT2 expression in the fasted state (Fig. 4B). This response depended on PGC-1α, as knockdown of PGC-1α blocked the SIRT1 induction of these genes in the fasted state (Fig. 4C).
Fig. 4.
SIRT1 controls hepatic expression of genes involved in lipid metabolism. Quantitative RT-PCR was used to analyze genes involved in fatty acid and cholesterol metabolism. (A and D) Fed, control shRNA (n = 5) or SIRT1 shRNA (n = 6); fasted for 20 h, control shRNA (n = 5) and SIRT1 shRNA (n = 5). (B and E) Fed, GFP (n = 6) or SIRT1 (n = 6); fasted for 19 h, GFP (n = 6) or SIRT1 (n = 6). (C and F) Double infection with GFP or SIRT1 overexpression and control or PGC-1α shRNA (each bar, n = 9) following a 20-h fast. All data are presented as the average ± SEM normalized to 36b4 expression. Significance was determined by Student's t test. GFP/control shRNA vs. SIRT1/SIRT1 shRNA: *, P < 0.05; **, P < 0.01. Control shRNA vs. PGC-1α shRNA: #, P < 0.05; ##, P < 0.01.
Because hepatic SIRT1 knockdown caused accumulation of total cholesterol in liver (Fig. 3A) and SIRT1 overexpression reduced liver cholesterol (Fig. 3B), we analyzed gene expression of enzymes involved in cholesterol transport, synthesis, and degradation. Hepatic gene expression of HMG-CoA reductase, a rate-limiting enzyme in cholesterol synthesis, was not altered by SIRT1 knockdown or overexpression (data not shown). However, SIRT1 did regulate several genes involved in cholesterol transport and degradation. Hepatic cholesterol levels are also tightly regulated by efflux-mediating cell membrane ATP binding cassette (ABC) transporters ABCA1 and ABCG1 as well as SR-B1. Hepatic expression of ABCA1 and SR-B1 was down-regulated by SIRT1 shRNA (Fig. 4D). Conversely, SIRT1 overexpression resulted in a fasting induction of SR-B1 and a trend toward induction of ABCA1, although not statistically significant (Fig. 4E). Another route for cholesterol import into hepatocytes is transport through the LDL receptor (LDLr) (17). Consistent with an increase in hepatic cholesterol levels, SIRT1 knockdown resulted in a fasting induction of LDLr gene expression (Fig. 4D). Bile acid synthesis from cholesterol is yet another pathway involved in cholesterol clearance and catabolism (37). A rate-limiting enzyme involved in bile acid synthesis, Cyp7A1, was decreased in fasted livers of SIRT1 shRNA mice (Fig. 4D) and induced by SIRT1 overexpression in the fasted state (Fig. 4E). The decreased expression of genes acting in cholesterol efflux and degradation is consistent with the accumulation of hepatic cholesterol in SIRT1 knockdown livers. SIRT1 overexpression in the fasted state caused an induction of the efflux and degradation pathways resulting in decreased in hepatic cholesterol content.
We also analyzed expression of two important transcriptional regulators of cholesterol transport and metabolism: LXRα and PGC-1β (19, 38). SIRT1 knockdown caused a slight yet highly significant decrease in LXRα expression in both fed and fasted livers. However, SIRT1 knockdown decreased expression of PGC-1β only the fasted state (Fig. 4D). These data suggest that the effects of SIRT1 knockdown on serum and hepatic cholesterol may be mediated through down regulation of LXRα and PGC-1β mRNA. However, SIRT1 overexpression was not capable of reversing these effects on PGC-1β and LXRα gene expression (Fig. 4E). Finally, we tested whether the effects of SIRT1 on cholesterol metabolism genes required PGC-1α. In contrast to the effects of SIRT1 on cholesterol levels, which were independent of PGC-1α (Fig. 3C), the SIRT1-mediated increases in SR-B1 and Cyp7a1 were totally dependent on PGC-1α (Fig. 4F). These data indicate that expression of SR-B1 and Cyp7A1 is not essential for changes in cholesterol levels. Although it is not clear exactly which genes mediate the effects of SIRT1 on cholesterol levels, compensatory effects such as a strong reduction of LDLr expression by PGC-1α knockdown may attempt to normalize cholesterol levels when efflux and degradation pathways are repressed (Fig. 4F).
Discussion
In response to nutritional challenges, metabolic gene expression needs to be precisely controlled to maintain glucose and lipid homeostasis. This control is accomplished by a network of transcription factors and coactivators, which are responsible for connecting the hormonal and nutrient signals to transcriptional regulation of metabolic pathways. In this context, we have found that SIRT1 has an important role mediating the liver's metabolic response to fasting. Hepatic knockdown of SIRT1 severely abrogates the fasting induction of gluconeogenic and fatty acid oxidation genes. Moreover, SIRT1 knockdown also reduced glycemia and serum cholesterol while causing accumulation of hepatic free fatty acids and cholesterol. We also found that SIRT1 contributes to the regulation of genes involved in triglyceride metabolism and cholesterol degradation and transport. Importantly, many of the effects we observed by SIRT1 knockdown were reversed with SIRT1 overexpression.
The coactivator PGC-1α is part of a transcriptional network that is involved in adaptation to nutrient stresses, through regulation of gluconeogenic and fatty acid β-oxidation gene expression (25, 39). We have previously shown that PGC-1α's ability to induce gluconeogenesis is largely regulated by acetylation (28, 40). Here, we extend these results by showing that in vivo knockdown and overexpression of SIRT1 is sufficient to alter endogenous acetylation of PGC-1α. We find that in the fasted liver the acetylation state of PGC-1α strongly correlates with repression/induction of gluconeogenic genes. Importantly, we show that when PGC-1α levels are reduced by shRNA knockdown, SIRT1 overexpression no longer reduces glucose tolerance or up-regulates gluconeogenic genes, suggesting that SIRT1 requires PGC-1α for these effects. Conversely, it seems that to a large extent that PGC-1α requires SIRT1 to stimulate glucose production. SIRT1 knockdown reduces the effect of PGC-1α overexpression in a PTT and on gluconeogenic gene expression. Moreover, the stimulatory effect of SIRT1 on PGC-1α is most likely mediated through deacetylation, as demonstrated by the greater induction of gluconeogenic genes and hyperglycemia caused by overexpression of the R13 PGC-1α acetylation mutant. These data combined suggest that the fasting induction, interaction, and deacetylation of PGC-1α by SIRT1 are an important regulatory component in the fasting induction of gluconeogenesis.
Oxidation of fatty acids and triglyceride synthesis are also important components of the liver's metabolic response to fasting and are activated by PGC-1α (19, 41). Consistent with SIRT1 activating PGC-1α upon fasting, expression of fatty acid oxidation and triglyceride genes were effected by SIRT1 knockdown and overexpression. Importantly, these changes in gene expression were also dependent on PGC-1α. These results further extend the role of SIRT1-PGC-1α to lipid metabolism in the fasted liver to control expression of key genes. One unexpected and new finding in our studies is the effect of SIRT1 on cholesterol metabolism and transport, an effect that seems to be independent of the fasting response. Intriguingly, whereas SIRT1 effects on systemic and hepatic cholesterol were PGC-1α independent, expression of genes that control cholesterol levels such as SR-B1 and Cyp7A1 required PGC-1α. A plausible explanation might involve compensatory mechanisms, such as decreased LDRr expression by SIRT1 and PGC-1α knockdown. Taken together, our results indicate that, although cholesterol levels are not dependent on PGC-1α, some of the effects on cholesterol metabolic gene expression require PGC-1α. It would be interesting to identify other factors that mediate the effects of SIRT1 in the liver and the extent to which PGC-1α is required for their activity.
We have previously reported induction of SIRT1 activity and protein in response to fasting in mouse liver and muscle and the role of pyruvate as an important nutrient signal in liver cells. Ablation of SIRT1 in cultured cells renders them unresponsive to nutrient fasting signals (28, 42). The fact that in vivo knockdown and overexpression of SIRT1 in liver had the most significant effect in the fasted state is consistent with our observations in vitro. This data provides evidence to suggest that SIRT1 is a nutrient sensor regulating metabolic pathways through deacetylation of PGC-1α and other targets. However, the specific nutrient and/or hormonal inputs that regulate SIRT1 are unknown at this time. SIRT1 may be able to respond to a broad range of signals through metabolic flux and NAD+/NADH ratios. Interestingly, this biological function is consistent with the hypothesized role of SIRT1 homologs in calorie restriction observed in other species (43). Taken together, our results indicate that SIRT1 plays a key role in coordinating metabolic responses to calorie restriction/nutrient deprivation. Using an in vivo model, we find that SIRT1 is required for the maintenance of glucose and lipid homeostasis in the liver.
Methods
Animal Experiments.
All animal experiments conformed to protocols approved by animal care and use committees at the Johns Hopkins School of Medicine and the Dana–Farber Cancer Institute. Experiments were performed in 6- to 8-week-old male BALB/c mice purchased from Harlan Laboratories (Indianapolis, IN). Adenovirus infections were performed by tail vein injection with 1 × 109 infectious particles per mouse (1.5 × 109 for double infections). Mice were killed 7–9 days after transduction. For detailed methods, see SI Methods.
GTT, ITT, and PTTs.
For the GTT, after a 5-h fast, mice received an i.p. injection of 2 g/kg dextrose (Sigma, St. Louis, MO) dissolved in PBS. For the ITT, after a 5-h fast, mice received an i.p. injection of 0.6 unit/kg insulin (Sigma) in PBS. For the PTT, after an 18-h fast, mice received an i.p. injection of 2 g/kg sodium pyruvate (Sigma) dissolved in PBS. Blood glucose concentrations were measure via tail bleed before and at times indicated after injection. All glucose measurements were made using an Ascenscia Elite XL glucometer (Bayer, Wuppertal, Germany).
Metabolite Measurements.
Serum true triglycerides and liver total triglycerides were measured using a colorimetric assay (TR0100; Sigma). Serum and liver free fatty acids (nonesterified fatty acids) were measured using the NEFA C kit (Wako, Osaka, Japan). Total serum and liver cholesterol was measured using cholesterol reagent (Pointe Scientific, Canton, MI). For detailed methods, see SI Methods.
For more information about PGC-1α acetylation, see SI Appendices 1 and 2.
Quantitative Real-Time PCR.
All gene expression was measured by quantitative real-time PCR. Expression was determined by ΔCT versus control 36B4. Primer sequences are available upon request.
Statistical Analysis.
All data presented is the average ± SEM, unless indicated otherwise. Significance was determined using two-tailed unpaired Student's t test. * denotes P < 0.05, and ** denotes P < 0.01.
Supplementary Material
Acknowledgments
We thank members of the P.P. Laboratory for insightful discussions and comments on this manuscript and Marc Montminy (Salk Institute, La Jolla, CA) for the PGC-1α antibody and shRNA adenovirus. J.T.R. is a recipient of a Mid-Atlantic American Heart Association Predoctoral Fellowship. This work was supported in part by an Ellison Medical Foundation New Scholar Award, the American Diabetes Association, the U.S. Department of Defense, and National Institutes of Health Grant R01 DK069966 (to P.P.).
Abbreviations
- ABC
ATP binding cassette
- CPT
carnitine palmitoyltransferase
- Cyp7A1
cytochrome P450 type 7A1
- FAS
fatty acid synthase
- G6Pase
glucose 6-phosphatase
- GK
glucokinase
- GTT
glucose-tolerance test
- HNF
hepatic nuclear factor
- ITT
insulin-tolerance test
- LDL
low-density lipoprotein
- LDLr
LDL receptor
- LPK
liver pyruvate kinase
- LXR
liver X receptor
- MCAD
medium chain acyl-CoA deyhydrogenase
- PGC
peroxisome proliferator-activated receptor coactivator
- Pepck
phosphoenolpyruvate carboxylase kinase
- PTT
pyruvate-tolerance test
- shRNA
small hairpin RNA
- SIRT1
sirtuin 1
- SR-B1
scavenge receptor B-1
- SREBP
sterol response element binding protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702509104/DC1.
References
- 1.Rutter GA. Curr Biol. 2000;10:R736–738. doi: 10.1016/s0960-9822(00)00737-5. [DOI] [PubMed] [Google Scholar]
- 2.Michael MD, Kulkarni RN, Postic C, Previs SF, Shulman GI, Magnuson MA, Kahn CR. Mol Cell. 2000;6:87–97. [PubMed] [Google Scholar]
- 3.Muoio DM, Newgard CB. Annu Rev Biochem. 2006;75:367–401. doi: 10.1146/annurev.biochem.75.103004.142512. [DOI] [PubMed] [Google Scholar]
- 4.Pilkis SJ, Granner DK. Annu Rev Physiol. 1992;54:885–909. doi: 10.1146/annurev.ph.54.030192.004321. [DOI] [PubMed] [Google Scholar]
- 5.Owen OE, Kalhan SC, Hanson RW. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 6.Bouche C, Serdy S, Kahn CR, Goldfine AB. Endocr Rev. 2004;25:807–830. doi: 10.1210/er.2003-0026. [DOI] [PubMed] [Google Scholar]
- 7.Wu C, Okar DA, Kang J, Lange AJ. Curr Drug Targets Immune Endocr Metabol Disord. 2005;5:51–59. doi: 10.2174/1568008053174769. [DOI] [PubMed] [Google Scholar]
- 8.Bailey CJ. Diabetes Care. 1992;15:755–772. doi: 10.2337/diacare.15.6.755. [DOI] [PubMed] [Google Scholar]
- 9.Radziuk J, Bailey CJ, Wiernsperger NF, Yudkin JS. Curr Drug Targets Immune Endocr Metabol Disord. 2003;3:151–169. doi: 10.2174/1568008033340298. [DOI] [PubMed] [Google Scholar]
- 10.Postic C, Dentin R, Girard J. Diabetes Metab. 2004;30:398–408. doi: 10.1016/s1262-3636(07)70133-7. [DOI] [PubMed] [Google Scholar]
- 11.den Boer MA, Berbee JF, Reiss P, van der Valk M, Voshol PJ, Kuipers F, Havekes LM, Rensen PC, Romijn JA. Arterioscler Thromb Vasc Biol. 2006;26:124–129. doi: 10.1161/01.ATV.0000194073.87647.10. [DOI] [PubMed] [Google Scholar]
- 12.Horton JD, Goldstein JL, Brown MS. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 14.Shin DJ, Campos JA, Gil G, Osborne TF. J Biol Chem. 2003;278:50047–50052. doi: 10.1074/jbc.M309736200. [DOI] [PubMed] [Google Scholar]
- 15.Zannis VI, Chroni A, Krieger M. J Mol Med. 2006;84:276–294. doi: 10.1007/s00109-005-0030-4. [DOI] [PubMed] [Google Scholar]
- 16.Oram JF, Heinecke JW. Physiol Rev. 2005;85:1343–1372. doi: 10.1152/physrev.00005.2005. [DOI] [PubMed] [Google Scholar]
- 17.Hobbs HH, Russell DW, Brown MS, Goldstein JL. Annu Rev Genet. 1990;24:133–170. doi: 10.1146/annurev.ge.24.120190.001025. [DOI] [PubMed] [Google Scholar]
- 18.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Lin J, Yang R, Tarr PT, Wu PH, Handschin C, Li S, Yang W, Pei L, Uldry M, Tontonoz P, et al. Cell. 2005;120:261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 20.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, et al. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 21.Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, et al. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Monks B, Ge Q, Birnbaum MJ. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto M, Accili D. Nat Med. 2006;12:33–34. doi: 10.1038/nm0106-33. 2006; discussion 34. [DOI] [PubMed] [Google Scholar]
- 24.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, et al. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 25.Lin J, Handschin C, Spiegelman BM. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Tontonoz P, Mangelsdorf DJ. Mol Endocrinol. 2003;17:985–993. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
- 27.Reddy JK, Rao MS. Am J Physiol Gastrointest Liver Physiol. 2006;290:G852–G858. doi: 10.1152/ajpgi.00521.2005. [DOI] [PubMed] [Google Scholar]
- 28.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 29.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, et al. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaeberlein M, McVey M, Guarente L. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tissenbaum HA, Guarente L. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 34.Rogina B, Helfand SL. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koo SH, Satoh H, Herzig S, Lee CH, Hedrick S, Kulkarni R, Evans RM, Olefsky J, Montminy M. Nat Med. 2004;10:530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- 36.Handschin C, Lin J, Rhee J, Peyer AK, Chin S, Wu PH, Meyer UA, Spiegelman BM. Cell. 2005;122:505–515. doi: 10.1016/j.cell.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 37.Goodwin B, Kliewer SA. Am J Physiol Gastrointest Liver Physiol. 2002;282:G926–G931. doi: 10.1152/ajpgi.00044.2002. [DOI] [PubMed] [Google Scholar]
- 38.Kalaany NY, Mangelsdorf DJ. Annu Rev Physiol. 2006;68:159–191. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- 39.Finck BN, Kelly DP. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM. Proc Natl Acad Sci USA. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bordone L, Guarente L. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.