Abstract
A surprisingly large population of mRNAs has been shown to localize to sensory axons, but few RNA-binding proteins have been detected in these axons. These axonal mRNAs include several potential binding targets for the La RNA chaperone protein. La is transported into axonal processes in both culture and peripheral nerve. Interestingly, La is posttranslationally modified in sensory neurons by sumoylation. In axons, small ubiquitin-like modifying polypeptides (SUMO)-La interacts with dynein, whereas native La interacts with kinesin. Lysine 41 is required for sumoylation, and sumoylation-incompetent LaK41R shows only anterograde transport, whereas WT La shows both anterograde and retrograde transport in axons. Thus, sumoylation of La determines the directionality of its transport within the axonal compartment, with SUMO-La likely recycling to the cell body.
Keywords: axonal transport, La/SSB, RNA localization, small ubiquitin-like modifying polypeptide
Localized protein synthesis in neuronal processes requires appropriate targeting of mRNAs, ribosomes, and translation factors to axons and dendrites (1). mRNA targeting can be highly selective, and specific mechanisms have evolved to package mRNAs for transport. Neuronal mRNAs are transported in granules that contain RNA-binding proteins and components of the translation machinery (2). Although the mRNAs from these granules are obviously used as templates for the translational machinery in neuronal processes, the fate of anterogradely transported RNA-binding proteins remains largely unknown.
The RNA-binding protein La was initially identified as an autoantigen in Sjögren's syndrome and systemic lupus erythematosus (3). This abundant protein has a well conserved N terminus that contains the “La motif” and an RNA recognition motif (RRM1) (4). Mammalian La contains a second RRM, RRM2. Much of the cellular La resides within the nucleus, where it interacts with RNA polymerase III transcripts assisting in RNA processing (3). La can shuttle between the nucleus and cytoplasm (5), and nuclear La moves to the cytoplasm upon cellular stress (6, 7). Cytoplasmic La can enhance viral RNA translation through internal ribosome entry sites (IRES), and it plays a role in IRES-dependent translation of cellular mRNAs (8–10). La also binds to and facilitates translation of other mRNAs containing a 5′-UTR terminal oligopyrimidine (TOP) element (11). Although La is thought to be ubiquitous, neuronal expression and localization have not been addressed. We have recently shown that adult rat sensory axons contain many different mRNAs, including the La target grp78/BiP (12). Here, we show that sensory axons contain several TOP mRNAs that are known targets for La. La is transported into axons, associating with kinesin during its anterograde transport. Axonal La is covalently modified by addition of small ubiquitin-like modifying polypeptides (SUMO). Sumoylated La (SUMO-La) binds to dynein, and the sumoylation at K41 is required for retrograde transport.
Results
La and Cellular mRNA Targets of La Extend into Regenerating Axons.
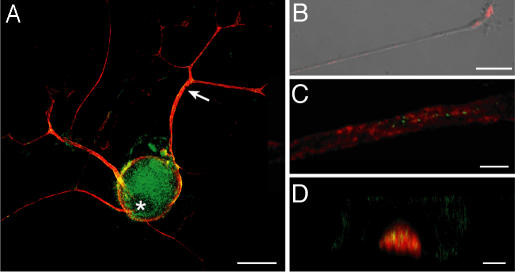
Several mRNAs encoding ribosomal proteins (RP) that contain 5′ terminal oligopyrimidine elements extend into axonal processes of dorsal root ganglion (DRG) neurons [supporting information (SI) Fig. 6]. Because La is known to bind to RP mRNAs in other cellular systems (13), we tested whether La is expressed in sensory neurons and whether, like the RP mRNAs, it localizes to distal neuronal processes. In DRG neurons, both nuclear and cytoplasmic La was visualized with mouse anti-La antibody (clone 44) (Fig. 1A) and human La antiserum (SI Fig. 6A). In the axonal processes that DRG neurons extend in vitro (14), La signals appeared rather granular (Fig. 1B and SI Fig. 7B). Similar granular signals were seen in axons of injured sciatic nerve (Fig. 1 C and D). The Schwann cells in these cultures were also immunoreactive for La, showing mostly nuclear signals (Fig. 1A).
Fig. 1.
La autoantigen extends into regenerating sensory axons. (A) Cultures of injury-conditioned DRGs were colabeled with mouse anti-La (green) and chick antineurofilament (NF) antibodies (red). La is seen in the cell body, including the nucleus, and axonal processes (arrow). (B) In a single xy image of a distal axon, mouse anti-La (red) merged with DIC shows a granular La immunoreactivity. (C and D) Sections of sciatic nerve at ≈1 cm proximal to the crush site (7 d after injury) were colabeled for mouse anti-La (green) and chick anti-NF (red). C shows a single xy plane taken through the center of the axon. D shows a single xz plane demonstrating intraaxonal La with less-aggregated signals in the surrounding Schwann cell cytoplasm. (Scale bars: A, 40 μm; B, 20 μm; C and D, 5 μm.)
La Is Sumoylated in Sensory Axons.
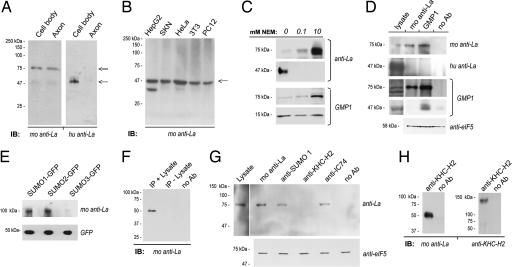
Immunoblotting was used to determine whether the axonal La antigenicity conformed to the expected molecular mass of rat La. The coding sequence of rat La predicts a protein of 47 kDa, which is consistent with molecular mass of human La (15). The mouse anti-La antibody detected a prominent band migrating at 75 kDa in DRG lysates (Fig. 2A). Although a band corresponding to the expected molecular mass of rat La was present, particularly for the whole DRG lysates vs. axons, this was overall a minor component. Reprobing DRG blots with human anti-La showed only a 47-kDa band (Fig. 2A). The mouse anti-La recognized a 47-kDa band in lysates prepared from nonneuronal cells using the same radioimmunoprecipitation assay (RIPA) buffer supplemented with a standard protease inhibitor mixture (Fig. 2B).
Fig. 2.
Neuronal La is sumoylated. (A) In immunoblots of cell body and axonal lysates from DRG cultures, immunoblotting with mouse anti-La shows a strong band migrating at ≈75 kDa (double arrow) and a faint band at ≈47 kDa (single arrow), whereas blotting with human anti-La only detects the lower band. (B) In blots prepared from HepG2, SKN, HeLa, NIH 3T3, and nondifferentiated PC12 cells lysed in standard RIPA buffer, mouse anti-La only detected a 47-kDa band (arrow). (C) Axoplasm from crushed sciatic nerve prepared in nondenaturing buffer supplemented NEM was used for immunoblotting. The mouse anti-La detected increasing amounts of 75-kDa La and decreasing amounts of 47-kDa La with increasing NEM concentration. The human anti-La showed a similar decrease in the 47-kDa La. Anti-SUMO1 (GMP1) showed increasing levels of a 75-kDa band with increasing NEM concentration. There was a slight decrease in the 17 kDa corresponding to free SUMO1 with increasing NEM concentration. (D) Axoplasm isolated in the presence of 10 mM NEM was used for immunoprecipitation with mouse and human anti-La or GMP1, as indicated. Mouse anti-La detects a 75-kDa band precipitated by GMP1. GMP1 recognized a sumoylated protein migrating at 75 kDa in the La precipitates, but nothing was detected at 47 kDa. The human anti-La did not recognize bands in the mouse anti-La or GMP1 precipitates. Supernatant from the precipitations probed with anti-eIF5 showed approximately equivalent levels of input protein for these samples. (E) Expression of SUMO-GFPs in DRG cultures followed by precipitation with rabbit anti-GFP shows an ≈100-kDa band for the SUMO1- and SUMO2-GFP-expressing cells when probed with mouse anti-La. The anti-La detects a faint band at ≈100 kDa in the SUMO3-GFP-expressing cells. Probing these blots with anti-GFP shows relatively equivalent expression of free SUMO-GFP. (F) Mouse anti-La immunoprecipitations were processed with or without lysate to test the specificity of the Trueblot reagent, which was used for G and H, for native vs. denatured mouse IgG. The latter would be present in both preparations, but antigen would be present only in the plus-lysate sample. Even with extended exposure, bands corresponding to La were detected only in the IP plus lysate sample. (G) DRG cultures were lysed in RIPA buffer supplemented with 10 mM NEM and used for immunoprecipitation with mouse anti-La, GMP1, anti-KHC-H2 (kinesin), and anti-IC74 (dynein) followed by immunoblotting for mouse anti-La. Seventy-five-kilodalton La-reactive bands (double arrow) are seen in the SUMO1 and dynein precipitates but not in the kinesin precipitates. Supernatant from the precipitations probed with anti-eIF5 showed approximately equivalent levels of input protein for these samples. (H) Axoplasm prepared in the absence of NEM and processed for precipitation with KHC2-H2 antibody. By immunoblotting, only 47-kDa La coprecipitates with kinesin heavy chain; reprobing with the anti-KHC2-H2 shows successful precipitation of kinesin heavy chain. Experiments performed in the presence of 10 mM NEM showed identical results (data not shown).
The slower mobility of La in DRG lysates could result from posttranslational modification(s). La can be phosphorylated (16), but the number of phosphates needed for a 20-kDa shift would substantially alter La's pI, and such was not seen (SI Fig. 8). Covalent addition of SUMO has been reported to shift the molecular mass of substrates by 20–30 kDa on SDS/PAGE (17). SUMO modifications are typically lost in cell lysates through constitutive activity of the Ulp cysteine proteases (17). Thus, for sumoylation to account for the molecular mass shift of La, SUMO-La must be particularly stable in cultured DRGs compared with other cellular preparations. To test this possibility, we asked whether the ratio of 47 to 75 kDa La could be altered in cell preparations with both 47- and 75-kDa La bands by treating with the Ulp protease inhibitor, N-ethylmaleimide (NEM). Axoplasm from sciatic nerve showed both 47- and 75-kDa La with standard protease inhibitors (Fig. 2C). Addition of increasing amounts of NEM decreased prevalence of the 47-kDa La and increased prevalence of the 75-kDa La. Probing the NEM-treated axoplasm with an antibody to SUMO1 (GMP1) showed multiple bands ranging from 17 to 150 kDa (data not shown). There was a prominent GMP1-reactive band at 75 kDa whose prevalence increased with increasing NEM concentration (Fig. 2C). To determine whether the 75-kDa GMP1-reactive band included SUMO-La, mouse anti-La and GMP1 antibodies were used for immunoprecipitation from axoplasm prepared in the presence of 10 mM NEM. The 75-kDa band precipitated by mouse anti-La was recognized by GMP1, and a 75-kDa band immunoreactive for mouse anti-La was detected in the GMP1 precipitates (Fig. 2D). Furthermore, mouse anti-La also recognized a single band in anti-GFP precipitates from PC12 cells transfected with SUMO-GFP constructs (Fig. 2E). This ≈100-kDa band approximates the molecular mass of SUMO-La plus GFP. These data argue that covalent addition of SUMO1 accounts for La's increased molecular mass in the neuronal preparations.
As shown in Fig. 2A, human anti-La did not appear to recognize the 75-kDa form of La. To test the possibility that this antiserum is selective for native La, PC12 cells were lysed in 20 μM NEM to generate approximately equal amounts of 47- and 75-kDa La (SI Fig. 7C). Probing these PC12 lysates with human anti-La showed only the 47-kDa band. Similarly, mouse anti-La recognized only a 47-kDa band in human anti-La precipitates. Finally, the human anti-La also did not recognize any bands in the GMP1 immunoprecipitates (Fig. 2D). Together, these data indicate this human anti-La is selective for native La.
Sumoylated La Binds Dynein but Not Kinesin.
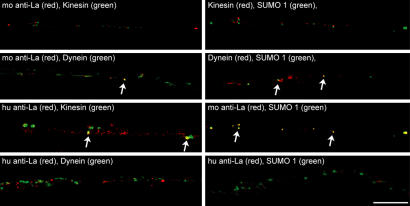
Sumoylation has been shown to target cytoplasmic proteins for nuclear localization (18). To test whether axonal SUMO-La is targeted for retrograde transport, we asked whether La shows any binding, directly or indirectly, to motor proteins. For this, anti-La, GMP1, anti-KHC-H2 (kinesin), and anti-IC74 (dynein) immunoprecipitates from DRG lysates were immunoblotted with mouse anti-La. Because this required using mouse antibodies both for precipitation and blotting, the Trueblot detection reagent (eBioscience, San Diego, CA) was used to avoid any overlapping signals from IgG heavy chain. Immunoprecipitation with and without any added lysate was used to test for specificity of this secondary antibody system for native mouse IgG. The Trueblot consistently did not detect the denatured mouse IgG (Fig. 2F). For coimmunoprecipitations, mouse anti-La detected a 75-kDa band in dynein and SUMO1 precipitates but not in kinesin precipitates (Fig. 2G). On the other hand, the human anti-La detected a 47-kDa La band in kinesin precipitates (Fig. 2H). By immunofluorescence, SUMO1 extended into axons of cultured DRGs (SI Fig. 9). Axonal SUMO1 and La also showed differential colocalization with motor proteins. Signals for the mouse anti-La overlapped with intraaxonal signals for dynein (Fig. 3). Although colocalization of the mouse anti-La signals with those for kinesin was less obvious, the human anti-La showed no dynein colocalization but did overlap with kinesin. SUMO1 immunoreactivity overlapped with dynein but not kinesin. The axonal SUMO1 also colocalized with mouse anti-La signals but showed no colocalization with human anti-La signals. Taken together, these data indicate that SUMO-La binds to dynein but not kinesin, and native La binds to kinesin but not dynein.
Fig. 3.
Regenerating axons contain SUMO that colocalizes with La and dynein. DRG cultures were colabeled with xenon-labeled GMP1, mouse anti-La, anti-KHC-H2, and anti-IC74, as indicated. Signals for human anti-La were detected by indirect immunofluorescence. Single xy planes of midaxon shaft of 24-h injury-conditioned DRG cultures are shown. Note that signals detected by the mouse anti-La show focal colocalization with dynein and SUMO1 (arrows). Conversely, the signals detected by the human anti-La, which does recognize SUMO-La, show focal colocalization with kinesin (arrows) but no overlap with SUMO1 or dynein. Signals for SUMO1 show focal colocalization with dynein (arrows) but no overlap with kinesin. (Scale bar: 10 μm.)
Sumoylation of La Is Required for Retrograde but Not Anterograde Transport.
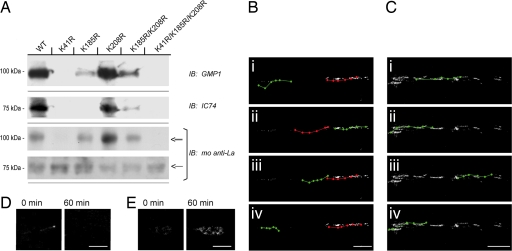
Human and rat La proteins are >80% homologous with the highest homology in the N-terminal half that contains the La motif and RRM1. SUMO plot analysis of La (www.expasy.org/tools) showed high-probability sumoylation sites (ΨKXE) in the La motif (K41), RRM1 (K185), and between RRM1 and RRM2 (K208). Lysine-to-arginine mutants were generated in a human La-GFP fusion construct (huLa-GFPWT) at K41, K185, and K208 (huLa-GFPK41R, huLa-GFPK185R, huLa-GFPK208R, huLa-GFPK185R/K208R, and huLa-GFPK41R/K185R/K208R) to determine whether human La is also sumoylated, and whether sumoylation affects its trafficking. Only huLa-GFP with intact K41 residue showed any discernable sumoylation (Fig. 4A). Neither the huLa-GFPK41R or huLa-GFPK41R/K185R/K208R mutants were recognized by anti-SUMO1. huLa-GFPK185R and huLa-GFPK185R/K208R showed decreased sumoylation compared with huLa-GFPWT and huLa-GFPK208R. No discernable dynein signal was detected in the huLa-GFPK41R or huLaGFPK41R/K185R/K208R immunoprecipitates. huLa-GFPK185R and huLa-GFPK185R/K208R showed decreased levels of coprecipitating dynein compared with the huLa-GFPWT and huLa-GFPK208R (Fig. 4A). Blotting with anti-La confirmed that huLa-GFP with intact K41 showed higher signals for the ≈100-kDa band than for the ≈75-kDa, corresponding the approximate molecular masses of SUMO-huLa-GFP and huLa-GFP, respectively. These data argue that K41 is required for La's sumoylation and interaction with dynein. K185 also can contribute to sumoylation competency and dynein interaction but less than that seen with K41.
Fig. 4.
Lysine 41 is critical for sumoylation and retrograde transport of La. (A and B) Naive DRG cultures were transfected with WT and mutant huLa-GFP, as indicated. Anti-GFP immunoprecipitates were used for immunoblotting. Sumoylated and native huLa-GFP, detected by the mouse anti-La, could be discerned by distinct migration (double and single arrows, respectively). GMP1 blots showed a prominent band in the huLa-GFPWT and huLa-GFPK208R with fainter bands detected in the huLa-GFPK185R and huLa-GFPK185R/K208R. These samples also showed more abundant SUMO-huLaGFP (≈100 kDa) than native huLa-GFP (≈75 kDa) when probed with the anti-La. The huLa-GFPK41R and huLa-GFPK41R/K185R/K208R mutants showed no detectable signals for SUMO1 and showed more abundant native huLa-GFP than SUMO-huLa-GFP based on molecular mass. A similar pattern was seen for coprecipitating dynein detected with the IC74 antibody (i.e., huLa-GFPWT, huLa-GFPK208R > huLa-GFPK185R, huLa-GFPK185R/K208R > huLa-GFPK41R, and huLa-GFPK41R/K185R/K208R). (B and C) DRG neurons transfected with huLa-GFPWT (B) and huLa-GFPK41R (C) were used for live cell imaging. Images were collected over a 60-min period and i–iv display movement of individual GFP aggregates over this course (anterograde, green; retrograde, red). Note that huLa-GFPWT shows both anterograde and retrograde motility, but the huLa-GFPK41R mutant shows only ograde motility (also see SI Movies 1 and 2). (D and E) Exposure-matched images of growth cones sampled at time 0 and 60 min for huLa-GFPWT (D) and huLa-GFPK41R (E) are shown. Note that only huLa-GFPK41R mutant accumulates in the growth cone. (Scale bars: B and C, 10 μm; D and E, 5 μm.)
We used live-cell imaging to more directly test the role of sumoylation in trafficking of La. The huLa-GFPWT extended into axons and appeared granular with bidirectional movement (Fig. 4B and SI Movie 1). Anterogradely moving huLa-GFPWT aggregates progressed at an average speed of 0.21 ± 0.04 μm/sec, whereas retrograde movement showed an average speed of 0.39 ± 0.08 μm/sec (P ≤ 0.05 for anterograde vs. retrograde). On the other hand, huLa-GFPK41R showed only anterograde movement in axons with an average speed 0.28 ± 0.02 μm/sec (Fig. 4C and SI Movie 2). Together, these studies argue that sumoylation of La is required for its retrograde transport. Many of the huLa-GFPWT particles periodically stalled over the observation period, but this appeared to be limited to particles moving anterogradely. Twenty-three percent (SD 5.8) of anterogradely moving huLa-GFPWT particles showed some stalling along the axon shaft, with stalled period extending for 36% (SD 5.6) of the observation period. No stalling was observed for huLa-GFPK41R. Interestingly, huLa-GFPK41R became concentrated in distal processes of the DRG neurons, but huLa-GFPWT did not (Fig. 4 D and E).
Sumoylated La Is Retrogradely Transported in Sciatic Nerve Axons.
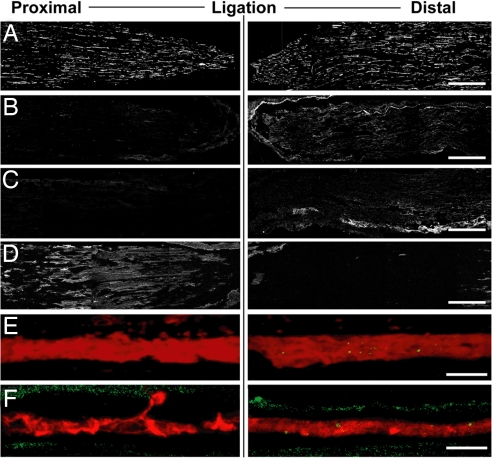
Nerve ligation was used to determine whether SUMO and La are retrogradely transported in vivo. Ligation successfully impeded both anterograde and retrograde axonal transport with accumulation of kinesin proximal to the ligation (Fig. 5 A and D). La was more intense in the nerve segments distal to the ligation (Fig. 5B); at high magnification, La was concentrated in the axons distal to the ligation (Fig. 5E). GMP1 signals were also stronger distal to the ligation (Fig. 5C); at high magnification, SUMO1 was seen in axons and Schwann cells, but the intraaxonal signals were consistently limited to segments distal to the ligation (Fig. 5F).
Fig. 5.
Sumoylated La accumulates distal to sciatic nerve ligation. Sections of ligated crushed sciatic nerve (3 d after injury and ligation) were processed for immunostaining and analyzed by confocal microscopy. Nerve segment proximal to the ligation is displayed on Left, and segment distal to the ligation is displayed on Right; crush site is Right. Montages of four free maximum projections are illustrated in each image of A–D (each projection was generated from 10 optical planes taken at 0.9 μm). E and F represent single optical planes through the central region of one representative axon from the corresponding regions. All image pairs are matched for laser power, photomultiplier tube voltage, offset, and processing. A displays the NF signal. Nerve sections stained with the mouse anti-La show a more prominent signal distal to the ligation site [B; also see merged NF (red) and anti-La (green) in E]. Staining with the GMP1 antibody showed a similar accumulation of SUMO1 distal to the ligation [C; also see merged NF (red) and GMP1 (green) in F]. Kinesin, detected by anti-KHC-H2 antibody, accumulated proximal to the lesion site (D). (Scale bars: A–D, 500 μm; E and F, 10 μm.)
Discussion
Only a few RNA-binding proteins are known to localize to axons. Our studies indicate that the La RNA chaperone shows both anterograde and retrograde transport in sensory axons. mRNA targets for La are also transported into these neuronal processes, including grp78/BiP and RP mRNAs (9, 12). The sumoylation of La we have shown here may provide a means for targeting La for nuclear transport from distal cytoplasmic locations, because only SUMO-La interacted with dynein. With the long distances that mRNAs and RNA-binding proteins must travel in neurons, sumoylation may provide a means to recycle some proteins to the nucleus by targeting these proteins for retrograde transport.
Sumoylation has recently been demonstrated for a few RNA-binding proteins (19–22). Sumoylation of hnRNP C decreases its nucleic acid binding (21). Sumoylation at K41 could affect La's interaction with target mRNAs, because this residue lies within the La motif, and deletions of the La motif alter La's affinity for target RNAs (3). In addition to interrupting its interaction with mRNAs, K41 sumoylation could alter La's interaction with small noncoding RNAs. La recognizes a general 3′UUU-OH motif in RNA polymerase III transcripts including tRNAs, 7S RNA of the signal recognition peptide (SRP), and Y RNA. RNA polymerase II transcripts that contain UUU-OH motifs (e.g., telomerase mRNA) are also targets for recognition by La (4). Although not demonstrated to date, axons must contain tRNAs for translation to occur. Given that locally synthesized EphB2 is inserted into membranes of developing axons (23), and some resident ER proteins are synthesized in DRG axons (12), SRP's 7S RNA is also likely transported into axons. Interestingly, recent findings in peripheral neuropathies suggest that some tRNA synthetases localize to axons (24, 25).
SUMO modifications have often gone undetected, because the SUMO-cleaving enzymes, Ulps, are thought to rapidly desumoylate conjugates during cell lysis. Further, only a small fraction of a given protein is thought to be sumoylated at any one time (17). Compared with cultures of nonneuronal cells, SUMO-La predominated over native La in DRG cultures. SUMO-La also appeared more abundant in the sciatic nerve axoplasm than in nonneuronal cells. Decreased activity of SUMO proteases may account for the abundance of SUMO-La in these preparations. Maintaining SUMO ligases and SUMO proteases in different compartments of neurons (e.g., distal axons vs. cell body) would be one means to restrict SUMO protease activity in these highly polarized cells. Although we cannot directly compare results from DRG cultures to the axoplasm preparations, it is curious that SUMO-La is so prevalent in the DRG cultures. The selective stalling of anterogradely transported huLa-GFPWT compared with sumoylation incompetent huLa-GFPK41R and retrogradely transported huLa-GFPWT may indicate that the balance of SUMO ligases and proteases in the cultured neurons favors sumoylation. Consistent with this, overexpression of Ubc9 ligase and SUMO in PC12 and U2-OS cells increases the overall prevalence of SUMO-La (data not shown).
Many known sumoylated proteins localize to the nucleus or nuclear pore (26). In mammals, nuclear SUMO substrates are often involved in transcriptional regulation, with sumoylation repressing their transactivating activities (17, 27). For example, the MEF2A transcription factor is sumoylated in developing cerebellar granular cells in an activity-dependent fashion (28). Although La shows both cytoplasmic and nuclear localization in the DRG neurons, SUMO1 and La did not colocalize in the nucleus, arguing that the predominant site of sumoylation of neuronal La is in the cytoplasm. In other cellular systems, sumoylation is needed for some cytoplasmic proteins to transit the nuclear pore (18). Thus, the axonal SUMO-La may be destined for delivery to the nucleus.
Neurons present a unique situation for transporting proteins from cytoplasm to the nucleus, because the distances separating distal cytoplasmic components are by far greater than in any other cell type. Discrete microtubule-based mechanisms have evolved to deliver macromolecules and vesicles over these long distances in neurons. For most proteins that show bidirectional transport, it is not clear what the signal is for changing their directionality (29). Sumoylation of La is evidence that covalent addition of small proteins can determine directionality of microtubule-based transport. It will be interesting to determine whether sumoylation destines other proteins for retrograde transport.
Materials and Methods
Nerve Crush and DRG Culture.
All animal experiments were approved by the Institutional Animal Care and Use Committee. Neurons were cultured from L4–6 DRGs of adult Sprague–Dawley rats. For injury conditioning, the sciatic nerve was crushed at midthigh level 7 d before culture (30). DRG dissociation and culture were performed as described (12). For the injury-conditioned DRG cultures, RNA synthesis was inhibited by using 80 μM 5,6-dichlorobenzimidazole riboside (30). The sciatic nerve was used for isolation of axoplasm or fixed in 4% paraformaldehyde and processed for cryosectioning. For sciatic nerve ligations, nerve was crushed twice at midthigh, and the proximal segment was ligated ≈1 cm from the crush site using 4·0 sutures. After 72 h, the ligated nerve was fixed in 4% paraformaldehyde and processed for cryosectioning.
For transfection of sensory neurons, dissociated naive DRGs were resuspended in electroporation buffer [123 mM NaCl/20 mM Hepes (pH 7.05)/5 mM KCl/0.7 mM Na2HPO4/6 mM glucose). One microgram of plasmid DNA was added, and cells were immediately placed on ice. Electroporation was done by using the ECM 830 device (BTX, Holliston, MA) set for one 200-ms pulse at 450 V in the “LV mode.” Cells were then transferred to DMEM/F12, 10% HS, 1% N1, and 3 mM EGTA and placed at 37°C, 5% CO2 for 1.5 h. Cells were centrifuged at 160 × g for 5 min, resuspended in standard media, and plated as above. Media was changed at 4 h after plating and twice daily thereafter. PC12 cells were electroporated with 3 μg of plasmid DNA by using the Amaxa (Gaithersburg, MD) Nucleofector per the manufacturer's instructions.
cDNA Expression Constructs.
huLa-GFP expression construct has been described (31). The SUMO-GFP constructs were provided by Ronald Hay (University of Dundee, Dundee, U.K.). Site-directed mutagenesis of huLa-GFP construct was performed by using the Quickchange kit (Stratagene, Cedar Creek, TX). Double and triple mutants were generated by sequential mutagenesis. All clones were verified by DNA sequencing.
Isolation of Axons.
Isolation of axonal and cell body preparations was performed as described (12). Purity of axonal preparations was tested by RT-PCR (see below).
mRNA Analyses.
RNA was extracted from the axonal and cell body preparations of DRG cultures by using RNAqueous Micro Kit (Ambion, Austin, TX). All RNA preparations were quantified by fluorimetry with RiboGreen (Molecular Probes, Eugene, OR). RT-PCR was performed as described (12), except that iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) was used for reverse transcription (RT). The RT reactions were diluted 10-fold and used for transcript-specific PCR with AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA). β-Actin mRNA amplification was used as a positive control. γ-Actin and MAP2 mRNA amplifications were used to check for any contamination of the axonal preparation with cell body or nonneuronal contents (12).
Immunofluorescence.
All steps were performed at room temperature unless otherwise indicated. For cultures, coverslips were rinsed in warm PBS and then fixed in methanol for 5 min at −20°C. Fixed coverslips and cryosections were rinsed in PBS, permeabilized in PBS containing 0.2% Triton X-100 for 15 min, and then blocked for 1 h in PBS containing 5% donkey and 5% rabbit sera. For standard immunolabeling, samples were incubated overnight at 4°C with primary antibodies diluted in blocking buffer. The following antibodies were used: chicken anti-NFH (1:2000; Chemicon, Temecula, CA), human anti-La (1:100; Immunovision, Springdale, AR; lot 4170), Clone 44 mouse anti-La (1:100; Transduction Laboratories, Lexington, KY), GMP1 mouse anti-SUMO1 (1:100; Zymed, San Francisco, CA), rabbit antiperipherin (1:500; Chemicon), mouse anti-IC74 (1:100; Chemicon), and mouse anti-KHC-H2 (1:100) (32). Cultures were rinsed in PBS and incubated for 1 h in secondary antibodies diluted in blocking buffer. The following secondary antibodies were used: FITC-conjugated donkey anti-chicken, TR-conjugated donkey anti-human, Cy5-conjugated donkey anti-rabbit, FITC-conjugated donkey anti-rabbit, and TR-conjugated donkey anti-mouse (Jackson ImmunoResearch, West Grove, PA).
For colabeling with multiple mouse antibodies, the Zenon Labeling kit was used to directly conjugate Alexafluor 488, 555, or 647 to mouse IgGs (Molecular Probes). Labeled IgGs were combined into a single staining solution and used for 1-h labeling (final concentration, 1 μg/ml). Samples were then rinsed with PBS and postfixed in 4% paraformaldehyde for 15 min. All samples were mounted by using Vectashield (Vector Laboratories, Burlingame, CA) with DAPI. Images were analyzed with a Leica (Exton, PA) TCS/SP2 microscope equipped with galvanometer stage.
Protein Isolation and Immunoprecipitations.
Protein lysates were prepared from cultures by lysis in RIPA buffer (0.1% SDS/50 mM Tris·Cl, pH 6.8/150 mM NaCl/0.5% Nonidet P-40/2 mM EDTA) supplemented with Protease Inhibitor mixture (Sigma, St. Louis, MO) for 20 min at 4°C. For samples treated with NEM, cells were rinsed in PBS plus NEM and then lysed in RIPA plus NEM. Lysates were cleared by centrifugation at 16,000 × g for 15 min, 4°C. Axoplasm was prepared as described by squeezing unfixed segments of nerve in ice-cold NTB (33). Protein content was normalized by Bradford assay (Bio-Rad). For immunoprecipitation, 500 μg of protein was mixed with 1 μg of primary antibody and incubated overnight at 4°C as indicated. Immunocomplexes were precipitated by using 35 μl of Protein A/G+ agarose (Santa Cruz Biotechnology, Santa Cruz, CA). After 1.5 h at 4°C, immunocomplexes were collected by centrifugation at 1,000 × g for 5 min. Pellets were washed five times with RIPA buffer (±NEM), resuspended in Laemmli sample buffer (Bio-Rad), and then boiled for 8 min.
Protein Electrophoresis and Immunoblotting.
For standard electrophoresis, denatured lysates or immunoprecipitates were resolved by 1D SDS/PAGE. For 2D gels, immunoprecipitates were precipitated overnight at −20°C with acetone and then resuspended in 8 M urea/2% CHAPS/50 mM DTT. Proteins were passively absorbed into immobilized pH gradient gel strips (pH 7–10; Amersham, Piscataway, NJ) by overnight rehydration. Strips were focused by 6- to 10-h linear ramp to 30,000 V/h. For the second dimension, strips were equilibrated in 6 M urea/0.375 M Tris·Cl (pH 8.8)/2% SDS/20% glycerol/2% DTT followed by the same buffer plus 2.5% iodoacetamide. Strips were then resolved by SDS/PAGE. All gels were immobilized onto PVDF membrane (Millipore, Billerica, WA) by electrophoretic transfer.
For immunoblotting, membranes were rinsed in TBS with 0.1% Tween 20 (TBST) and then blocked in TBST/5% nonfat dry milk. Blots were incubated overnight at 4°C in the following antibodies: mouse anti-La (1:500; Transduction Laboratories), human anti-La (1:500), mouse GMP1 anti-SUMO1 (1:500; Zymed), rabbit anti-SUMO1 (1:250; Abcam, Cambridge, MA), mouse anti-IC74 (1:500; Chemicon), mouse anti-KHC-H2 (1:500), mouse anti-GFP (1:2,000; Roche, New York, NY), and rabbit anti-GFP (1:1,000; Abcam). Blots were rinsed several times in TBS and then incubated with for 1 h with anti-mouse IgG Trueblot (1:1,000, eBioscience), HRP-conjugated anti-mouse (1:5,000; Cell Signaling Technology, Boston, MA), or HRP-conjugated anti-human IgGs (1:400; Jackson ImmunoResearch) diluted in the blocking buffer. Blots were washed for 30 min in TBS and developed with ECLplus (Amersham).
Time-Lapse Imaging.
DRG cultures transfected with the huLa-GFP cDNAs were used for time-lapse imaging 3 d after transfection. Imaging was performed at 37°C in an environmental chamber on TCS/SP2 confocal microscope. Z stacks were recorded at 5-min intervals to avoid bleaching the GFP signals. GFP fluorescent granules were tracked by using the Volocity Classification software package (Improvision, Lexington, MA). Objects were tracked through the image sequence of a 3D volume of three xy planes taken at 1 μm in the z axis. The centroid position of at least 10 mobile objects in midaxon shaft was tracked in each of three separate live-cell imaging experiments. The normalized centroid position of each tracked object was then used to determine distance traveled over time and time stalled.
Acknowledgments
We are grateful to Dr. Kirk Czymmek for assistance with live cell imaging and Dr. Ronald T. Hay for providing GFP-SUMO expression plasmids. This work was supported by National Institutes of Health Grant NS041596 (to J.L.T.), U.S.–Israeli Binational Science Foundation Grant 2003–2008 (to J.L.T.), Christopher Reeve Foundation Grant TB2-0602-2 (to J.L.T.), the Dr. Miriam and Sheldon Adelson Medical Research Foundation, the Nemours Foundation, and Deutsche Forschungsgemeinschaft Grant DFG HE 2814/3-2.
Abbreviations
- DRG
dorsal root ganglion
- huLa-GFP
human La-GFP
- NF
neurofilament
- RP
ribosomal protein
- RRM
RNA recognition motif
- SUMO
small ubiquitin-like modifying polypeptides
- SUMO-La
sumoylated La protein
- RIPA
radioimmunoprecipitation assay
- NEM
N-ethylmaleimide.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611562104/DC1.
References
- 1.Twiss JL, van Minnen J. J Neurotrauma. 2006;23:295–308. doi: 10.1089/neu.2006.23.295. [DOI] [PubMed] [Google Scholar]
- 2.Hirokawa N. J Neurosci. 2006;26:7139–7142. doi: 10.1523/JNEUROSCI.1821-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolin SL, Cedervall T. Annu Rev Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 4.Maraia RJ, Bayfield MA. Mol Cell. 2006;21:149–152. doi: 10.1016/j.molcel.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Fok V, Friend K, Steitz JA. J Cell Biol. 2006;173:319–325. doi: 10.1083/jcb.200601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachmann M, Chang S, Slor H, Kukulies J, Muller WE. Exp Cell Res. 1990;191:171–180. doi: 10.1016/0014-4827(90)90002-r. [DOI] [PubMed] [Google Scholar]
- 7.Baboonian C, Venables PJ, Booth J, Williams DG, Roffe LM, Maini RN. Clin Exp Immunol. 1989;78:454–459. [PMC free article] [PubMed] [Google Scholar]
- 8.Holcik M, Korneluk RG. Mol Cell Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YK, Back SH, Rho J, Lee SH, Jang SK. Nucleic Acids Res. 2001;29:5009–5016. doi: 10.1093/nar/29.24.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trotta R, Vignudelli T, Candini O, Intine RV, Pecorari L, Guerzoni C, Santilli G, Byrom MW, Goldoni S, Ford LP, et al. Cancer Cell. 2003;3:145–160. doi: 10.1016/s1535-6108(03)00020-5. [DOI] [PubMed] [Google Scholar]
- 11.Cardinali B, Carissimi C, Gravina P, Pierandrei-Amaldi P. J Biol Chem. 2003;278:35145–35151. doi: 10.1074/jbc.M300722200. [DOI] [PubMed] [Google Scholar]
- 12.Willis DE, Li KW, Zheng J-Q, Smit AB, Kelly TK, Merianda TT, Sylvester J, van Minnen J, Twiss JL. J Neurosci. 2005;25:778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crosio C, Boyl PP, Loreni F, Pierandrei-Amaldi P, Amaldi F. Nucleic Acids Res. 2000;28:2927–2934. doi: 10.1093/nar/28.15.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng J-Q, Kelly T, Chang B, Ryazantsev S, Rajasekaran A, Martin K, Twiss J. J Neurosci. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semsei I, Troster H, Bartsch H, Schwemmle M, Igloi G, Bachmann M. Gene. 1993;126:265–268. doi: 10.1016/0378-1119(93)90378-g. [DOI] [PubMed] [Google Scholar]
- 16.Broekhuis CH, Neubauer G, van der Heijden A, Mann M, Proud CG, van Venrooij WJ, Pruijn GJ. Biochemistry. 2000;39:3023–3033. doi: 10.1021/bi992308c. [DOI] [PubMed] [Google Scholar]
- 17.Johnson ES. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 18.Hay RT. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Li T, Evdokimov E, Shen RF, Chao CC, Tekle E, Wang T, Stadtman ER, Yang DC, Chock PB. Proc Natl Acad Sci USA. 2004;101:8551–8556. doi: 10.1073/pnas.0402889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, Mann M, Lamond AI. J Biol Chem. 2004;279:33791–33798. doi: 10.1074/jbc.M404201200. [DOI] [PubMed] [Google Scholar]
- 21.Vassileva MT, Matunis MJ. Mol Cell Biol. 2004;24:3623–3632. doi: 10.1128/MCB.24.9.3623-3632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellis DJ, Dehm SM, Bonham K. Gene. 2006;379:68–78. doi: 10.1016/j.gene.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 23.Brittis PA, Lu Q, Flanagan JG. Cell. 2002;110:223–235. doi: 10.1016/s0092-8674(02)00813-9. [DOI] [PubMed] [Google Scholar]
- 24.Seburn KL, Nangle LA, Cox GA, Schimmel P, Burgess RW. Neuron. 2006;51:715–726. doi: 10.1016/j.neuron.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 25.Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, et al. Nat Genet. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 26.Gill G. Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 27.Gill G. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 29.Vale RD. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 30.Smith DS, Skene P. J Neurosci. 1997;17:646–658. doi: 10.1523/JNEUROSCI.17-02-00646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horke S, Reumann K, Schweizer M, Will H, Heise T. J Biol Chem. 2004;279:26563–26570. doi: 10.1074/jbc.M401017200. [DOI] [PubMed] [Google Scholar]
- 32.King-Smith C, Bost-Usinger L, Burnside B. Cell Motil Cytoskeleton. 1995;31:66–81. doi: 10.1002/cm.970310108. [DOI] [PubMed] [Google Scholar]
- 33.Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]