Abstract
Glioblastoma multiforme (GBM) is the most aggressive brain tumor in adults and remains incurable despite multimodal intensive treatment regimens. EGFRvIII is a truncated extracellular mutant of the EGF receptor (EGFR) commonly found in GBMs that confers enhanced tumorigenic behavior. To gain a molecular understanding of the mechanisms by which EGFRvIII acts, we have performed a large-scale analysis of EGFRvIII-activated phosphotyrosine-mediated signaling pathways and thereby have identified and quantified 99 phosphorylation sites on 69 proteins. Distinct signaling responses were observed as a function of titrated EGFRvIII receptor levels with the phosphatidylinositol 3-kinase pathway being dominant over the MAPK and STAT3 pathways at a high level of EGFRvIII expression. Within this data set, the activating phosphorylation site on the c-Met receptor was found to be highly responsive to EGFRvIII levels, indicating cross-activation of the c-Met receptor tyrosine kinase by EGFRvIII. To determine the significance of this finding, we devised a combined treatment regimen that used a c-Met kinase inhibitor and either an EGFR kinase inhibitor or cisplatin. This regimen resulted in enhanced cytotoxicity of EGFRvIII-expressing cells compared with treatment with either compound alone. These results suggest that the clinical use of c-Met kinase inhibitors in combination with either EGFR inhibitors or standard chemotherapeutics might represent a previously undescribed therapeutic approach to overcome the observed chemoresistance in patients with GBMs expressing EGFRvIII.
Keywords: mass spectrometry, mutant EGF receptor, signal transduction, tyrosine phosphorylation
Glioblastoma multiforme (GBM) is the most aggressive form of adult human brain tumor, with median survival of <12 months (1). This dismal prognosis is due in part to the lack of therapeutic agents available to eliminate the diffuse glioma infiltrate that remains in the brain after surgical resection. Molecular profiling of genetic lesions in these tumors holds the promise of stratifying tumors into categories amenable to targeted treatment modalities. One such example is the application of EGF receptor (EGFR)-targeted therapeutic agents to treat GBMs that overexpress EGFR. Of these tumors, about half express the deletion mutant EGFRvIII, a truncated extracellular mutant of EGFR lacking exons 2–7, including the extracellular ligand-binding domain. Although EGFRvIII is incapable of binding the EGF family of ligands, it has been shown to be constitutively tyrosine-phosphorylated at ≈10% relative to ligand-stimulated wild-type (WT) EGFR (2). This truncated mutant receptor has been exclusively found in tumors, suggesting selection for EGFRvIII during the process of tumorigenesis. Clinical studies have demonstrated a correlation between EGFRvIII expression and poor prognosis for patients with GBM, indicating that it may be important in driving tumorigenic behavior in GBMs.
Although much work has been done over the past decade to elucidate pathways involved in EGFRvIII receptor signaling, the global map of the signaling network(s) it activates remains incomplete, making it difficult to assess downstream components involved in EGFRvIII-mediated transformation. Here, we used a previously described mass spectrometry (MS)-based phosphoproteomics approach (3) to quantitatively map cellular signaling events activated by the EGFRvIII receptor as a function of titrated receptor levels. This systems-level strategy provides insights into the biology of the EGFRvIII receptor and has identified the c-Met receptor as a cotarget for treatment of EGFRvIII-expressing tumors.
Results
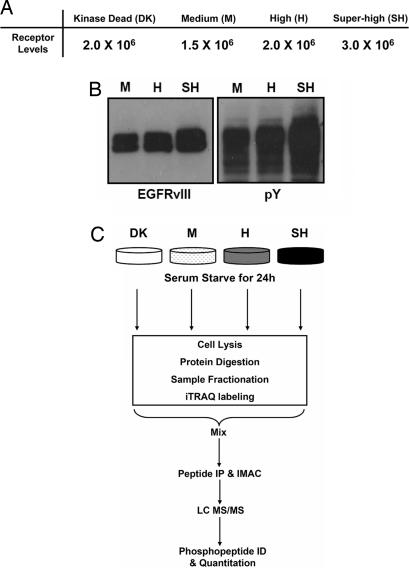
To determine the effect that EGFRvIII receptor expression level has on phosphotyrosine-mediated cellular signaling networks, U87MG glioblastoma cell lines expressing differential levels of EGFRvIII were isolated by FACS sorting of transduced populations (Fig. 1A) and then analyzed by MS to identify and quantify tyrosine phosphorylation sites on cellular signaling proteins. Western blot and FACS analysis (data not shown) confirm the differing expression levels of EGFRvIII and quantify relative levels of tyrosine phosphorylation across the three cell lines [Fig. 1B and supporting information (SI) Fig. 5]. A previously derived U87MG cell line expressing 2 million copies of a kinase-dead (DK) EGFRvIII receptor was used as a control (4); we have recently shown that tumorigenic potential increases with increased EGFRvIII receptor levels (5).
Fig. 1.
Cell lines and experimental strategy. (A) EGFRvIII expression levels in retrovirally transfected U87MG cell lines. (B) Western blot of U87MG cell lines expressing titrated levels of EGFRvIII. Cells were serum-starved for 36 h, lysed, and probed for EGFRvIII or phosphotyrosine levels. (C) Outline of MS-based experimental strategy.
To identify the signaling pathways constitutively activated downstream of the EGFRvIII receptor while minimizing confounding signaling associated with serum and cell culture media, the four cell lines were serum-starved for 24 h before cell lysis and sample preparation. Peptides from the four samples were stable isotope labeled, mixed, and tyrosine-phosphorylated peptides were immunoprecipitated with a pan-specific antiphosphotyrosine antibody (Fig. 1C). After immunoprecipitation, phosphorylated peptides were further enriched by immobilized metal affinity chromatography and analyzed by liquid chromatography tandem MS. In total, quantitative phosphorylation profiles were generated for 99 phosphorylation sites on 69 proteins across the four cell lines. Two biological replicates were performed with an average SD of 15% for tyrosine phosphorylated peptides that appear in both analyses (SI Table 1).
Quantitative Effects of Titrated EGFRvIII Levels on Receptor Phosphorylation and Major Downstream Signaling Pathways.
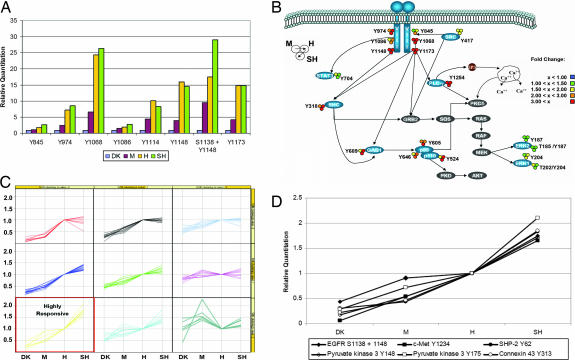
We identified eight phosphorylation sites on EGFRvIII and determined their quantitative phosphorylation profiles as a function of increasing EGFRvIII receptor levels (Fig. 2A). Interestingly, phosphorylation levels on Y845 and Y1086 increased proportionally with EGFRvIII expression level, whereas phosphorylation of Y974, Y1068, Y1114, Y1148, and Y1173 increased >2-fold from the U87-M (1.5 million copies EGFRvIII per cell) to U87-H (2 million copies EGFRvIII per cell) cells. This large increase in the latter cluster of EGFRvIII autophosphorylation sites indicates a threshold of EGFRvIII expression; once EGFRvIII exceeds this threshold expression level, activation of the receptor and resulting autophosphorylation increases significantly. For these sites, increasing receptor expression beyond 2 million copies per cell did not increase the level of phosphorylation, suggesting a saturation point. However, the level of phosphorylation on many sites downstream from the receptor continued to increase past this saturation point, indicating that negative-feedback mechanisms might be responsible for the lack of increase in EGFR autophosphorylation levels in the U87-SH cells.
Fig. 2.
Effect of EGFRvIII receptor levels on downstream signaling networks. (A) Relative quantification of EGFRvIII phosphorylation sites across the four cell lines. Phosphorylation levels are normalized relative to that of the DK cell line. (B) Visualization of the fold change in phosphorylation levels in the canonical EGFR signaling cascades as a function of titrated EGFRvIII levels. (C) Clustering analysis of phosphotyrosine protein networks using self-organizing maps (SOMs). Each column within the matrix components represent the relative phosphorylation level in the -DK, -M, -H, and -SH U87MG cell lines normalized against the U87H cell line. Optimal SOM architecture was a 3 × 3 matrix, because smaller matrices tended to cluster dissimilar phosphorylation profiles. (D) Protein phosphorylation sites found within the highly responsive cluster.
Mapping the phosphorylation data to canonical EGFR signaling cascades (Fig. 2B) indicates that EGFRvIII favors the utilization of different downstream pathways compared with WT EGFR. We have previously demonstrated that EGF stimulation of human mammary epithelial cells expressing WT EGFR led to a dramatic increase in the active form of Erk1, Erk2, and STAT3 within 5 min of stimulation (6). In contrast, increasing EGFRvIII receptor expression levels had little effect on the phosphorylation levels of these proteins. Moreover, our previous temporal analysis of WT EGFR signaling indicated that activation of this receptor led to a modest increase in the tyrosine phosphorylation levels on phosphatidylinositol 3-kinase (PI3K) and its upstream adaptor protein GAB1. However, increasing EGFRvIII receptor levels dramatically increased the phosphorylation levels on these proteins by >3-fold, indicating that the PI3K pathway is highly active in EGFRvIII-overexpressing cells. These data are consistent with previous studies in which EGFRvIII has been found to activate the PI3K pathway (7). Preferential activation of this pathway by EGFRvIII (in addition to its constitutive activation) may be a reason for its observed tumorigenic properties in vivo, because PI3K signaling has been implicated in promoting cell proliferation, survival, and migration.
c-Met Receptor Tyrosine Kinase Activation Is Highly Responsive to EGFRvIII Receptor Levels.
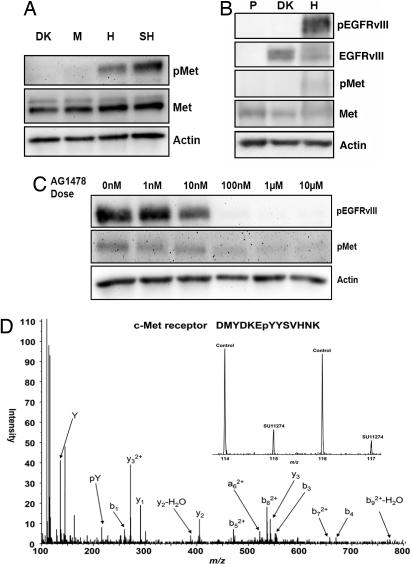
To characterize relationships within the data set, a self-organizing map was used to identify clusters of tyrosine phosphorylation sites with similar profiles (Fig. 2C). One such cluster consisted of phosphorylation sites that significantly increased as a function of increasing EGFRvIII expression level. Sites in the cluster include Y1234, the activating phosphorylation site on the catalytic loop of the c-Met receptor tyrosine kinase (6-fold increase) and an uncharacterized phosphorylation site (Y62) on tyrosine phosphatase SHP-2 (10-fold increase), a protein known to be downstream of the c-Met receptor (Fig. 2D). Phosphorylation of the c-Met receptor was confirmed by Western blot analysis of the four cell lines both in vitro and in vivo (Fig. 3 A and B); similar results were obtained by using c-Met immunoprecipitations followed by Western blotting with antiphosphotyrosine antibodies (results not shown). EGFRvIII-dependent phosphorylation of c-Met Y1349 was also detected in the U87H cell lines by Western blot (data not shown). To ensure that c-Met phosphorylation was not unique to U87MG cells, tet-inducible EGFRvIII expressing U373MG glioblastoma cell lines were tested. EGFRvIII-mediated activation of the c-Met receptor was also observed in these cells (SI Fig. 6).
Fig. 3.
c-Met receptor activation and kinase inhibition. (A) Western blot of specific phosphorylation sites on the c-Met receptor (Y1230/Y1234/Y1235) across the four different cell lines in vitro after 24-h serum starvation. (B) Western blot of c-Met receptor phosphorylation levels of in vivo parental (P), DK, or EGFRvIII high-expressing U87MG-derived xenografts. (C) Western blot of U87-H cell line subjected to 1 h AG1478 dose escalation after 24-h serum starvation. (D) Comparison of the quantification of the phosphorylation levels for c-Met Y1234 upon treatment with either DMSO (control) or 10 μM c-Met kinase inhibitor SU11274 for 1 h after 24-h serum starvation. Two biological replicates were performed and peak areas for iTRAQ marker ions enable quantification of phosphorylation for each condition.
Increased phosphorylation of the c-Met activation site led us to postulate that the EGFRvIII receptor was constitutively activating the c-Met receptor pathway. Mapping of our phosphoproteomic data to previously described c-Met signaling pathways (8) confirmed that many of the known components downstream of the c-Met receptor were activated at least 3-fold as a function of increasing EGFRvIII expression levels (SI Fig. 7). Many of the activated downstream components of the c-Met receptor overlap with the downstream targets of EGFRvIII (Fig. 2B). It is well established that receptor tyrosine kinases share common downstream signaling components. The activation of a common set of downstream signaling events may represent an integrated signaling cascade resulting from the coactivation of both the c-Met and EGFRvIII receptors.
As a complementary approach to demonstrate that c-Met receptor activation was a direct consequence of EGFRvIII receptor activation, we treated U87-H cells with AG1478, an EGFR kinase inhibitor that has some preference for EGFRvIII. Western blot analysis revealed a dose-dependent decrease in EGFRvIII phosphorylation levels accompanied by a concomitant decrease in the phosphorylation status of c-Met (Fig. 3C). Because the U87MG cell line has previously been shown to express the c-Met ligand hepatocyte growth factor (HGF) (9), we sought to determine whether this EGFRvIII-mediated c-Met activation was ligand-dependent. Measurement of HGF secretion did not reveal any appreciable trends across the four cell lines (SI Fig. 8A). Treating the U87-H cells with anti-HGF also did not affect c-Met phosphorylation levels, indicating that c-Met activation was not ligand-mediated (SI Fig. 8B). Further studies are required to fully elucidate the mechanism of EGFRvIII-mediated activation of c-Met.
Combined Inhibition of the EGFRvIII and c-Met Receptors Enhanced Cytotoxic Effects of an EGFR Kinase Inhibitor.
To determine the biological consequence of the c-Met activation, we treated U87-H cells with the c-Met kinase inhibitor, SU11274 (10). Quantitative mass spectrometric analysis of the c-Met receptor Y1234 phosphorylation site confirmed a decrease in c-Met receptor phosphorylation upon treatment of the U87-H cell line with SU11274 (Fig. 3D). In addition, mass spectrometric analysis of SU11274-treated U87-H cells indicated this drug did not affect EGFRvIII receptor tyrosine phosphorylation on multiple sites (data not shown).
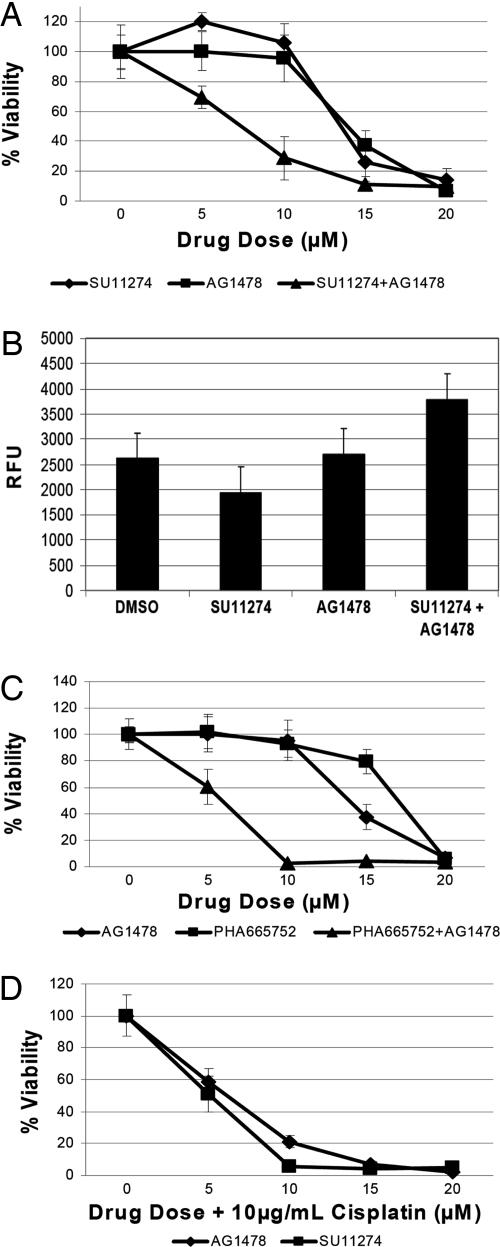
Because of the observed coactivation of EGFRvIII and c-Met receptors, we hypothesized that cotreatment of EGFRvIII-expressing cells with EGFR and c-Met kinase inhibitors may have an additive effect on cell viability and death. Treatment of U87-H cells with either AG1478 or SU11274 followed a similar profile in which cell viability was decreased only at high inhibitor doses. In contrast, the combination of a constant dose of 5 μM AG1478 with increased dosing of SU11274 led to a significant decrease in cell viability and increase in cell death (Fig. 4 A and B) at low inhibitor doses. To test for the possibility that off-target effects of SU11274 may have been responsible for decreased cell viability, we performed similar analyses with another c-Met inhibitor, PHA665752 (11), and found it also demonstrated similar enhanced effects upon cotreatment of the U87-H with AG1478 (Fig. 4C). These results suggest that the biological responses to the c-Met inhibitors were driven by on-target effects against c-Met itself or, perhaps, c-Met and another kinase which is a shared off-target molecule.
Fig. 4.
Dose–response of U87-H cell line upon treatment with kinase inhibitors or cisplatin. (A) Dose–response of U87-H cells to AG1478, SU1127, or a combination of SU11274 and 5 μM AG1478 over 72 h after 24-h serum starvation. Viability was measured by using the metabolic dye WST-1. Combination treatment significantly enhanced cytotoxicity at 10 μM SU11274 (P < 0.001). (B) Apoptosis measured by caspase 3/7 cleavage upon drug treatment over 24 h after 24-h serum starvation. Concentration of drugs used was 10 μM SU11274, 10 μM AG1478, or a combination of 10 μM SU11274 and 5 μM AG1478. Combination treatment significantly enhanced apoptosis (P < 0.01). (C) Dose–response of U87-H to AG1478, PHA665752, or a combination of PHA665752 and 5 μM AG1478 over 72 h after 24-h serum starvation. Combination treatment significantly enhanced cytotoxicity at 10 μM PHA665752 (P < 0.0001). (D) Viability of U87-H cells in response to a combination treatment of 10 μg/ml cisplatin with either AG1478 or SU11274.
c-Met Kinase Inhibition Overcomes Chemoresistance Conferred by EGFRvIII.
EGFRvIII confers chemoresistance to classical chemotherapeutics such as cisplatin through modulation of Bcl-XL and caspase 3 expression levels. Correspondingly, human glioblastoma xenografts expressing EGFRvIII are resistant to cisplatin unless cisplatin is coadministered with the EGFR kinase inhibitor, AG1478 (12, 13). Activation of the c-Met receptor has also previously been shown to confer resistance to a wide variety of chemotherapeutics (14). We therefore hypothesized that the observed chemoresistance of EGFRvIII-expressing tumors may be due in part to EGFRvIII-mediated activation of the c-Met receptor. To test this hypothesis, we treated U87-H cells with increasing dose of the c-Met kinase inhibitor SU11274 in combination with a constant 10 μg/ml dose of cisplatin. As demonstrated in Fig. 4D, combination treatment resulted in a dramatic decrease in cell viability when compared with treatment with cisplatin alone (SI Fig. 9). This result suggests that c-Met may play a significant role in the chemoresistance of EGFRvIII-positive tumors.
Discussion
Here we report a large-scale analysis of phosphotyrosine-mediated signaling pathways downstream of the EGFRvIII receptor. In this analysis, 99 phosphorylation sites on 69 proteins were identified and quantified, including eight phosphorylation sites on EGFRvIII. Although phosphorylation sites on EGFRvIII may not be qualitatively different from those observed in WT, quantitative differences in the levels of phosphorylation at each individual site may have functional implications on resultant downstream signaling pathways and biological response. EGFRvIII expression appears to have both a threshold and a saturation level. Between the threshold and saturation levels, EGFRvIII autophosphorylation increases disproportionately to changes in EGFRvIII expression, whereas above the saturation level, autophosphorylation is largely unaffected by further increases in receptor expression. These data highlight the need for additional functional analysis by site-directed mutagenesis to uncover the biological consequence of altered phosphorylation levels of these sites in the context of different EGFRvIII expression levels.
Although EGFRvIII autophosphorylation levels appear to saturate, many downstream protein phosphorylation sites continue to increase in the U87-SH cells relative to the U87-H cells, suggesting that negative-feedback mechanisms (e.g., phosphatases) decrease receptor phosphorylation when EGFRvIII expression exceeds a critical saturating level (Fig. 2 C and D). Our analysis in this study provides a systematic demonstration of the importance of oncogene dosage in the selection and propagation of downstream cellular signaling pathways. For instance, pathway analysis of the phosphoproteomic data set indicates that cells that highly overexpress EGFRvIII preferentially use the PI3K pathway over the MAPK and STAT3 pathways. This result provides a possible mechanistic basis for the success of PI3K and mTOR small molecule inhibitors in combination with EGFR kinase inhibitors in the treatment of EGFRvIII-expressing cells and xenografts (15, 16). Our data also suggest that therapeutic approaches targeting the MAPK and STAT3 pathways for the treatment of tumors that express high levels of EGFRvIII would be predicted to be ineffective. Quantitative determination of such functional threshold limits for other cancer genes may represent a means to determine the relative order and dominance of oncogenes and their resultant cellular signaling pathways in human tumors. The ability of mass spectrometry-based network analysis to provide a mechanistic understanding linking oncogene expression levels to dysregulated signaling events in cancer highlights its utility in aiding in the selection of optimal cancer therapeutic targets for use in the clinic.
Clustering analysis of our phosphoproteomic data revealed that phosphorylation of the activation site on c-Met increased as a function of EGFRvIII expression level. Constitutive activation of c-Met by EGFRvIII is reminiscent of the constitutively active Tpr-Met fusion mutant of the c-Met receptor, which exhibits a more potent signaling potential than transient receptor activation stimulated by HGF ligand binding (17). c-Met has been implicated in the progression of a wide variety of cancers, including glioma and lung and breast cancer. An analysis of 32 glioma tumors showed that all of the tumors examined were positive for c-Met expression (18). Furthermore, glioma cell lines expressing c-Met were highly proliferative, motile, and invasive upon treatment with HGF, suggesting that c-Met may play an important role in glioma tumorigenicity. Amplification of c-Met and HGF has been associated with highly invasive and metastatic tumors and poor prognosis in lung cancer, and overexpression of c-Met has recently been linked to the development of gefitinib-resistant lung cancer (19). In cell lines derived from these tumors, a correlation between c-Met overexpression and ErbB3 activation has been demonstrated along with enhanced cytotoxicity after combined treatment with an EGFR kinase inhibitor and a c-Met kinase inhibitor. Although we have also observed enhanced effects of combining EGFR and c-Met kinase inhibitors, it is important to note that the underlying mechanisms appear to be significantly different. For instance, gefitinib-resistant lung cancer relies on the overexpression of c-Met to activate HER3, whereas EGFRvIII overexpression drives c-Met phosphorylation and activation regardless of c-Met expression levels in glioblastoma cell lines. In addition, glioblastoma tumors have been shown to express very low levels of HER3, indicating that the sensitivity of EGFRvIII-expressing cells to c-Met kinase inhibition is not due to the proposed reduction in c-Met-mediated activation of the HER3/PI3K/Akt pathway observed in the gefitinib-resistant cell lines (20, 21). Based on these two studies, it is tempting to speculate about potential synergy with other receptor tyrosine kinase inhibitors, especially given that other receptors show increased phosphorylation as a function of increased EGFRvIII expression (see SI Table 1).
There is a wide variety of approaches to regulate c-Met receptor activation, including the use of anti-HGF monoclonal antibodies and c-Met small-molecule kinase inhibitors. Our data indicate that constitutive c-Met activation in EGFRvIII-overexpressing cells is not mediated by autocrine HGF secretion but instead appears to be due to the direct signaling effects of the EGFRvIII receptor. This mechanism precludes the utilization of anti-HGF antibodies to suppress c-Met activation levels in EGFRvIII-expressing tumors. However, we have demonstrated that c-Met kinase inhibitors provide enhanced cytotoxicity when combined with EGFRvIII kinase inhibitors in EGFRvIII-expressing glioblastoma cells. Because most kinase inhibitors are known to inhibit multiple kinases, it is therefore possible that enhanced cell killing is associated with off-target effects in addition to inhibition of c-Met. However, SU11274 and PHA665752 have significantly different structures and should have different specificities, thereby decreasing the likelihood of similar off-target effects. It is important to note that our observations were made in the U87MG cell line, which contains secondary genetic lesions commonly found to occur in human GBM patients, namely loss of PTEN and Ink4A/Arf. PTEN is a tumor suppressor protein with both phosphoinositide and phosphotyrosine phosphatase activities and is commonly mutated in many advanced cancers including lung and prostate carcinomas. Correspondingly, it has been previously demonstrated that clinical response to EGFR inhibitors such as erlotinib and gefitinib in human glioblastoma patients was significantly associated with the coexpression of PTEN and EGFRvIII in tumors and in vitro in U87MG cell lines transfected to coexpress both EGFRvIII and PTEN (22). Because PTEN mutation is seen in 30–44% of high-grade gliomas (1), a large proportion of GBM patients are refractory to EGFR kinase inhibitor therapy. Our in vitro data suggest that cotreatment of EGFRvIII-overexpressing tumors with both EGFR and c-Met kinase inhibitors may overcome this chemoresistance even in PTEN-null tumors. Assaying for the expression of EGFRvIII and c-Met in human gliomas may guide the combined use of these inhibitors in the clinic.
Chemoresistance of diffuse lesions in glioblastoma patients results in recurrence after surgical resection for almost all patients (1). Here we have demonstrated that cotreatment of U87-H cells with cisplatin and a c-Met kinase inhibitor led to a dose-dependent decrease in cell viability similar to cotreatment with cisplatin and AG1478, an EGFR kinase inhibitor. This result raises the possibility that c-Met activation may account for a significant proportion of EGFRvIII-mediated chemoresistance. In fact, it is plausible to suspect that many of the tumor-associated phenotypes previously attributed to the EGFRvIII receptor may be due in part to cross-activation of c-Met or other receptor tyrosine kinases (RTKs). Activation of multiple RTKs by EGFRvIII may potentiate a multitude of additional tumorigenic properties, each arising either from the independent activity of individual activated receptors or from an integrated signal arising from the combinatorial activation of multiple receptors. In our analysis, in addition to the activation of the c-Met receptor, we also observed increased phosphorylation of Axl and EphA2 RTKs. It will be important to test the simultaneous inhibition of multiple RTKs, because this may represent a therapeutic strategy to overcome the multifaceted clinical features seen in GBM.
EGFRvIII-mediated phosphorylation and activation of c-Met was uncovered through network analysis of EGFRvIII signaling pathways in U87MG cell lines by MS. Cotreatment with c-Met kinase inhibitors and cisplatin or c-Met kinase inhibitors and EGFR kinase inhibitors demonstrated enhanced cytotoxicity in U87-H cells. It is important to extend these studies to murine xenograft models and eventually to other clinical models to evaluate the efficacy of this cotreatment in treating tumors in vivo. Our results highlight the potential of using unbiased network analysis to identify novel therapeutic targets in signaling networks. Further analyses of other GBM cell lines and human tumors may highlight additional proteins and key signaling nodes, which may serve as targets to treat this devastating disease.
Methods
Cell Culture, Retrovirus Infection, and Transfection.
The human glioblastoma cell lines, U87MG and U373MG, and their engineered derivatives were cultured in DMEM with 10% FBS/2 mM glutamine/100 units/ml penicillin/100 mg/ml streptomycin in 95% air/5% CO2 atmosphere at 37°C. U87MG cells expressing EGFRvIII or DK cells were selected in 400 μg/ml G418 and maintained, as described (23). For expression of tetracycline-regulated EGFRvIII and DK, U373 glioma cells were transfected with pRev-tet-off (Invitrogen, Carlsbad, CA) by the calcium phosphate method (24) and selected in 400 μg/ml G418. Individual tetracyclin-controlled transactivator (tTA) expressing clones were analyzed for GFP expression, as expressed from transiently transfected pTRE-GFP, in the presence and absence of 1 μg/ml doxycycline (dox). A clone (c.16) demonstrating robust expression of GFP in the absence of dox was subsequently cotransfected with pBABE-puro and pTRE-EGFRvIII-IRES-GFP or pTRE-DK-IRES-GFP, and stable populations were obtained by selection in 1 μg/ml puromycin. Induction of EGFRvIII-IRES-GFP and DK-IRES-GFP was achieved upon growth in dox-free media.
Xenografts.
Cells (1 × 106) were suspended in 0.1 ml of PBS and injected into the right flanks of nude mice. Tumor volumes were defined as (longest diameter) × (shortest diameter)2 × 0.5. All of the procedures were approved by the animal care and use committee of the University of California at San Diego.
Sample Preparation, Peptide Immunoprecipitation, and MS.
U87MG cells were maintained in DMEM supplemented with 10% FBS. Cells (1.5 × 106, per 10-cm plate) were seeded for 24 h, then washed with PBS and incubated for 24 h in serum-free media. Cells were lysed and further processed and labeled with the iTRAQ reagent, as described (3). For each of the two biological replicates performed, lysate from three 10-cm plates were pooled together. Peptide immunoprecipitation was performed as described (3), with the following exceptions: 10 μg of protein G Plus-agarose beads (Calbiochem, La Jolla, CA) were incubated with 12 μg of antiphosphotyrosine antibody (pTyr100 (Cell Signaling Technology, Beverly, MA) in 200 μl of immunoprecipitation buffer (100 mM Tris/100 mM NaCl/1% Nonidet P-40, pH 7.4) for 8 h at 4°C. Immobilized metal affinity chromatography (IMAC) was performed to enrich for phosphorylated peptides and remove nonspecifically retained nonphosphorylated peptides. Peptides retained on the IMAC column were eluted with 250 mM sodium phosphate (pH 8.0) and analyzed by electrospray ionization liquid chromatography tandem MS on a QqTof (QSTAR XL Pro, Applied Biosystems, Foster City, CA), as described (3).
Phosphopeptide Sequencing, Quantification, and Clustering.
MS/MS spectra were extracted, searched, and quantified by using ProQuant (Applied Biosystems). Phosphorylation sites and peptide sequence assignments were validated by manual confirmation of raw MS/MS data. Peak areas of iTRAQ marker ions (m/z 114, 115, 116, and 117) were normalized with values from the iTRAQ marker ion peak areas of nonphosphorylated peptides in supernatant of the immunoprecipitation. Each condition was normalized against the U87H cell line to obtain fold changes across all four conditions. Final normalized data sets were loaded into Spotfire (Spotfire, Somerville, MA) and the self-organizing map algorithm was used to cluster the phosphorylation sites.
Immunoblot Analysis.
Cells were lysed in lysis buffer (20 mmol/liter Tris·HCl/150 mmol/liter NaCl/1 mmol/liter EDTA/1% Triton X-100/2.5 mmol/liter sodium PPi/1 mmol/liter β-glycerophosphate) containing protease and phosphatase inhibitors after the indicated treatment. Primary antibodies used were anti-EGFR pY1173, anti-c-Met (Santa Cruz Biotechnology, Santa Cruz, CA), antiphosphotyrosine 4G10, anti-c-Met pY1230/1234/1235 (Upstate Biotechnology, Lake Placid, NY), anti-EGFR, and anti-actin (Cell Signaling Technologies). Secondary antibody used was goat anti-rabbit antibody (Upstate Biotechnology).
Kinase Inhibitor Treatment.
Cells were serum-starved for 24 h before being treated with the indicated dose of either AG1478 (A.G. Scientific, San Diego, CA) or SU11274 (Calbiochem) for 1 h. Cells were then lysed as described above for either immunoblotting or mass spectrometric analysis.
Cell Viability Assays.
Four thousand cells were seeded per well in a 96-well plate. Twenty-four hours later, the cells were serum-starved for 24 h before addition of fresh serum-free media containing AG1478, SU11274, PHA665752 (a kind gift from Eli Lilly, Indianapolis, IN) or cisplatin (Sigma–Aldrich, St. Louis, MO) at the indicated doses and combinations. After 72 h, cell viability was measured by using the WST-1 reagent (Roche Applied Sciences, Indianapolis, IN), following the manufacturer's recommendations.
Apoptosis Assay.
Ten thousand cells were seeded per well in a 96-well plate. Twenty-four hours later, the cells were serum-starved for 24 h before addition of fresh serum-free media containing AG1478 and SU11274 at the indicated dose and combinations. After 24 h of drug treatment, caspase 3/7 activity was measured by using Apo-ONE Homogeneous Caspase-3/7 Assay (Promega, Madison, WI), following the manufacturer's recommendations. Details of additional methods, including flow cytometry, HGF Elisa, and anti-HGF treatment, are described in SI Text.
Supplementary Material
Acknowledgments
We thank members of the F.M.W. laboratory for helpful discussions and Eli Lilly for the kind gift of PHA665752. We also thank Rachel Takara for protein expression analysis and Maria Del Mar Inda for helpful discussions. This work was supported by National Cancer Institute (NCI) Grant U54-CA112967 and National Institutes of Health (NIH) Grant P50-GM68762 (to F.M.W.), an NCI integrative cancer biology program graduate fellowship (to P.H.H.), an award from the Goldhirsh Foundation (to F.B.F.), and NIH Grant P01-CA95616 (to W.K.C. and F.B.F.). W.K.C. is a fellow of the National Foundation for Cancer Research.
Abbreviations
- EGFR
EGF receptor
- RTK
receptor tyrosine kinase
- GBM
glioblastoma multiforme
- HGF
hepatocyte growth factor
- PI3K
phosphatidylinositol 3-kinase
- DK
kinase dead.
Footnotes
Conflict of interest statement: P.H.H. and F.M.W. are part of a patent application on methods for treating cancers associated with constitutive EGFR signaling.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705158104/DC1.
References
- 1.Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 2.Cavenee WK. Carcinogenesis. 2002;23:683–686. doi: 10.1093/carcin/23.5.683. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Mol Cell Proteom. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, Huang CM, Gill GN, Wiley HS, Cavenee WK. J Biol Chem. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 5.Johns TG, Perera RM, Vernes SC, Vitali AA, Cao DX, Cavenee WK, Scott AM, Furnari FB. Clin Cancer Res. 2007;13:1911–1925. doi: 10.1158/1078-0432.CCR-06-1453. [DOI] [PubMed] [Google Scholar]
- 6.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Proc Natl Acad Sci USA. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moscatello DK, Holgado-Madruga M, Emlet DR, Montgomery RB, Wong AJ. J Biol Chem. 1998;273:200–206. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- 8.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 9.Chattopadhyay N, Butters RR, Brown EM. Brain Res Mol Brain Res. 2001;87:100–108. doi: 10.1016/s0165-3806(00)00154-1. [DOI] [PubMed] [Google Scholar]
- 10.Sattler M, Pride YB, Ma P, Gramlich JL, Chu SC, Quinnan LA, Shirazian S, Liang C, Podar K, Christensen JG, Salgia R. Cancer Res. 2003;63:5462–5469. [PubMed] [Google Scholar]
- 11.Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, Chen J, Wang X, Ruslim L, Blake R, et al. Cancer Res. 2003;63:7345–7355. [PubMed] [Google Scholar]
- 12.Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ. Proc Natl Acad Sci USA. 1998;95:5724–5729. doi: 10.1073/pnas.95.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagane M, Narita Y, Mishima K, Levitzki A, Burgess AW, Cavenee WK, Huang HJ. J Neurosurg. 2001;95:472–479. doi: 10.3171/jns.2001.95.3.0472. [DOI] [PubMed] [Google Scholar]
- 14.Bowers DC, Fan S, Walter KA, Abounader R, Williams JA, Rosen EM, Laterra J. Cancer Res. 2000;60:4277–4283. [PubMed] [Google Scholar]
- 15.Wang MY, Lu KV, Zhu S, Dia EQ, Vivanco I, Shackleford GM, Cavenee WK, Mellinghoff IK, Cloughesy TF, Sawyers CL, Mischel PS. Cancer Res. 2006;66:7864–7869. doi: 10.1158/0008-5472.CAN-04-4392. [DOI] [PubMed] [Google Scholar]
- 16.Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, Stokoe D, Shokat KM, Weiss WA. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peschard P, Park M. Oncogene. 2007;26:1276–1285. doi: 10.1038/sj.onc.1210201. [DOI] [PubMed] [Google Scholar]
- 18.Koochekpour S, Jeffers M, Rulong S, Taylor G, Klineberg E, Hudson EA, Resau JH, Vande Woude GF. Cancer Res. 1997;57:5391–5398. [PubMed] [Google Scholar]
- 19.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 20.Andersson U, Guo D, Malmer B, Bergenheim AT, Brannstrom T, Hedman H, Henriksson R. Acta Neuropathol. 2004;108:135–142. doi: 10.1007/s00401-004-0875-6. [DOI] [PubMed] [Google Scholar]
- 21.Schlegel J, Stumm G, Brandle K, Merdes A, Mechtersheimer G, Hynes NE, Kiessling M. J Neurooncol. 1994;22:201–207. doi: 10.1007/BF01052920. [DOI] [PubMed] [Google Scholar]
- 22.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 23.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furnari FB, Huang HJ, Cavenee WK. Cancer Res. 1998;58:5002–5008. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.