Abstract
Recently identified hepatitis C virus (HCV) isolates that are infectious in cell culture provide a genetic system to evaluate the significance of virus–host interactions for HCV replication. We have completed a systematic RNAi screen wherein siRNAs were designed that target 62 host genes encoding proteins that physically interact with HCV RNA or proteins or belong to cellular pathways thought to modulate HCV infection. This includes 10 host proteins that we identify in this study to bind HCV NS5A. siRNAs that target 26 of these host genes alter infectious HCV production >3-fold. Included in this set of 26 were siRNAs that target Dicer, a principal component of the RNAi silencing pathway. Contrary to the hypothesis that RNAi is an antiviral pathway in mammals, as has been reported for subgenomic HCV replicons, siRNAs that target Dicer inhibited HCV replication. Furthermore, siRNAs that target several other components of the RNAi pathway also inhibit HCV replication. MicroRNA profiling of human liver, human hepatoma Huh-7.5 cells, and Huh-7.5 cells that harbor replicating HCV demonstrated that miR-122 is the predominant microRNA in each environment. miR-122 has been previously implicated in positively regulating the replication of HCV genotype 1 replicons. We find that 2′-O-methyl antisense oligonucleotide depletion of miR-122 also inhibits HCV genotype 2a replication and infectious virus production. Our data define 26 host genes that modulate HCV infection and indicate that the requirement for functional RNAi for HCV replication is dominant over any antiviral activity this pathway may exert against HCV.
Keywords: antivirals, miR-122, RNAi, HCVcc-siRNA
Hepatitis C virus (HCV) has a notable ability to establish persistent infections in ≈70% of cases, resulting in 130 million chronically infected people throughout the world (1). This prevalence has spurred considerable interest in the study of HCV–host interactions, on both cellular and molecular levels. The inability to grow HCV in cell culture led some groups to focus on the identification of cellular proteins that interact with individual HCV proteins or RNA elements, resulting in the accumulation of a large number of putative HCV–host interactions. Unfortunately, the significance of most of these with respect to the HCV life cycle is currently unknown (reviewed in ref. 2). Over the past 6 years, cell culture systems have been developed that enable the characterization of HCV replication and entry (3–6). This effort recently culminated in the development of cell culture systems that reproduce the entire viral life cycle (7–9). A number of virus–host interactions have been characterized by using these experimental systems. For example, CD81 has been demonstrated to play a role in HCV entry (10–12).
Sequence-specific gene silencing of RNAi is ideal for assessing the genetic phenotypes associated with virus–host interactions. We have previously shown that siRNAs are highly effective at silencing either host or viral RNAs in cells that contain replicating HCV, demonstrating the potential of RNAi as a tool to study HCV–host interactions (12–16). The goal of this study is to define host cofactors involved in HCV replication. We first identified 10 host proteins that interact with NS5A by using the two-hybrid system and copurification approaches. These were combined with other published HCV–host interactions to provide a sample set in which we could evaluate both the significance of these proteins for HCV replication and the utility of siRNA screens for identifying host genes and pathways that modulate HCV replication. siRNAs targeting 26 of the 62 host genes tested, including the RNAi ribonuclease Dicer, modulate the production of infectious HCV by at least 3-fold. Interference with multiple components of the RNAi pathway involved in miRNA biogenesis or the liver-specific miRNA miR-122 resulted in an inhibition of HCV replication. Thus, the RNAi pathway and miR-122 in particular are required for optimal HCV replication and infectious virus production, consistent with recent HCV replicon data from Jopling et al. (17).
Results
Identification of HCV NS5A-Interacting Proteins.
This study presents RNAi analysis of the significance of 62 host genes in HCV replication and infectious virus production. The majority of these genes have been published to interact with HCV RNA or proteins [supporting information (SI) Table 2]. Additionally, we identified a number of host interactions with HCV NS5A that were subsequently included in this analysis. Two approaches were used to identify NS5A-interacting proteins. The first approach involved the yeast two-hybrid system; details are provided in Materials and Methods (18). A LexA DNA-binding domain-NS5A (1a H77) fusion protein was used to a HeLa cDNA library whose translatable products are fused with an acidic transcriptional activation domain. Nine cDNAs encoding three unique proteins that interacted with NS5A were identified from a library of 108 cDNA clones. They are (i) the nuclear transport protein IPO4, (ii) the receptor recycling protein VPS35, and (iii) hsp90-associated protein 1, HSPA1A. These interactions were specific, inasmuch as the identified clones interacted with HCV NS5A, but not with the irrelevant bicoid protein or the closely related bovine viral diarrhea virus NS5A.
Our second approach involved the identification of cellular kinases that bind to NS5A in a complex that phosphorylates NS5A in vitro. The Kinetworks KPKS screen (Kinexus; Kinetworks, Vancouver, BC, Canada) is a method that can detect and quantify 75 protein kinases in a complex mixture of proteins based on recognition by specific antibodies. Purified preparations of GST and GST-NS5A were prepared and subjected to this analysis. Twelve candidate protein serine/threonine kinases that specifically associated with GST-NS5A were identified. Seven protein kinases match with the characterized profile of the NS5A kinase, including kinase inhibitor profiles, pH optimum, and divalent metal ion preferences (19). These kinases were identified as CDK6, ERK6/MAPK12, RAF1, AKT1, PDPK1, GSK3α, and GSK3β. The summary of NS5A–protein interaction data is shown in SI Table 2. Interestingly, the putative yeast homologues of four of these genes, AKT1, PDPK1, GSK3α, and GSK3β, have been reported to phosphorylate NS5A in vitro (20).
A Systematic RNAi Screen Identified Host Genes That Modulate HCV Replication.
We next tested the significance of 62 host genes for HCV replication. These host genes encode proteins that physically interact with HCV RNA or proteins, including the NS5A-interacting proteins identified above, or alternatively, that belong to signaling pathways thought to modulate RNA virus replication. A list of host genes, sites of HCV interaction, and associated references is available in SI Table 3. Huh-7.5 cells were transfected with at least two different siRNAs per gene (described in SI Table 4) and then infected with HCV over a sliding window, beginning at 24, 48, or 72 h after transfection. After 48 h of infection, cellular supernatants were collected for titration of infectious virus, whereas intracellular HCV RNAs were quantified by quantitative real-time RT-PCR. Cell viability assays measuring intracellular ATP levels were taken from parallel samples at the time of harvesting (SI Fig. 4). Changes in HCV RNA and virus levels were then calculated for each siRNA.
The fold change in HCV RNA or virus levels after silencing of the target gene, relative to the median value of all genes tested, is shown in Table 1. Twenty-six of the 62 genes that were targeted modulate HCV infectious virus production >3-fold. Real-time RT-PCR assays (described in SI Table 5) were developed for these genes to measure the expression levels of each siRNA's target RNA. The percent inhibition of target gene expression 2 days after siRNA transfection is shown in SI Fig. 5. For each siRNA, the target gene expression decreased at least 60%, with an average inhibition of ≈85%, (see SI Fig. 5). Thus, each siRNA inhibited the RNA accumulation of its intended target gene.
Table 1.
Changes in HCV replication after siRNA targeting of host gene RNAs
| siRNA* | Virus† | HCV RNA | siRNA | Virus | HCV RNA |
|---|---|---|---|---|---|
| HCV | < −230 | < −10000 | MAPK12 | −2.0 + 0.5••• | −1.3 + 0.8 |
| DDX3X | −42 + 19•• | −1600 + 800 | NCL | −1.8 + 0.4•• | 4.5 + 1.6 |
| EIF2S3 | −30 + 3.7••• | −55 + 34 | CDC2 | −1.8 + 0.5••• | 1.8 + 0.3 |
| STAT3 | −13 + 4.4••• | −8.3 + 2.4 | EIF2AK3 | −1.8 + 0.4•• | −2.0 + 0.1 |
| CD81 | −11 + 0.6•• | −6.1 + 2.6 | IPO4 | −1.7•• | 1.9 + 0.5 |
| ELAVL1 | −9.1 + 2.8••• | −3.3 + 0.4 | HNRPL | −1.5 + 1.8•• | −1.6 + 0.4 |
| VAP-ABC‡ | −8.7 + 1.7••• | −5.9 + 0.3 | XBP1 | −1.4 + 1.8••• | −1.5 + 0.7 |
| DICER1 | −7.5 + 2.5••• | −3.1 + 0.7 | CSNK2A1 | −1.3 + 0.1••• | 1.4 + 0.2 |
| HSPBP1 | −6.4 + 0.7•• | −5.7 + 2.3 | TP53 | −1.3•• | 1.7 + 0.6 |
| GRB2 | −6.3 + 0.9••• | −1.3 + 0.3 | EIF2B3 | −1.2 + 0.1••• | −3.5 + 0.6 |
| HM13 | −6.2 + 1.8•• | −10 + 4.0 | CALR | −1.2 + 1.8•• | 2.4 + 0.1 |
| RAF1 | −5.6 + 1.8•• | −3.6 + 3.1 | SCD | −1.2 + 2.3••• | −1.1 + 0.5 |
| EIF2AK2 | −5.5 + 0.7••• | −1.9 + 0.7 | VPS4A | −1.1 + 1.3••• | −1.4 + 1.1 |
| PSMA7 | −5.4 + 1.7•• | −1.3 + 0.8 | AKT1 | 1.0 + 0.1••• | 1.7 + 0.5 |
| SRCAP | −4.9 + 1.4••• | −1.4 + 0.5 | SSB | 1.0 + 0.3••• | −2.0 |
| PTBP1 | −4.7 + 1.6••• | −2.1 + 1.0 | CDK2 | 1.1 + 0.5••• | 1.3 + 0.6 |
| GAPDH | −4.6 + 0.5••• | −2.2 + 0.3 | ISG15 | 1.1 + 0.2•• | −1.2 + 0.5 |
| EIF4E | −4.4 + 0.8••• | 3.6 + 1.3 | SCARB1 | 1.1•• | 2.7 + 1.5 |
| VPS35 | −4.4 + 1.1••• | −2.3 + 0.4 | USP18 | 1.1 + 0.2•• | 3.5 + 2.4 |
| RANBP5 | −4.4 + 1.1••• | −4.1 + 0.8 | PRMT1 | 1.2 + 0.6••• | 1.9 + 0.1 |
| HNRPC | −3.9 + 0.9••• | −3.0 + 0.2 | PCBP1 | 1.2 + 0.6••• | −2.3 + 0.1 |
| ACTN1 | −3.9 + 0.7•• | −4.0 + 1.6 | RPL3 | 1.3 + 0.0••• | 2.3 + 0.8 |
| RELA | −3.4 + 0.8•• | −3.5 + 0.7 | PRMT5 | 1.3 + 0.0••• | 1.8 + 0.5 |
| MAPK1 | −3.2••• | 2.0 + 0.8 | LSM1 | 1.3 + 0.6••• | −7.5 + 1.9 |
| PCBP2§ | −3.1•••, 3.4•• | −1.8, 1.8 | ISGF3G | 1.5•• | 2.7 + 3.0 |
| RPL22 | −2.9 + 0.3•• | −6.4 + 0.3 | PRKACA | 1.5••• | 2.9 + 0.1 |
| PDPK1 | −2.8 + 0.6••• | −4.2 + 2.2 | DNAJC14 | 1.5 + 1.1••• | 1.6 + 0.2 |
| AHSA1 | −2.5 + 0.5••• | −3.2 + 1.0 | HSPCA | 1.7 + 0.4•• | −1.6 + 0.4 |
| CDK6 | −2.4 + 1.0•• | 2.1 + 1.2 | ADI1 | 2.0 + 1.1••• | 1.2 + 1.0 |
| SEC11L1 | −2.4 + 0.4••• | −2.5 + 0.7 | SNX1 | 2.4 + 1.3•• | 2.9 + 0.1 |
| CANX | −2.2 + 0.7•• | −1.4 + 0.1 | ATF6 | 6.8•• | 3.8 + 0.4 |
Dots represent number of days between siRNA treatment and initial infection that produced the most extreme viral phenotype.
*Names refer to the gene name of the siRNA target (HUGO nomenclature).
†Values represent fold change in HCV levels plus SEM in specific siRNA-treated cells compared with controls. Values are based on the geometric mean of two replicate experiments. A negative value reflects a decrease in relative HCV levels.
‡VAP-ABC siRNAs target VAPA, VAPB, and VAPC.
§PCBP2 siRNAs produce an early increase in HCV levels, followed by a decrease.
A few trends are apparent from the data. First, changes in HCV infectious virus production generally parallel the changes in HCV RNA levels for most siRNAs. This correlation is expected for defects in a stage of the virus life before the packaging and release of infectious virus. Second, there is no correlation between the effect of siRNAs on HCV replication and cell viability, suggesting that most phenotypes do not result from changes in cellular physiology. In total, 26 genes modulate HCV replication by the following criteria: (i) Introduction of siRNAs targeting the genes alter infectious HCV production by >3-fold and (ii) a decrease in target gene RNA accumulation after siRNA treatment. These genes and their regions of HCV interaction are shown in SI Fig. 6.
RNAi Is Required for Efficient HCV Infection.
We extended these studies for one of the 26 “hits” from the initial screen, the RNAi enzyme Dicer. Dicer was included in this screen, because a number of groups postulated that RNAi might be an innate antiviral defense in mammals (21). We initially hypothesized that if RNAi were antiviral against HCV, then silencing Dicer would increase HCV replication. However, we observed the opposite phenotype: silencing Dicer inhibited HCV replication ≈7-fold. Conflicting data have been published on this topic using HCV genotype 1 replicons. The microRNA miR-122 was reported to be required for HCV replication (17); however, Dicer siRNAs, which would interfere with miR-122 biogenesis, were subsequently reported to enhance HCV subgenomic replication (22). We decided to reexamine the role of RNAi in HCV replication by using the infectious HCV genotype 2a isolate.
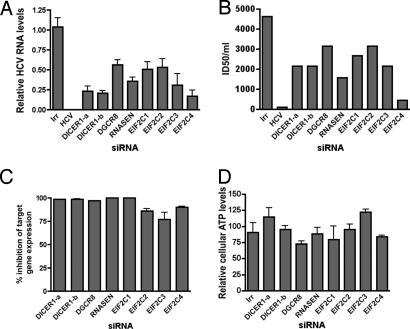
We investigated the role of RNAi in the HCV life cycle by silencing multiple components of the RNAi pathway and testing the effects on HCV replication and infectious virus production as above. Additionally, different siRNAs that target distinct Dicer sequences were used to minimize the possibility of nonspecific off-target effects that can be associated with RNAi experiments. All siRNAs tested reduced target RNA accumulation by at least 80% with no appreciable effects on cell viability (Fig. 1 C and D). We found that siRNAs targeting genes involved in miRNA biogenesis (DICER1, Drosha/RNASEN, and DGCR8) and the RISC effector complex (EIF2C1–4) inhibited HCV replication (Fig. 1 A and B).
Fig. 1.
Silencing RNAi machinery inhibits HCV replication. siRNAs targeting genes in the RNAi pathway, HCV, or an irrelevant sequence (IRR) were individually introduced into Huh-7.5 cells, which were subsequently infected with HCV. (A) Relative HCV RNA levels were quantified by real-time RT-PCR for HCV and 18S RNA. Values are averages of two sets of triplicates ± SEM. (B) Infectious HCV in cellular supernatants were quantified by limiting dilution and expressed as ID50/ml. (C) Percent inhibition of target RNA levels after siRNA treatment compared with irrelevant treated samples. Values are the levels of target RNA/GAPDH RNA. (D) Cell viability (ATP levels) after siRNA treatment at the time of virus harvesting. ATP levels are normalized to the median of treated samples.
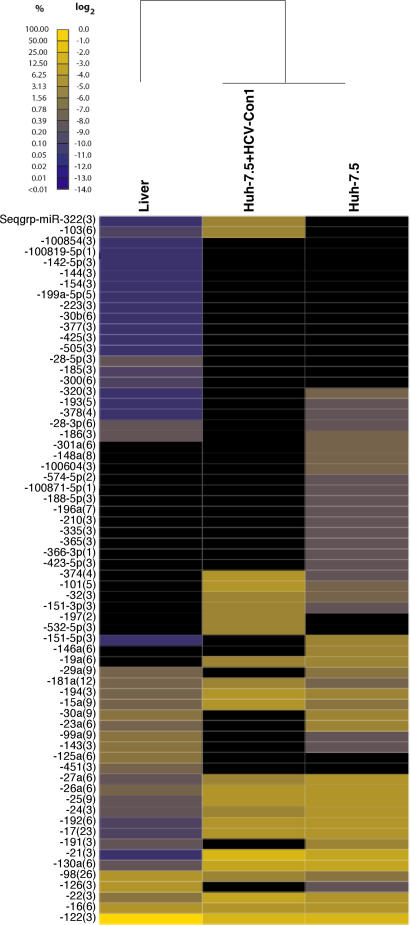
We next examined the miRNA environment associated with HCV infection to identify miRNAs that are expressed within the environment of HCV replication and to test whether HCV replication interfered with miRNA biogenesis. Total RNA from human liver, Huh-7.5 cells, and Huh-7.5 cells containing replicating genomic HCV-Con1 was electrophoretically separated and ≈21-nt RNAs were gel-purified, cloned, sequenced, and annotated. The relative abundances of cloned miRNAs are plotted in Fig. 2. Seventy-one distinct miRNAs, including four previously unidentified miRNAs (miR-100604, -100819, -100854, and -100871; SI Table 6) were isolated in liver cells. Strikingly, miR-122 was the most frequently cloned miRNA, representing 72% of total miRNAs. It was also the most highly expressed miRNA in Huh-7.5 cells and Huh-7.5 + HCV cells, representing 23% and 15% of the cloned miRNAs, respectively.
Fig. 2.
Relative miRNA expression profile of liver, Huh-7.5 cell line, and Huh-7.5 with replicating genomic HCV-Con1. miRNAs of human, mouse, and rat were aligned in sequence alignments, and sequence groups were built (specified in SI Table 7). The number of miRNAs in one sequence group is indicated in brackets. The clone count of sequence groups relative to all miRNA clones obtained from liver and Huh7 cell lines is depicted in the specified color code. Tissues were hierarchically clustered based on the miRNA profiles. miRNA expression patterns in the hepatocellular carcinoma cell lines cluster together and separately from liver, as indicated by lines at the top.
Some differences between Huh-7.5 cells that harbor replicating HCV and the parental Huh-7.5 cell line with respect to the expression of low-abundance miRNAs include higher expression of the miR-322, 197, 532–5p, and 374 miRNA families and the absence of miR-146a, 30a, 23a, and 191 families in cells with replicating HCV. The miRNA expression patterns match with previous Northern blot analysis, showing that the levels of miR-122, -21, and -130 are unaffected by HCV replication (13). Thus, HCV replication does not grossly affect miRNA biogenesis.
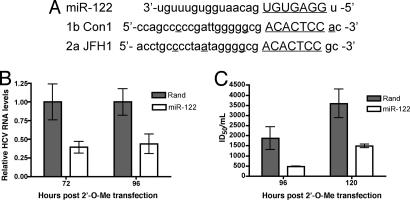
A recent study implicated miR-122 in the modulation of HCV replication by using genotype 1 replicons (17). The miR-122-binding site is conserved between genotype 1 and 2a, suggesting that miR-122 might have a similar function in the production of infectious genotype 2a HCV (Fig. 3A). We tested this possibility by transfecting 2′-O-methyl oligos targeting either a random sequence or the miR-122 sequence into Huh-7.5 cells and measuring the effects on HCV replication. miR-122 depletion consistently inhibited HCV replication and infectious virus production over a number of time points (Fig. 3 B and C). These data demonstrate that the requirement for miR-122 in HCV replication is conserved between genotype 1b replicons and infectious genotype 2a HCV. Thus, the requirement of the RNAi pathway for infectious HCV production likely reflects a need for miR-122 expression.
Fig. 3.
Interfering with miR-122 inhibits HCV replication. (A) Alignment of miR-122-binding sites in HCV 5′ NTR of genotype 1b Con1 and genotype 2a JFH-1. (B and C) 2′-O-methylated RNAs targeting a random sequence (Rand) or miR-122 (miR-122) were introduced into Huh7.5 cells, infected with HCV, and harvested at the indicated time points, and HCV RNA (B) and infectious virus (C) was quantified. Values are averages of triplicates ± SEM. P value < 0.001.
Discussion
Host genes modulate viral infection and are an underappreciated target for antiviral therapy. We identified 26 human genes that modulate HCV replication and implicated the RNAi pathway itself as a key regulator of HCV replication. Significant host proteins that interact with the structural genes include CD81, the dead box helicase DDX3X, the cellular proteases signal peptidase (SEC11L1), and signal peptide peptidase (HM13). CD81 is a tetraspanin that promotes HCV entry via its interaction with HCV E2. DDX3X binds to the core protein; its role in the HCV life cycle is currently unknown (23). DED1, a yeast homolog of DDX3X, is also required for the replication of brome mosaic virus, another positive-strand RNA virus (24). The functional conservation of DDX3X among diverse viruses and the degree of HCV inhibition after the silencing of DDX3X suggest that DDX3X plays a significant role in HCV replication. Signal peptidase proteolyses the HCV structural genes, whereas signal peptide peptidase further proteolyses the core protein into the mature core (25).
Cellular proteins that interact with HCV nonstructural genes include AFT6, a component of the unfolded protein response (UPR), which is induced by NS4B (26). The UPR is a cellular pathway that is activated by various stresses to the endoplasmic reticulum, including HCV replication. Interestingly, the silencing of ATF6 results in an increase in HCV replication and virus production and is also associated with increased cell viability. This suggests that the UPR becomes induced in our experiments, and that the ATF6 pathway limits viral replication. NS5A-interacting proteins that modulate HCV replication include VAP-A/B and VPS35, which function in vesicular trafficking (27–30). VAPA can bind to NS5B as well as hypophosphorylated NS5A and has been proposed to serve as a scaffold for the replication complex within lipid microdomains (31–34). The cytoskeletal component ACTN1 also binds to NS5B and may be involved in replicase localization. PDPK1, RAF1, and EIF2AK2 are kinases that bind to NS5A. It is unclear whether these kinases phosphorylate NS5A or alternatively impact HCV replication indirectly through their various (i.e., Ras/MAPK) signaling pathways. Similarly, GRB2 binds to NS5A and is a key component of RAS signaling. The roles of RANBP5, SRCAP, and AHSA1 in HCV replication remain unclear.
Some proteins identified in our screen bind to HCV RNA sequences that are associated with translation and/or RNA synthesis. EIF2S3 binds to the 5′ nontranslated region (5′ NTR) of HCV and has been reported to modulate HCV translation (35). PTBP1 binds to both 5′ and 3′ NTRs, suggesting a possible interaction between the 5′ and 3′ termini of HCV RNA (35–37). RPL22 and GAPDH interact specifically with the 3′ NTR (38). PTBP1, ELAVL1, and HNRNPC bind to the 3′ termini of both (+) and (−) strands, suggesting they may be components of the RNA synthesis initiation complexes (39, 40).
Cellular stress response pathways also modulate HCV replication, including the UPR, dsRNA activation of IFN signaling (PKR/EIF2AK2), oxidative stress (NF-κB/RELA, STAT3), and heat shock (Hsp70/HSPA1A). Numerous groups have observed the activation of these pathways during HCV replication, so it is likely that these pathways are responding, at least in part, to HCV infection (41).
RNAi silencing of VAPA, VAPB, PTBP1, SSB (La), EIF2B3, ELAVL1, PSMA7, and RAF1 inhibit replication of HCV genotype 1 replicons (32, 42–44). Our data with infectious genotype 2a HCV correlated with that published for each of these genes. SSB (La) and EIF2B3 siRNAs displayed a small degree of inhibition of HCV RNA accumulation, whereas the remaining genes are listed as “hits” in this screen. These are likely to play important roles in HCV replication, given that similar results were found by different groups using different siRNA sequences, silencing approaches, HCV genotypes, and replication systems.
In this paper, the RNAi pathway was demonstrated to be required for optimal HCV replication. The function of miR-122 in HCV replication, however, remains unclear. Genetic evidence exists for an HCV-specific function of miR-122. HCV RNAs that carry a mutation within the 5′ NTR of the miR-122-binding site do not replicate but can be rescued by expression of a miR-122 variant containing a complementing mutation (17). miR-122 did not appear to greatly influence HCV translation in the replicon system, suggesting that miR-122 may regulate viral RNA synthesis or the trafficking of viral RNAs to subcellular compartments. Alternatively, HCV may interfere with the capacity of miR-122 to silence its endogenous substrates. In support of this hypothesis, HCV replication is associated with an increase in expression of cholesterol biosynthesis genes that are regulated by miR-122 (45–48).
The conserved expression of distinct miRNAs was identified by miRNA profiling of liver and Huh-7.5 cells, the most abundant of these being miR-122. miR-122 is expressed at high levels in Huh-7.5 cells but not in other transformed liver cells, correlating with susceptibility of the cell type to HCV replication (17, 49). However, HCV can be adapted to replicate in a number of cell types that do not express detectable miR-122, suggesting that miR-122 may not be required for HCV replication in different cellular environments (50). Although we cannot rule out a role for other identified miRNAs in HCV replication, the sheer abundance of miR-122 in liver (72% of total miRNAs) suggests that this interaction predominates.
Mounting evidence suggests that RNAi is not a robust antiviral pathway in mammals (51). HCV core protein overexpressed outside of its genomic context was reported to inhibit Dicer. However, we previously showed that replicating HCV did not inhibit RNAi, either with siRNAs or shRNAs targeting viral or cellular RNAs, or by altering the expression of endogenous miRNAs (13, 22). If RNAi were indeed an antiviral pathway against HCV, one would expect that HCV replication would activate the RNAi machinery and small viral siRNAs would be generated. To test this hypothesis, we identified ≈2,000 small RNAs from Huh-7.5 cells containing actively replicating HCV; however, no HCV siRNAs were identified (13). Thus, although replicating HCV was shown to be susceptible to transfected HCV-specific siRNAs and not inhibitory to RNAi, replicating HCV did not appear to be susceptible to endogenous RNAi. Similar studies with other viruses revealed a surprising dependence on the RNAi machinery: HCMV, HHV8, MHV68, SV40, and HIV encode viral miRNAs (13, 52–57).
Previous studies of subgenomic HCV replicons yielded conflicting data; miR-122 was required for viral replication, yet Dicer was reported to inhibit the replication of transfected HCV RNAs (17, 22). This suggested that the RNAi machinery may have two interactions with HCV, a microRNA interaction and an antiviral siRNA interaction. These data indicate that the requirement of functional RNAi for replication is dominant over any antiviral activity this pathway may exert against HCV. These results also implicate the RNAi machinery as a potential antiviral target to limit HCV replication.
Materials and Methods
Cells and Virus.
Huh-7.5 cells are a subline derived from Huh-7 hepatoma cells that are highly permissive for the initiation of HCV replication (58). Huh-7.5 cells containing the HCV-Con1 replicon [Con1/Fl-neo(S2204I)] were used for miRNA profiling (59). Cells were maintained in DMEM supplemented with nonessential amino acids and 10% FBS, whereas replicon cell media also contained 0.75 mg/ml G418. HCV FL-J6/JFH is a full-length genotype 2a sequence that produces the full replication cycle in cell culture (9). It is a chimera containing the JFH-1 5′ nontranslated region (60), the J6 core through NS2 genes (61), and the JFH-1 NS3 through 3′ nontranslated region.
RNAi Assay.
The primary screen included 116 siRNAs that were designed to target 58 genes (two siRNAs per gene), whereas four other host genes were targeted by gene-specific siRNA smart pools (Dharmacon, Lafayette, CO; SI Table 4). RNAi assays were performed as described (14). Briefly, 2.5 × 106 Huh7 cells in 0.4 ml of PBS pH 7.4 were electroporated with 1 nanomole of siRNA for five pulses of 870 V for 99 μs with 1-s intervals on a BTX (Holliston, MA) 820 electroporator. Cells were plated and infected at 24, 48, or 72 h after electroporation with a multiplicity of 0.5 infectious HCV particles per cell for 6 h, rinsed with media, then maintained for 2 days at 37°C. Rand and miR-122 2′O-methyl RNAs were described previously (17).
HCV Quantification.
Virus titers were determined by limiting dilution analysis as described (9). Viral RNA was quantified by real-time RT-PCR analysis as described (62). Cellular RNAs were quantified by using ABI TaqMan assays, as recommended (Applied Biosystems, Foster City, CA). For SYBR green assays of cellular genes, 0.75 μg of DNaseI-treated total RNA was reverse-transcribed with SuperScript II (Invitrogen, Carlsbad, CA) and oligo(dT) for 1 h at 42°C, then heat-inactivated at 80°C for 20 min. One-twentieth of the cDNA mix was mixed with an equal volume of 2× SYBR Green Master Mix (Applied Biosystems) and the appropriate primers. PCR conditions were: 50°C, 2 min; 95°C, 10 min (95°C, 15 sec; 55°C, 1 min) × 40 cycles (63). Results were analyzed with SDS 1.7 software (Applied Biosystems). Relative HCV RNA levels are the quantity of HCV normalized to 18S RNA levels. Relative cellular gene RNA levels are the quantity of the specific gene RNA normalized to GAPDH RNA levels.
NS5A Two-Hybrid Analysis.
The LexA DNA-binding domain (pEG-202) was fused to the NS5A gene from the genotype 1a H77 strain. It was used to screen a HeLa cDNA library that is fused with a B42 acidic transcriptional activation domain (pJG4/5). Plasmids were transformed into the Saccharomyces cerevisiae strain EGY48, and interactors were assayed for growth in minimal dropout media lacking uracil, histidine, tryptophan, and LacZ expression. Nine cDNAs encoding three unique proteins that specifically interact with NS5A were identified from a library of 108 HeLa cDNA clones.
NS5A Kinase Complex Isolation.
The mammalian expression and isolation of GST and GST-NS5A was performed as described, with minor modifications (19). Briefly, BHK21 cells in 150-mm dish were infected with 10 pfu per cell of vTF7–3 virus for 1 h in PBS plus 1% FBS, washed, then transfected with 170 μl of lipofectamine (Invitrogen), and 30 μg of plasmid DNA in 5 ml of Opti-MEM. After 24 h, cells were lysed in 5 ml of NETN buffer (50 mM Tris•HCl, pH 7.5/120 mM NaCl/1 mM EDTA/0.5% Nonidet P-40) supplemented with 5 mM DTT/1 μg/ml aprotinin/1 μg/ml leupeptin/20 μg/ml PMSF. Lysates were clarified at 16,000 × g for 10 min at 4°C and incubated with 500 μl of a 1:1 slurry of glutathione agarose in lysis buffer for 30 min. Agarose beads were briefly centrifuged at 3,000 × g, washed, then resuspended in a final volume of 250 μl of kinase buffer (50 mM Tris•HCl, pH 7.5/5 mM MnCl2/5 mM DTT). NS5A-associated kinase activity was assayed as described (19). Agarose beads with associated proteins were then resuspended in SDS/PAGE sample buffer, heated to 95°C for 5 min, and centrifuged, then supernatants were removed and sent to Kinexus for the KPKS kinase screen.
Total RNA Isolation, Small RNA Library Preparation, Sequencing, and Sequence Annotation.
Small RNA cloning was performed as described (64) with the following modifications. Samples were cloned by using preadenylated 3′ adapter oligonucleotides and T4 RNA ligase Rnl2 as described (13) (indicated by A, SI Table 7). Radiolabeled 19- and 24-nt oligonucleotide size markers containing a PmeI site were added to total RNA before gel electrophoresis. 3′-Adapter ligation products were gel-purified and ligated to a 5′-oligonucleotide by using T4 RNA ligase 1 (NEB) plus ATP. After reverse transcription and a first PCR amplification step, the PCR product was digested with PmeI to eliminate the size markers. After a second PCR step, which introduced nonpalindromic restriction sites, the PCR product was digested with BanI, concatemerized by using T4-DNA ligase, and ligated into pCR2.1 (Invitrogen). Colonies were screened by PCR for the presence and size of inserts and sequenced.
Annotation used GenBank (www.ncbi.nih.gov/GenBank/index.html), a human tRNA database (http://lowelab.ucsc.edu/GtRNAdb), sn/snoRNA databases (www-snorna.biotoul.fr/index.php, http://noncode.bioinfo.org.cn, and www.imb.uq.edu.au), a miRNA database (http://microrna.sanger.ac.uk/sequences/index.shtml), the repeat element annotation of version 17 of the human genome assembly, version 6 of the mouse genome assembly, and version 3 of the rat genome assembly from http://genome.cse.ucsc.edu.
Supplementary Material
Acknowledgments
Two-hybrid system reagents were provided by Roger Brent. We thank Kathleen Hefferon for critical reading and editing of the manuscript. This work was funded by the Public Health Service (National Institutes of Health Grants CA57973, CA85883, and AI40034), the Ellison Medical Foundation, and the Greenberg Medical Research Institute (C.M.R). C.M.R. is an Ellison Medical Foundation Senior Scholar in Global Infectious Diseases. G.R. is supported by the American Cancer Society (Grant PF-02-016-01-MBC), the National Institute of Diabetes and Digestive and Kidney Diseases/Digestive Disease Research Core Center (Grant P30 DK42086), and Susan and David Sherman. T.L.T. was supported by the Charles Revson Foundation for Biomedical Research and the National Institutes of Health/National Institute of Allergy and Infectious Diseases Ruth L. Kirschstein National Research Service award (5F32 AI51820-03). B.D.L. is supported by the National Cancer Institute Howard Temin Award (CA107092).
Abbreviations
- HCV
hepatitis C virus
- UPR
unfolded protein response.
Footnotes
Conflict of interest statement: C.M.R. has equity in Apath, LLC, which has a commercial license for the Huh-7.5 cell line and certain HCVcc derivatives.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704894104/DC1.
References
- 1.Wkly Epidemiol Rec. 1997;72:341–344. Anonymous. [PubMed] [Google Scholar]
- 2.Tellinghuisen TL, Rice CM. Curr Opin Microbiol. 2002;5:419–427. doi: 10.1016/s1369-5274(02)00341-7. [DOI] [PubMed] [Google Scholar]
- 3.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 4.Blight KJ, Kolykhalov AA, Rice CM. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 5.Bartosch B, Dubuisson J, Cosset FL. J Exp Med. 2003;197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. Proc Natl Acad Sci USA. 2003;100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, et al. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 10.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset FL. J Biol Chem. 2003;278:41624–41630. doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- 11.Cormier EG, Tsamis F, Kajumo F, Durso RJ, Gardner JP, Dragic T. Proc Natl Acad Sci USA. 2004;101:7270–7274. doi: 10.1073/pnas.0402253101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Randall G, Higginbottom A, Monk P, Rice CM, McKeating JA. J Virol. 2004;78:1448–1455. doi: 10.1128/JVI.78.3.1448-1455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 14.Randall G, Grakoui A, Rice CM. Proc Natl Acad Sci USA. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Randall G, Rice CM. Virus Res. 2004;102:19–25. doi: 10.1016/j.virusres.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 16.Randall G. Hepatology. 2005;41:1220–1222. doi: 10.1002/hep.20731. [DOI] [PubMed] [Google Scholar]
- 17.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 18.Mendelsohn AR, Brent R. Curr Opin Biotechnol. 1994;5:482–486. doi: 10.1016/0958-1669(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 19.Reed KE, Xu J, Rice CM. J Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coito C, Diamond DL, Neddermann P, Korth MJ, Katze MG. J Virol. 2004;78:3502–3513. doi: 10.1128/JVI.78.7.3502-3513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gitlin L, Andino R. J Virol. 2003;77:7159–7165. doi: 10.1128/JVI.77.13.7159-7165.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Kato N, Jazag A, Dharel N, Otsuka M, Taniguchi H, Kawabe T, Omata M. Gastroenterology. 2006;130:883–892. doi: 10.1053/j.gastro.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 23.Owsianka AM, Patel AH. Virology. 1999;257:330–340. doi: 10.1006/viro.1999.9659. [DOI] [PubMed] [Google Scholar]
- 24.Noueiry AO, Chen J, Ahlquist P. Proc Natl Acad Sci USA. 2000;97:12985–12990. doi: 10.1073/pnas.240460897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussy P, Langen H, Mous J, Jacobsen H. Virology. 1996;224:93–104. doi: 10.1006/viro.1996.0510. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Y, Gao B, Ye L, Kong L, Jing W, Yang X, Wu Z, Ye L. J Microbiol. 2005;43:529–536. [PubMed] [Google Scholar]
- 27.Haft CR, de la Luz Sierra M, Bafford R, Lesniak MA, Barr VA, Taylor SI. Mol Biol Cell. 2000;11:4105–4116. doi: 10.1091/mbc.11.12.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar AJ, Polak JM. Biochem Biophys Res Commun. 2000;277:622–630. doi: 10.1006/bbrc.2000.3727. [DOI] [PubMed] [Google Scholar]
- 29.Zhang P, Yu L, Gao J, Fu Q, Dai F, Zhao Y, Zheng L, Zhao S. Genomics. 2000;70:253–257. doi: 10.1006/geno.2000.6380. [DOI] [PubMed] [Google Scholar]
- 30.Weir ML, Klip A, Trimble WS. Biochem J. 1998;333:247–251. doi: 10.1042/bj3330247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans MJ, Rice CM, Goff SP. Proc Natl Acad Sci USA. 2004;101:13038–13043. doi: 10.1073/pnas.0405152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamamoto I, Nishimura Y, Okamoto T, Aizaki H, Liu M, Mori Y, Abe T, Suzuki T, Lai MM, Miyamura T, Moriishi K, Matsuura Y. J Virol. 2005;79:13473–13482. doi: 10.1128/JVI.79.21.13473-13482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao L, Aizaki H, He JW, Lai MM. J Virol. 2004;78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tu H, Gao L, Shi ST, Taylor DR, Yang T, Mircheff AK, Wen Y, Gorbalenya AE, Hwang SB, Lai MM. Virology. 1999;263:30–41. doi: 10.1006/viro.1999.9893. [DOI] [PubMed] [Google Scholar]
- 35.Kruger M, Beger C, Li QX, Welch PJ, Tritz R, Leavitt M, Barber JR, Wong-Staal F. Proc Natl Acad Sci USA. 2000;97:8566–8571. doi: 10.1073/pnas.97.15.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ali N, Siddiqui A. J Virol. 1995;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito T, Lai MM. Virology. 1999;254:288–296. doi: 10.1006/viro.1998.9541. [DOI] [PubMed] [Google Scholar]
- 38.Petrik J, Parker H, Alexander GJ. J Gen Virol. 1999;80:3109–3113. doi: 10.1099/0022-1317-80-12-3109. [DOI] [PubMed] [Google Scholar]
- 39.Spangberg K, Wiklund L, Schwartz S. Virology. 2000;274:378–390. doi: 10.1006/viro.2000.0461. [DOI] [PubMed] [Google Scholar]
- 40.Ito T, Lai MM. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tardif KD, Waris G, Siddiqui A. Trends Microbiol. 2005;13:159–163. doi: 10.1016/j.tim.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Yamada O, Sakamoto T, Yoshida H, Iwai T, Matsushita Y, Shimamura H, Araki H, Shimotohno K. Virology. 2004;320:135–143. doi: 10.1016/j.virol.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 43.Korf M, Jarczak D, Beger C, Manns MP, Kruger M. J Hepatol. 2005;43:225–234. doi: 10.1016/j.jhep.2005.02.046. [DOI] [PubMed] [Google Scholar]
- 44.Burckstummer T, Kriegs M, Lupberger J, Pauli EK, Schmittel S, Hildt E. FEBS Lett. 2006;580:575–580. doi: 10.1016/j.febslet.2005.12.071. [DOI] [PubMed] [Google Scholar]
- 45.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, Chisari FV. Proc Natl Acad Sci USA. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kapadia SB, Chisari FV. Proc Natl Acad Sci USA. 2005;102:2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 49.Chang J, Nicholas E., Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 50.Zhu Q, Guo JT, Seeger C. J Virol. 2003;77:9204–9210. doi: 10.1128/JVI.77.17.9204-9210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cullen BR. Nat Immunol. 2006;7:563–567. doi: 10.1038/ni1352. [DOI] [PubMed] [Google Scholar]
- 52.Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. Nature. 2006;442:82–85. doi: 10.1038/nature04836. [DOI] [PubMed] [Google Scholar]
- 53.Samols MA, Hu J, Skalsky RL, Renne R. J Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 55.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Proc Natl Acad Sci USA. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bennasser Y, Le SY, Yeung ML, Jeang KT. Retrovirology. 2004;1:43. doi: 10.1186/1742-4690-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 58.Blight KJ, McKeating JA, Rice CM. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blight KJ, McKeating JA, Marcotrigiano J, Rice CM. J Virol. 2003;77:3181–3190. doi: 10.1128/JVI.77.5.3181-3190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. Gastroenterology. 2003;125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 61.Yanagi M, Purcell RH, Emerson SU, Bukh J. Virology. 1999;262:250–263. doi: 10.1006/viro.1999.9889. [DOI] [PubMed] [Google Scholar]
- 62.Randall G, Chen L, Panis M, Fischer AK, Lindenbach BD, Sun J, Heathcote J, Rice CM, Edwards AM, McGilvray ID. Gastroenterology. 2006;131:1584–1591. doi: 10.1053/j.gastro.2006.08.043. [DOI] [PubMed] [Google Scholar]
- 63.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 64.Peffer S, Lagos-Quintana M, Tuschl T. Cloning of Small RNA Molecules. New York: Wiley; 2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.