Abstract
The influence of donor and recipient KIR genotype on the outcome of hematopoietic cell transplantation between HLA-matched siblings was investigated. Transplants were divided into four groups according to the combination of group A and B KIR haplotypes in the transplant donor and recipient. Overall survival of myeloid patients varied with KIR genotype combination. Best survival was associated with the donor lacking and the recipient having group B KIR haplotypes; poorest survival was associated with the donor having and the recipient lacking group B KIR haplotypes. The latter combination was also associated with increased relapse and acute GVHD. However, its detrimental effects were seen only for transplants where the recipient and donor were homozygous for the C1 KIR ligand and therefore lacked the C2 ligand. Presence of the Bw4 ligand was also associated with increased acute GVHD. In contrast presence of both KIR3DL1 and its cognate Bw4 ligand was associated with decreased non-relapse mortality. Analysis of the KIR genes individually revealed KIR2DS3 as a protective factor for chronic GVHD. The results suggest how simple assessments of KIR genotype might inform the selection of donors for hematopoietic cell transplantation.
Keywords: NK cell, transplantation, haplotype, killer-immunoglobulin-like receptor (KIR), HLA class I
Introduction
Lymphocyte-mediated alloreactions can greatly affect the outcome following hematopoietic cell transplantation (HCT). T cells in the graft can kill residual leukemic cells, thus facilitating engraftment and preventing relapse. Alternatively, they can attack the recipient's tissues causing life-threatening graft-versus-host disease (GVHD). The strength and clinical impact of these T-cell alloreactions correlates with the extent of HLA-disparity between donor and recipient. NK cells can also initiate alloreactions following HCT (reviewed in [1]). The study of T-cell depleted, haploidentical transplantation and HLA-mismatched, unrelated transplantation shows that NK cell mediated alloreactions can confer clinical benefit [2-4]. For these transplants, certain HLA class I differences can activate donor-derived, alloreactive NK cells that improve survival [2] by decreasing relapse and acute GVHD (aGVHD ) [4]. The underlying cause of these alloreactions is the inability of recipient HLA class I molecules to engage inhibitory killer immunoglobulin-like receptors (KIR) expressed by donor-derived NK cells.
The KIR gene family is in the leukocyte receptor complex (LRC) of human chromosome 19 [5, 6]. KIR differ in the number of extracellular immunoglobulin-like domains, which determines ligand-binding specificity, and in the length of the cytoplasmic tail. In general, the long-tailed KIR (designated L) are inhibitory receptors and the short-tailed KIR (designated S) are activating receptors. An exception, KIR2DL4, has potential for both activating and inhibitory function [7-10]. Best characterized are four inhibitory long-tailed KIR with specificity for polymorphic determinants of HLA-A (KIR3DL2) [11, 12], HLA-B (KIR3DL1) [13, 14] and HLA-C (KIR2DL1 and KIR2DL2/3) [15-17]. Whereas a minority of HLA-A and B allotypes function as KIR ligands, every HLA-C allotype is a ligand for either KIR2DL1 or KIR2DL2/3. These two groups of KIR ligands, called C2 and C1, respectively, are distinguished by lysine (C2) or asparagine (C1) at position 80 of HLA-C [18]. Although ligands for the activating short-tailed KIR are poorly defined, weak affinity of KIR2DS1 for C2 and KIR2DS2 for C1 has been reported [19-22].
KIR are expressed by NK cells and subpopulations of γδ and αβ T cells [23]. Within populations of KIR-expressing lymphocytes, individual cells express different numbers and combinations of KIR [24]. This ‘variegated’ expression produces a repertoire of cells having different requirements for activation. In healthy individuals NK cells become tolerant of autologous HLA class I through expression of an inhibitory receptor, usually a KIR or CD94:NKG2A, that engages self HLA class I [24]. NK cell alloreactions, both in vitro and in the transplant recipient, involve NK cell subpopulations expressing inhibitory KIR that cannot engage an HLA class I molecule of the allogeneic target. Expression of the KIR locus is a coordinated process, which starts with KIR2DL4, the only gene to be ubiquitously expressed, and spreads to the other KIR genes [25, 26]. For them the frequency of cellular expression of any one gene is influenced by all the other KIR genes as well as by the expression of CD94:NKG2A and the HLA class I genotype [27]. Consistent with this strictly coordinated regulation of the KIR locus is its organization as a compact array of KIR genes that contains little unique sequence [6].
KIR genes vary from one person to another, and the extent of human KIR diversity rivals that of the HLA genes [28]. Three components contribute to the diversity: KIR haplotypes differ in gene content; KIR genes are polymorphic; and KIR haplotypes associate randomly to form KIR genotypes. Consequently, unrelated individuals rarely have identical KIR genotype and the majority of HCT involves donors and recipients of different KIR genotype [29]. Despite the complexity, KIR haplotypes divide simply into two functionally distinctive groups [28], (reviewed in [30]). Group A haplotypes have a fixed content of seven KIR genes and two pseudogenes, and are diversified through allelic polymorphism. The genes include those specifying inhibitory receptors for each of the four KIR ligands, as well as KIR2DL4, KIR3DL3 (inhibitory receptor of unknown specificity and function) and KIR2DS4 (activating receptor of unknown specificity and function). The group B haplotypes are diversified by both gene content and allelic polymorphism. Although all genes of the group A haplotype are represented in the group B haplotypes, what distinguishes group B haplotypes is presence of a variable number of KIR genes that are not components of the group A haplotypes: KIR2DS1, 2DS2, 2DS3, 2DS5, 3DS1 and 2DL5. Most of the activating KIR are associated only with the group B haplotypes, whereas both group A and group B haplotypes have comparable complements of inhibitory receptors. Thus the group A and B KIR haplotypes provide distinct and complementary functions in the biology of NK cells and T cells.
Evidence for such distinct and complementary functions comes from studies to correlate KIR factors with parameters of human health and disease, in which the observed associations have mostly been with individual or groups of activating KIR (reviewed in [31]). In HCT a variety of KIR associations have been observed. Increased aGVHD was correlated with donor KIR2DS3 in HLA-matched unrelated transplants and with donors having more than four activating KIR in haploidentical transplants [32, 33]. In contrast, DeSantis reported increased overall survival and decreased aGVHD when the donor had more KIR than the recipient [34], a situation in which most of the additional KIR would have been of activating type. More recently, Sun et al correlated increased aGVHD in HLA-matched unrelated transplantation for AML when the recipient had more inhibitory KIR than the donor and/or when the donor had more activating KIR than the recipient [35]. Finally, decreased relapse in HLA-matched transplantation for AML, CML and ALL has been correlated to donors having KIR2DS1 and 2DS2 [36].
Because of the extensive linkage disequilibrium in the KIR locus it is inherently difficult to separate effects due to the individual KIR genes from those of the haplotype [31]. To avoid this issue and to simplify the analysis we examined 202 HLA-matched transplant patients to determine what influence group A and group B KIR haplotypes in the patient and donor, as well as HLA genotype had on outcome parameters such as survival, relapse, non-relapse mortality and GVHD.
Materials and Methods
Patients
Two hundred and two transplants performed at the Stanford University Medical Center between March 1995 and March 2005 were analyzed. Patients received HLA-identical allografts from sibling donors for myeloid leukemia (acute myelogenous leukemia [AML], chronic myelogeneous leukemia [CML], myelodysplastic syndrome [MDS], myeloproliferative disease [MPD] and myelofibrosis), lymphoid leukemia (acute lymphoblastic leukemia [ALL], chronic lymphoblastic leukemia [CLL], non-Hodgkins lymphoma [NHL] and Hodgkins disease [HD]), multiple myeloma (MM), or non-malignant disease (severe aplastic anemia [SAA] and paroxysmal nocturnal hemoglobinuria [PNH]). All but ten patients were in either partial or complete remission at the time of transplant. Patient characteristics are given in Table 1. Clinical information was obtained by reviewing data stored in the Stanford transplantation database and by examination of medical records. All patient samples were collected with written consent from the patients and were approved for use by the Stanford Institutional Review Board.
Table 1.
Characteristics of transplant patients. Abbreviations: AML, acute myelogenous leukemia; CML, chronic myelogenous leukemia; MDS, myelodysplastic syndrome; MPD, myeloproliferative disease; ALL, acute lymphocytic leukemia; CLL, chronic lymphocytic leukemia; NHL, non-Hodgkin's lymphoma. Standard risk disease was defined as patients in CR1 (complete remission) for AML, ALL, CLL, Hodgkin's disease, NHL, and multiple myeloma; CP (chronic phase) for CML; RARS and RA (refractory anemia with and without ringed sideroblasts) for MDS. Advanced risk disease was defined as CR>1 or not in remission for AML and ALL; CP>1, AP (acclerated phase), BP (blast phase) for CML; relapsed or never in remission for CLL; RAEB (refractory anemia with excess blasts) for MDS; CR>1, relapse or never in remission for Hodgkin's disease; PR (partial remission) or chemotherapy resistant for NHL; relapsed or refractory for multiple myeloma; all myelofibrosis patients as only patients with advanced disease were eligible for allogeneic hematopoietic cell transplantation. Myeloablative conditioning regimens included either busulphan plus cyclophosphamide alone or in combination with etoposide; cyclophosphamide and anti-thymocyte globulin; carmustine, etoposide and cyclophosphamide; or fractionated total body irradiation alone or in combination with cyclophosphamide. Reduced intensity conditioning regimens consisted of fludarabine plus total body irradiation, or anti-thymocyte globulin plus total lymphoid irradiation.
| No. of patients n=202 |
|||
|---|---|---|---|
| Disease: | Standard | Advanced | |
| Myeloid leukemia: (n=113) | Risk | Risk | |
| AML | 57 | 34 | 23 |
| CML | 28 | 16 | 12 |
| MDS | 21 | 6 | 15 |
| MPD | 2 | 0 | 2 |
| Myelofibrosis | 5 | 0 | 5 |
| Lymphoid leukemia: (n=61) | |||
| ALL | 21 | 7 | 14 |
| CLL | 6 | 2 | 4 |
| NHL | 33 | 7 | 26 |
| Hodgkin disease | 1 | 0 | 1 |
| Multiple myeloma: | 24 | 23 | 1 |
| Other Diseases: | 4 | N/A | N/A |
| Patient Age: | 45 (18-70) | ||
| Graft product: | |||
| Bone marrow | 23 | ||
| Peripheral blood progenitor cells | 179 | ||
| Conditioning: | |||
| Myeloablative | 126 | ||
| Reduced intensity | 76 |
KIR and HLA genotyping
Genomic DNA from patients and donors was prepared from peripheral blood mononuclear cells prior to transplantation using a QIAamp Blood kit (Qiagen, Valencia, CA). The presence or absence of 11 KIR genes (KIR2DL1, 2DL2, 2DL3, 2DL5, 3DL1, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5 and 3DS1) was determined by PCR-SSP using the method reported by Gomez-Lozano and Vilches [37]. KIR2DL4, 3DL2 and 3DL3 were not analyzed because these genes are present in all individuals [28]. Neither was typing for the KIR2DP1 and 3DP1 pseudogenes performed. For KIR2DS4, full-length alleles were distinguished from alleles containing a 22bp deletion in exon 4, as described by Yawata et al [38].
HLA-A and HLA-B antigens, including Bw4 and Bw6, were determined by PCR-SSP analysis in the Stanford Histocompatibility Laboratory using commercially available reagents. The KIR ligands C1 (HLA-Casn80) and C2 (HLA-Clys80) were discriminated by PCR-SSP using a modification of the method described by Frohn and colleagues [39]. Initial amplification of exons 2 and 3 of HLA-C was performed in a 25μL reaction volume containing 100ng of genomic DNA, Taq polymerase buffer (Invitrogen, Carlsbad, CA), 50 mM MgCl2, 2.5mM each of four deoxyribonucleoside triphosphates, 20 pmol of each primer and 5 units of Taq polymerase (Invitrogen, Carlsbad, CA). The sense and antisense primers were HLA-Cexon2F 5'-AGGCTCCCACTCCATGAGGTA-3' and HLA-Cexon3R 5'-CTTCCCGTTCTCCATGTATC-3', respectively. An initial 5 minute denaturation was followed by ten cycles of 20s denaturation at 95°C; annealing of 45s at 62°C; extension of 120s at 72°C; followed by a final 10 minute extension at 72°C. PCR was carried out in a Perkin Elmer GeneAmp PCR System 9600 thermocycler (Perkin Elmer, Boston, MA). The HLA-C exon 2-3 PCR product was then used as a DNA template for the C1/C2 PCR-SSP amplification [39]. All 202 donor-recipient pairs were HLA-A, B and DR identical, and all but two were identical for the HLA-C allotype.
Statistical Analysis
The endpoints examined in this study included overall survival, defined as time to death from any cause; disease-free survival (DFS), defined as time to relapse, censored at non-cancer death; non-relapse mortality (NRM), defined as time to death from all causes in the absence of relapse; acute graft-versus-host-disease (aGVHD), defined as development of grade II-IV GVHD during the first 100 days post-transplantation; and chronic GVHD (cGVHD), defined as GVHD occurring in patients after d100 post-transplantation. Only patients still alive at d100 and for which cGVHD information had been collected were included in the cGVHD analysis (n=143). All 202 transplant patients were included in analysis of all endpoints examined (overall survival, DFS and NRM) with the exception of aGVHD where information was unavailable for a single myeloid leukemia patient (n=201).
To examine fully the impact of KIR in hematopoietic cell transplants, a hierarchy of hypotheses was examined. We began by examining all 202 transplant pairs, then examined the patients by disease group (myeloid, lymphoid, AML and CML). All the above groups were then examined by conditioning regimen (myeloablative and non-myeloablative, myeloablative only and non-myeloablative only). For each of the above groups, we investigated correlations with the following parameters: KIR matched versus mismatched; KIR haplotype (AA versus Bx); individual activating KIR (2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DS1); number of activating KIR (0-2 versus >3; or 0-3 versus >4); HLA-type (HLA-A, B and C); number of donor and recipient KIR (i.e. where donor had less, equal number or more KIR than the recipient); donor HLA-C type and presence of KIR2DS2 in the recipient; and situations where the door had a KIR for which the recipient had no ligand. We report any correlations with a p-value less than 0.05.
As the number of patients examined in this study is small, no attempts were made to correct the p-values using Bonferroni inequality when multiple comparisons were performed on the same data set. Probability of overall survival, DFS, NRM, aGVHD and cGVHD were calculated by the method of Kaplan-Meier and compared using the log-rank test. When p-values were less than p≤ 0.05, we performed post-hoc analysis using Cox proportional hazard modeling to determine pairwise comparison of curves. The overall type I error rate was controlled by the initial test.
For Cox proportional hazard modeling, both univariate and multivariate stepwise analysis (phreg, SAS software version 9.1) were performed to test the influence of KIR, alone and in combination with different transplant variables (disease stage at transplantation [standard versus advanced risk, as defined in Table 1], patient age at transplantation [older/younger than age 50; older/younger than age 40], pre-transplant conditioning regimen [myeloablation versus reduced intensity conditioning; irradiation alone versus chemotherapeutic agents alone], and donor graft product [bone marrow versus peripheral blood progenitor cells]) on all clinical endpoints. Any correlations with p ≤ 0.05 are reported.
Results
The reported benefit of alloreactive NK cells for haploidentical transplant recipients [2-4] stimulated retrospective immunogenetic analysis in the HLA-mismatched transplant setting, where alloreactions due to KIR-ligand mismatch were expected to occur [32, 33]. Unexpectedly, KIR and HLA factors were also discovered to influence the outcome of HLA-matched transplantation [40, 41]. These studies have usually considered the effects of individual KIR genes. With its compact structure and co-ordinated expression [25, 26], the KIR gene family can be considered to function as a single entity. This hypothesis suggested the alternative approach that we explore here, one in which the effects of KIR haplotype and genotype on transplant outcome are considered. The effects of KIR and HLA genotype were evaluated for 202 patients of mixed disease type who received an HLA-matched transplant from a sibling donor. When correlations with clinical outcome neared or reached significance, the analysis was narrowed to subgroups of patients with particular disease, for example myeloid versus lymphoid malignancies. As such, a hierarchy of hypotheses was examined. Only correlations where p ≤ 0.05 are described.
Donor-recipient KIR genotype combination is a risk factor for transplant outcome
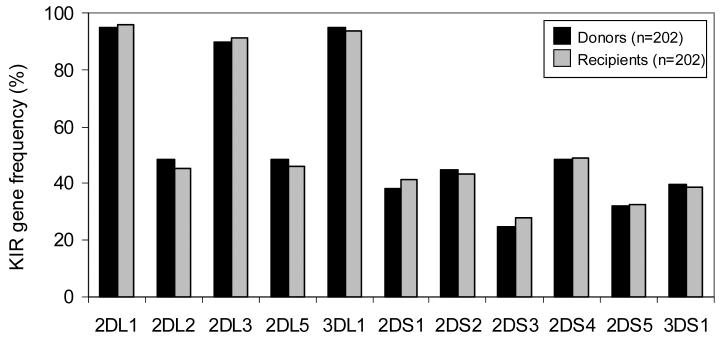
KIR gene content was determined by PCR-SSP typing of 202 transplant recipients and their HLA-matched sibling donors. Comparison of KIR gene frequencies in donors versus recipients revealed no significant differences (Figure 1); as was also seen when defined subgroups of the transplant pairs were similarly analyzed, for example the 113 transplants for myeloid disease (data not shown). For each donor and recipient the KIR gene content was used to infer group A and B KIR haplotypes and to assign each person to one of two genotypes. Individuals having only genes of the group A KIR haplotypes (3DL3, 2DL1, 2DL3, 2DL4, 3DL1, 2DS4 and 3DL2) were considered to be homozygous for group A haplotypes and assigned the KIR genotype AA. All other individuals had one or more B haplotype specific genes (2DS1, 2DS2, 2DS3, 2DS5, 3DS1 and 2DL5) and therefore had either one (AB heterozygotes) or two (BB homozygotes) B haplotypes. Such individuals were assigned the KIR genotype Bx. Approximately 28% of the donors (56/202) and 30% of the recipients (61/202) had the simple AA KIR genotype, the remainder having at least one B haplotype. These percentages are consistent with KIR genotype frequencies observed in other caucasian populations [42-45].
Figure 1.
Donors (n=202) and recipients (n=202) have similar KIR gene frequencies. KIR2DL4, 3DL2 and 3DL3 were not typed for in this panel as these genes are present in all individuals.
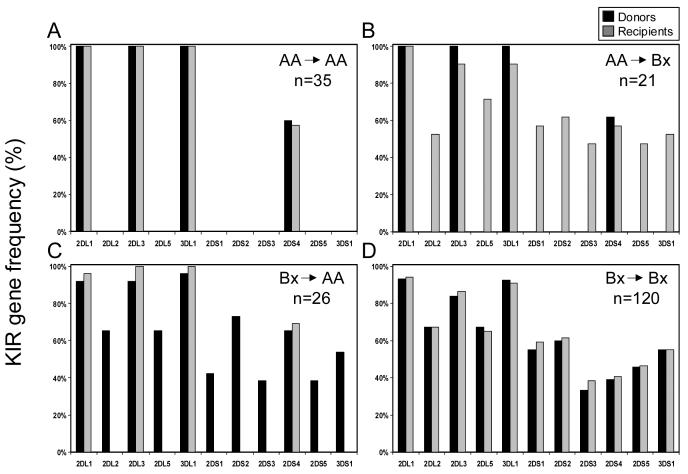
Having assigned the AA or Bx genotype to each donor and recipient, we were then able to assign each transplant to one of four kinds according to the combination of the donor and recipient KIR genotypes. Figure 2 shows the differences between these four groups: AA donor, AA recipient (Figure 2A); AA donor, Bx recipient (Figure 2B); Bx donor, AA recipient (Figure 2C) and Bx donor, Bx recipient (Figure 2D). We then tested the hypothesis that differences in outcome correlate with donor-recipient KIR genotype combination.
Figure 2.
KIR gene frequencies of 202 leukemia patients divided according to the KIR genotype of the donor and recipient. A. Donor-AA/recipient-AA: donors and patients have identical KIR gene content and similar KIR gene frequencies. B. Donor-AA/recipient-Bx: recipients have more activating KIR than their donors. C. Donor-Bx/recipient-AA: donors have more activating KIR than their recipients. D. Donor-Bx/recipient-Bx: donors and recipients have similar KIR gene frequencies.
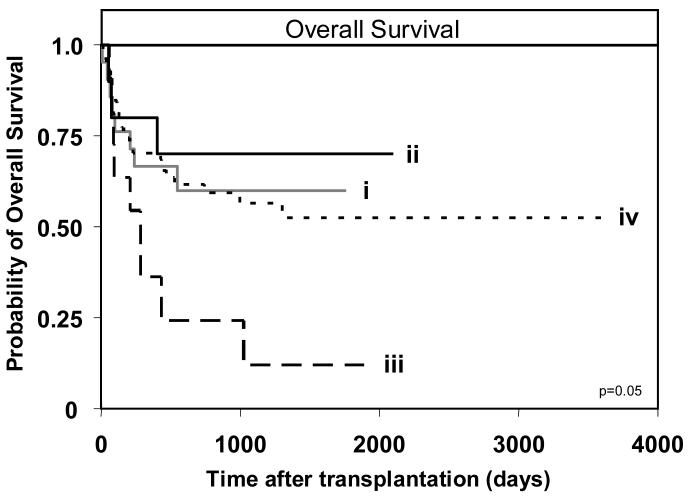
Analysis of overall survival revealed no significant difference when all 202 patients were compared, although a trend towards poorer survival was observed in AA patients receiving a Bx graft (data not shown). However, when only the 113 myeloid leukemia patients were considered, an effect was observed (Figure 3). Best survival was achieved by Bx patients receiving an AA graft, followed by AA patients receiving an AA graft and then Bx patients receiving a Bx graft. Transplants involving AA recipients and Bx donors had the poorest survival, being 3.8 times more likely to have poorer survival than transplants in which both donor and recipient were AA (p=0.046). These results pointed to the combination of KIR haplotypes in the donor and recipient having an influence on the outcome of HCT. Notably, that group B KIR haplotypes in the donor's genotype can be detrimental to the outcome of transplantation for myeloid leukemia, whereas group B haplotypes in the recipient's genotype can be beneficial.
Figure 3.
Myeloid leukemia patients (n=113) of AA KIR genotype that receive a transplant from a donor with a Bx KIR genotype (BB or AB) have poorest overall survival. Kaplan-Meier analysis of overall survival for patients with the donor-recipient KIR genotype combinations: (i) donor-AA/recipient-AA (n=8/21), (ii) donor-AA/recipient-Bx (n=3/10), (iii) donor-Bx/recipient-AA (n=9/11), and (iv) donor-Bx/recipient-Bx (n=29/71). The equality of all four curves was tested (d.f.=3, p=0.05). Pairwise comparisons of the curves was then performed by Cox proportional hazard modeling, where the overall type I error rate was controlled by the initial test. Comparison of survival between patients with donor-AA/recipient-Bx and: donor-AA/recipient-AA (p=0.59); donor-Bx/recipient-AA (p=0.046; hazard ratio=3.8, 95% CI=1.02-5.45); and donor-Bx/recipient-Bx (p=0.5). KIR genotype ‘Bx’ indicates that the individual has at least one ‘B’ haplotype; the second haplotype can either be an ‘A’ or ‘B’. The (n=) refers to the number of patients in each respective group who experienced the clinical endpoint being analyzed.
When donor genotype alone was considered as a risk factor, no significant difference in survival was seen for patients receiving grafts from AA versus Bx donors (p=0.36) (Supplemental Figure 1A). Similar consideration of recipient genotype alone revealed no significant survival difference between AA and Bx patients (p=0.18) (Supplemental Figure 1B). In contrast, when the KIR genotypes of both the donor and recipient were taken into account, Cox proportional hazard modeling showed a correlation between the two, such that recipient AA genotype was protective (hazard ratio=0.5, p=0.02) while donor Bx genotype was a risk factor (hazard ratio=2.1, p=0.05) for survival. Thus, it is the combination of donor and recipient KIR genotypes that influences survival, not the individual genotype of either the donor or recipient.
Cox proportional hazard modeling also showed that the combination of a Bx donor with an AA recipient was an independent risk factor for survival (hazard ratio=2.6, p=0.009), as were an advanced stage of disease at the time of transplantation (hazard ratio=3.4, p=0.0001) and patient age greater than 50 years (hazard ratio=2.4, p=0.003). Both disease stage and patient age have been well documented as prognostic factors affecting transplant outcome [46]. In addition, an effect due to the type of graft was found in this model, with peripheral blood cells being a risk factor (hazard ratio=4.2, p=0.02) compared to bone marrow cells. Although unexpected as a risk factor in this transplant setting, grafts derived from peripheral blood have been correlated with increased cGVHD [47], which can reduce survival. In contrast the conditioning regimen that patients received prior to transplantation was not a significant risk factor for survival in this model.
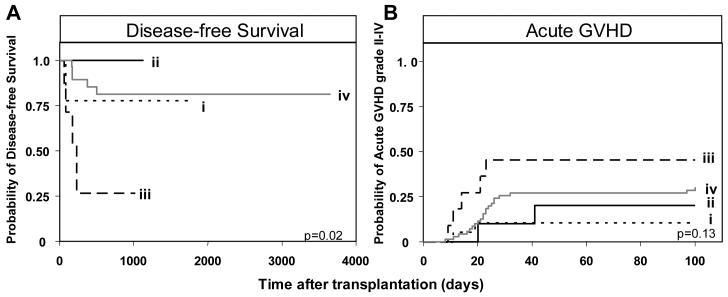
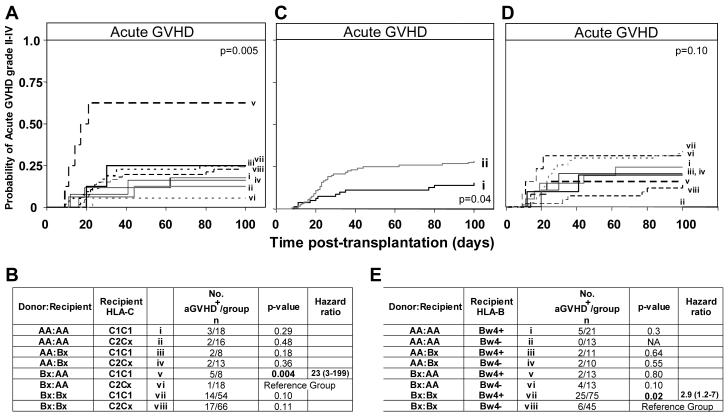
The combination of Bx donor and AA recipient was also a risk factor for relapse and aGVHD grade II-IV following transplantation for myeloid leukemia (Figure 4). Eight of the 59 myeloablative transplants performed for AML/MDS involved this combination of KIR genotypes and four of them relapsed. In contrast, none of the five Bx patients grafted from an AA donor relapsed (Figure 4A). Of the 112 patients transplanted for myeloid leukemia, the incidence of aGVHD in the four KIR genotype combinations was 46% (5/11) for Bx-donor/AA-recipient, 10% (2/20) for AA-donor/AA-recipient, 20% (2/10) for AA-donor/Bx-recipient, and 30% (21/71) for Bx-donor/Bx-recipient (Figure 4B). AA-recipients given Bx grafts were 5.8 times more likely to develop clinically relevant aGVHD (grade II-IV) than AA recipients given an AA graft (p=0.04).
Figure 4.
Patients with an AA KIR genotype receiving a transplant from a donor with a Bx genotype have poorer clinical outcome following HLA-matched sibling transplantation for myeloid disease. A. Kaplan-Meier analysis of disease-free survival in myeloablative AML/MDS patients (n=59) with the KIR genotypes (i) donor-AA/recipient-AA (n=2/12), (ii) donor-AA/recipient-Bx (n=0/5), (iii) donor-Bx/recipient-AA (n=4/8) and (iv) donor-Bx/recipient-Bx (n=5/34). The equality of all four curves was tested (d.f.=3, p=0.02). B. Kaplan-Meier analysis of acute GVHD grade II-IV in myeloid patients (n=112) with different KIR genotype combinations. The equality of all four curves was tested (d.f.=3, p=0.13). Pairwise comparisons of the curves was then performed by Cox proportional hazard modeling, where the overall type I error rate was controlled by the initial test. Comparison of aGVHD between (i) donor-AA/recipient-AA (n=2/20) patients and: (ii) donor-AA/recipient-Bx (n=2/10, p=0.5); (iii) donor-Bx/recipient-AA (n=5/11, p=0.04; hazard ratio=5.8, 95% CI=1.1-30.3); and (iv) donor-Bx/recipient-Bx (n=21/71, p=0.14). ‘Bx’ and ‘(n=)’ are defined in Figure 2.
Our results indicate that presence of activating KIR in the donor graft leads to both increased relapse, a host-versus-graft effect, and increased aGVHD, a graft-versus-host effect, responses which are seemingly contradictory and therefore difficult to reconcile. One possibility is that more activating KIR expressed by donor-derived NK cells and/or T cells in the graft leads to alloaggression of the graft towards the host, which in turn could impair the reconstitution of a functioning immune system. This could prevent an effective graft versus leukemic response and increase graft versus host effects, leading to the decreased disease-free survival, increased aGVHD and poorer survival observed in our study.
Recipient HLA class I genotype combines with donor-recipient KIR genotypes to influence transplant outcome
Because HLA-C provides the dominant ligands for KIR, and HLA-C type has figured in several clinical correlations, including beneficial NK-cell alloreactions following haploidentical transplantation [3, 4], we investigated whether HLA-C type had an influence on outcome in the transplants studied here. A key difference between the C1 and C2 ligands and their cognate inhibitory KIR is that C2 produces stronger inhibition than C1. For this reason we divided all transplants into two groups according to whether the recipients (and their HLA-identical donors) had C2 or not. Recipients lacking C2 are C1 homozygotes and were designated as ‘C1C1’; recipients having C2 include both C2C2 homozygotes and C1C2 heterozygotes and were designated as ‘C2Cx’.
Strikingly, the combination of Bx donor and AA recipient was found to be a risk factor for aGVHD II-IV only in C1C1 transplants and not C2Cx transplants (hazard ratio=23, p=0.004) (Figure 5A,B). Indeed, when HLA-C type was examined without consideration of KIR genotype, there was a slight trend for C1C1 homozygosity to be a risk factor for GVHD (hazard ratio=1.6, p=0.17), which was not the case for C2C2 homozygosity (hazard ratio=1.0, p=0.99) (Supplemental Figure 2). Consideration of HLA-C in combination with either the donor or the recipient KIR genotypes also supported the correlation observed with all three factors. Thus recipient-AA/C1C1 was a risk factor compared to recipient-AA/C2Cx (hazard ratio=4.4, p=0.03) (Supplemental Figure 3A) and a slight, but similar, trend was seen for donor-Bx/C1C1 (hazard ratio=2.5, p=0.09) (Supplemental Figure 3B). Furthermore, in the myeloid leukemia patients a trend to poorer overall survival was observed for the combination of Bx donor, AA recipient and C1C1 (p=0.08).
Figure 5.
Influence of HLA-C and HLA-B genotype on acute GVHD grade II-IV in all patients with different donor/recipient KIR genotype combinations. A. Kaplan-Meier analysis of grade II-IV acute GVHD in patients who have been grouped according to donor-recipient KIR and recipient HLA-C genotype combination. The equality of all 8 curves was tested (d.f.=7, p=0.005). B. Pairwise comparisons of the curves was then performed by Cox proportional hazard modeling, where the overall type I error rate was controlled by the initial test. Comparison of the probability of aGVHD between donor-Bx/recipient-AA/recipientC2Cx and all other KIR/HLA genotype combinations are shown. C. Kaplan-Meier analysis of aGVHD grade II-IV in patients who are (i) Bw4− (Bw6/Bw6 homozygous) (n=12/81) compared to (ii) patients who have at least one Bw4 allele (Bw4/Bw4 homozygotes or Bw4/Bw6 heterozygotes) (n=34/120, p=0.04; hazard ratio=2.0, 95% CI=1.04-3.9). D. Kaplan-Meier analysis of aGVHD in patients who have been grouped according to donor-recipient KIR and recipient HLA-B genotype combination. The equality of all 8 curves was tested (d.f=7, p=0.1). E. Pairwise comparisons of the curves was then determined by Cox proportional hazard modeling, where the overall type I error rate was controlled by the initial test. Comparison of the probability of aGVHD between donor-Bx/recipient-Bx/recipient-Bw4− and all other KIR/HLA genotype combinations are shown. ‘Bx’ is as defined in Figure 2, ‘C2Cx’ indicates that the individual has at least one C2 haplotype. The second haplotype can either be C1 or C2. Ratios were considered significant when p ≤ 0.05 (bold). In panels B and E, the patient group differing only by HLA type from the group showing the greatest effect was chosen as the reference group to which all other groups were compared.
Given the correlation observed with the HLA-C KIR ligands, we extended the analysis to HLA-B, for which the Bw4 epitope is a much stronger ligand for KIR3DL1 than the alternative Bw6. Assessment of recipient (and donor) HLA-B type revealed that aGVHD was more prevalent in the presence of Bw4 than in its absence (hazard ratio=2.0, 0=0.04) (Figure 5C). Thus the correlation for HLA-B is the opposite of that seen for HLA-C, in that the stronger inhibitory ligand is associated with aGVHD. When patients with the four donor-recipient KIR genotype combinations were divided according to HLA-B type, increased aGVHD was observed for the combination of Bx donor, Bx recipient and HLA-Bw4 positive (hazard ratio=2.9, p=0.02) as compared to the same combination when Bw4 was absent (when the recipient was Bw6-Bw6 homozygous). No difference in aGVHD was observed when Bw4 was present or absent in Bx-donor, AA-recipient transplants (p=0.1), the combination which showed increased aGVHD when the recipient was C1C1 (Figure 5D,E).
In summary, we observe a clear effect of HLA-C genotype upon the influences of KIR genotype on transplant outcome. In the presence of C2, the stronger inhibitor, the detrimental effects of the donor-Bx/recipient AA genotype combination are reduced. These data are consistent with a mechanism in which the HLA-C molecules of the recipient can inhibit alloreactive, donor-derived, KIR-expressing cells that contribute to aGVHD and relapse, and that C2 performs this function better than C1. The influence of HLA-B is weaker than that of HLA-C and in the opposite direction. Here the stronger Bw4 ligand increases the likelihood of aGVHD, indicating that HLA-B and –C affect the development of aGVHD through different mechanisms.
Recipient's lack of Bw4 ligands for donor inhibitory KIR3DL1 correlates with non-relapse mortality
Observing that interaction between KIR and HLA genotypes affected the outcome of transplantation, we tested whether this could be attributed to the recipient's lack of ligands for donor inhibitory KIR. From assessment of the recipient's HLA type for the A3/A11, Bw4, C1 and C2 ligands and of the donor's KIR type for their cognate inhibitory KIR (3DL2, 3DL1, 2DL2/3 and 2DL1, respectively), patients were divided as to whether they lacked an HLA ligand for an inhibitory KIR present in the donor. For ninety two percent of the transplants (186/202) there were one or more missing ligands (Table 2).
Table 2.
Matching of recipient HLA class I ligands to donor inhibitory KIR.
| All donor- recipient pairs (n=202) |
AML/MDS donor- recipient pairs (n=78) |
|
|---|---|---|
| N % | N % | |
| Recipient has all ligands for Donor inhibitory KIR | 16 (8) | 6 (8) |
| Recipient A3 or 11− / Donor 3DL2+ | 44 (22) | 19 (24.5) |
| Recipient Bw4− / Donor 3DL1+ | 10 (5) | 4 (5) |
| Recipient C1− / Donor 2DL2/3+ | 5 (2) | 1 (1.25) |
| Recipient C2− / Donor 2DL1+ | 16 (8) | 8 (10) |
| Recipient A3/11− and Bw4− / Donor 3DL2+ and 3DL1+ | 18 (9) | 5 (6.5) |
| Recipient A3/11− and C1− / Donor 3DL2+ and 2DL2/3+ | 16 (8) | 5 (6.5) |
| Recipient A3/11− and C2− / Donor 3DL2+ and 2DL1+ | 26 (13) | 14 (18) |
| Recipient Bw4− and C1− / Donor 3DL1+ and 2DL2/3+ | 0 (0) | 0 (0) |
| Recipient Bw4− and C2− / Donor 3DL1+ and 2DL1+ | 13 (6) | 4 (5) |
| Recipient A3/11−- and Bw4− and C1− / Donor 3DL2+ and 3DL1+ and 2DL2/3+ | 8 (4) | 1 (1.25) |
| Recipient A3/11− and Bw4− and C2− / Donor 3DL2+ and 3DL1+ and 2DL2/3+ | 30 (15) | 11 (14) |
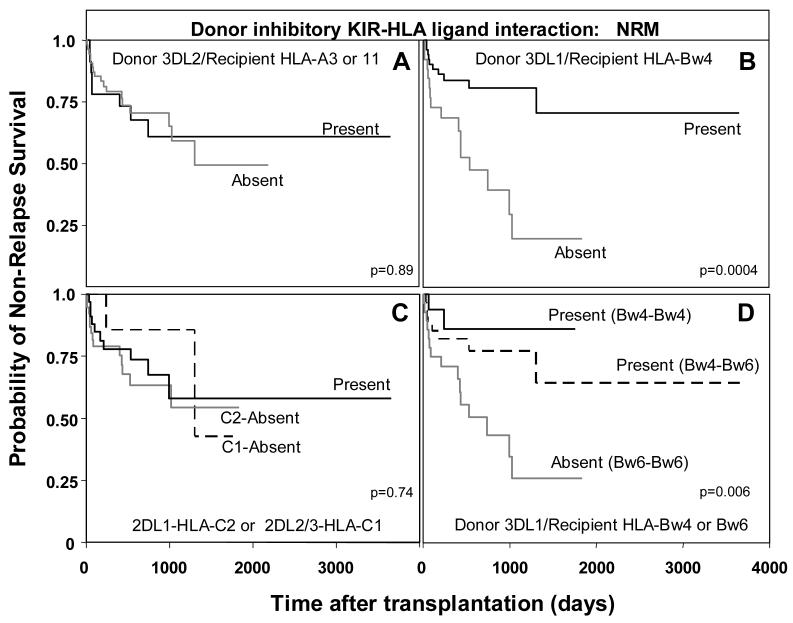
All 202 transplants were first analyzed as a group. Apart from the above mentioned association of the Bw4-KIR3DL1 interaction with aGVHD, no significant correlation was observed between any transplant endpoint (survival, DFS, NRM, aGVHD and cGVHD) and the presence or absence of each separate KIR-ligand interaction (3DL2-HLA-A3 or A11; 3DL1-HLA-Bw4; 2DL2/3-HLA-C1; or 2DL1-HLA-C2). Restricting analysis to the transplants for AML and MDS, revealed that patients lacking an HLA-Bw4 ligand for donor KIR3DL1 were at increased risk for NRM (hazard ratio=3.6, p=0.002) (Figure 6B). This was also observed when survival was examined as an endpoint, although to a lesser degree (hazard ratio=1.95, p=0.05). Strikingly, no significant difference in risk for NRM was observed when patients lacked HLA-A3/11 for donor KIR3DL2 (Figure 6A) or HLA-C1 and C2 ligands for donor KIR2DL2/3 and 2DL1 (Figure 6C). Cox proportional hazard modeling indicated that lack of the KIR3DL1-HLABw4 interaction in patients was an independent risk factor for NRM (hazard ratio=4.2, p=0.001), along with advanced disease stage (hazard ratio=3.9, p=0.003). In addition, irradiation as part of the pre-transplant conditioning regimen was a risk factor for NRM (hazard ratio=3.9, p=0.003), while patient age and marrow ablation were not significant.
Figure 6.
Increased non-relapse mortality in AML/MDS patients (n=78) when the recipient lacks the Bw4 ligand for donor KIR3DL1. A. Kaplan-Meier analysis of non-relapse survival in AML/MDS patients (n=78) when: (A) the donor KIR3DL2-HLA-A3 or A11 ligand interaction is present (n=8/23) or absent (n=17/55, p=0.89); (B) when donor KIR3DL1-HLA-Bw4 ligand interaction is present (n=10/52) or absent (n=15/26, p=0.002; hazard ratio=3.6, 95% CI=1.6-8.4); and (C) when donor KIR2DL2/3-HLA-C1 or KIR2DL1-HLA-C2 interactions are present (n=10/33) or absent (C1 absent n=2/7, p=0.5; C2 absent n=13/38, p=0.76). D. Kaplan-Meier analysis of non-relapse survival in AML/MDS patients when donor KIR3DL1-HLA-B interactions are divided according to whether the donor is Bw4 homozygous or Bw4-Bw6 heterozygous. The equality of all three curves was tested (d.f.=2, p=0.006). Pairwise comparisons of all curves were then determined by Cox proportional hazard modeling, where the overall type I error rate was controlled by the initial test. Interactions between KIR3DL1 and HLA-Bw4 homozygous patients (n=2/16) were compared to interactions between KIR3DL1 and Bw4-Bw6 heterozygous patients (n=8/34, p=0.39), and to patients lacking the 3DL1-HLA-Bw4 ligand interaction (n=15/28, 0=0.03; hazard ratio=5.2, 95% CI=1.2-22.8).
Thus, the interaction between HLA-Bw4 and KIR3DL1 on donor-derived lymphocytes cells appears protective for non-relapse survival. This protection was observed regardless of whether the recipient was Bw4 homozygous (hazard ratio=0.19, p=0.03) or Bw4-Bw6 heterozygous (hazard ratio=0.38, p=0.03) (Figure 6D). Therefore, the presence of Bw6 itself is not detrimental for transplant outcome. Instead, it is the absence of the KIR3DL1-Bw4 interaction that reduces survival. Patients in the NRM group died from a variety of causes including infection, aGVHD, graft failure, and organ failure. The role of KIR-expressing cells in NRM could be in response to infection and in alloreactivities and autoreactivities that impair tissues and organs due to lack of tolerance and other dysregulation of the immune system. The inhibition provided by the KIR3DL1-Bw4 interaction could be such that it reduces the detrimental alloreactivities and autoreactivities while retaining an effective response to infection.
Donor KIR2DS3 is protective against cGVHD
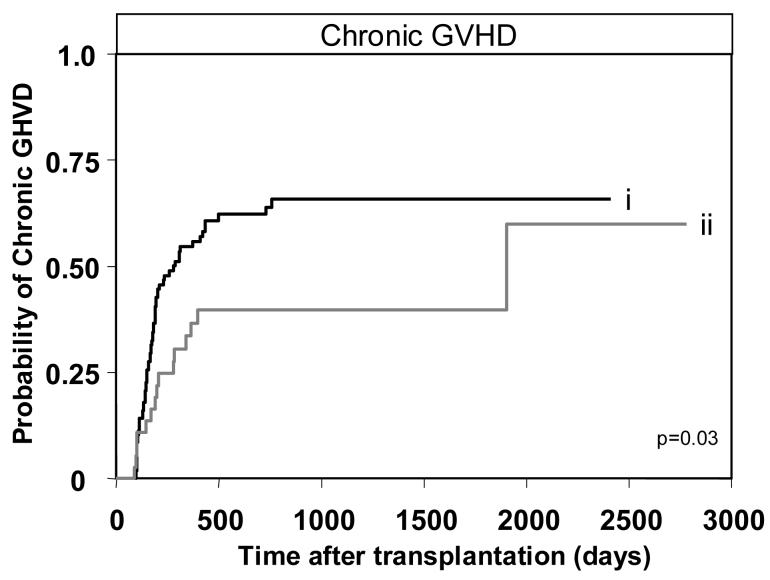
Having determined the detrimental effect of donor-derived group B haplotype genes on transplant recipients having AA KIR genotypes and lacking C2, we then tested for correlation with the activating KIR genes that characterize the group B haplotypes (KIR2DS1,2,3,5 and KIR3DS1). Also included in the analysis was KIR2DS4 which is a component of all A haplotypes and some B haplotypes. For each gene univariate analysis was performed for the transplant endpoints: survival, DFS, NRM, aGVHD and cGVHD. The one significant correlation identified in this analysis was that presence of KIR2DS3 in the donor genotype was protective against cGVHD (n=143, hazard ratio=0.5, p=0.03) (Table 3, Figure 7). Cox proportional hazard modeling to examine the effect of donor KIR2DS3, after adjustment for transplant co-variables such as disease stage at transplantation, recipient age, source of graft, pre-transplant conditioning and myeloablation, showed that donor KIR2DS3 was an independent prognostic factor for cGVHD. In addition to donor KIR2DS3 the only other significant parameter to emerge from the analysis was that peripheral blood as source for the graft was an independent risk factor for cGVHD, as was previously reported [47]. Increased protection against cGVHD was also observed in transplants for lymphoid leukemia when the donor had KIR2DS1 and in transplants for myeloid leukemia when the donor had KIR2DS2, although neither trend reached significance (data not shown). In summary our results suggest that while group B haplotypes are correlated with aGVHD the individual genes can provide protection against cGVHD.
Table 3.
Correlations between donor activating KIR and clinical outcome. Clinical endpoints examined included overall survival (OS), disease-free survival (DFS), non-relapse mortality (NRM), acute graft-versus-host disease (aGVHD) and chronic graft-versus-host disease (cGVHD). Hazard ratios were measured by univariate Cox proportional hazard modeling. 95% confidence intervals are shown in brackets. Ratios were considered significant then p ≤ 0.05 (bold italics, boxed).
| OS | DFS | NRM | aGVHD | cGVHD | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hazard ratio |
p | Hazard ratio |
p | Hazard ratio |
p | Hazard ratio |
p | Hazard ratio |
p | |
| KIR2DS1 | 1.2 (0.8-2.0) |
0.35 | 1.0 (0.6-1.8) |
0.87 | 1.2 (0.6-2.1) |
0.63 | 0.9 (0.5-1.7) |
0.76 | 0.9 (0.5-1.4) |
0.57 |
| KIR2DS2 | 0.8 (0.5-1.3) |
0.44 | 0.9 (0.5-1.5) |
0.64 | 0.8 (0.5-1.5) |
0.55 | 1.0 (0.6-1.8) |
0.99 | 1.0 (0.3-3.2) |
0.98 |
| KIR2DS3 | 1.0 (0.6-1.7) |
0.91 | 0.7 (0.3-1.3) |
0.23 | 1.1 (0.6-2.1) |
0.79 | 0.9 (0.5-1.8) |
0.84 |
0.5 (0.3-0.9) |
0.03 |
| KIR2DS4 | 0.8 (0.5-1.3) |
0.31 | 1.2 (0.7-2.1) |
0.51 | 0.7 (0.4-1.4) |
0.34 | 0.7 (0.4-1.3) |
0.26 | 1.2 (0.8-1.9) |
0.43 |
| KIR2DS5 | 1.2 (0.7-1.9) |
0.5 | 1.2 (0.6-2.0) |
0.70 | 1.0 (0.5-1.9) |
0.97 | 0.9 (0.5-1.6) |
0.66 | 1.0 (0.6-1.5) |
0.84 |
| KIR3DS1 | 1.2 (0.7-1.9) |
0.46 | 0.9 (1.5-1.6) |
0.75 | 1.1 (0.6-2.0) |
0.74 | 1.1 (0.6-1.9) |
0.86 | 1.0 (0.6-1.6) |
0.97 |
Figure 7.
Presence of KIR2DS3 in the donor protects against chronic GHVD. Kaplan-Meier analysis and Cox proportional hazard modeling of chronic GVHD in all patients (n=143) transplanted from (i) donors with KIR2DS3 absent (n=63/106) compared to (ii) donors with KIR2DS3 present (n=15/37, p=0.03; hazard ratio=0.5, 95% CI=0.3-0.9).
Discussion
This study examined the influence of KIR and HLA genotypes on the outcome of HLA-matched sibling-donor transplantation for several malignant diseases. We found that the presence of activating KIR in the donor graft promotes immune reactivity while the presence of inhibitory KIR in the recipient promotes immune tolerance. The most significant effects were seen for myeloid disease, where the combination of donor and recipient KIR genotypes was shown to affect survival, and the incidence of aGVHD and relapse. The poorest clinical outcome was associated with presence of one or two group B KIR haplotypes in the donor and their absence in the patient (Bx donor-AA recipient). Conversely, the best outcome was associated with the absence of group B haplotypes in the donor and their presence in the recipient (AA donor-Bx recipient). This suggests that the presence of activating KIR in the graft promotes graft versus host responses while discouraging graft versus tumor effects.
The results of Sun et al fit with this hierarchy [35]. Their AML patients showed increased aGVHD when the patient had a greater number of inhibitory KIR than the donor and/or when the donor had a greater number of activating KIR than the recipient. The KIR combination associated with poorest outcome is also that with the greatest potential for facilitating alloreactions in the graft-versus-host direction, whereas the KIR combination associated with best outcome is that with least potential for such alloreactions. Because NK cells, αβ T cells and γδ T cells can express KIR, the possible involvement of all three cells in the observed effects should be considered.
The effects of donor-recipient KIR combination on outcome were shown to depend upon HLA class I type. From analysis of all the transplants, the combination of Bx donor-AA recipient was shown to be a risk factor for aGVHD only when in combination with homozygosity for HLA-C allotypes having the C1 ligand for KIR2D. Because the donor and recipient were HLA identical in the transplants studied, it is not known if the role of C1 is effected by cells of donor or recipient genotype, or both. Study of different human populations has revealed an inverse relationship between frequencies of AA KIR genotypes and HLA-C2 [48], suggesting there is a selective disadvantage to this KIR-HLA combination. Consequently, this would lead to an association of A haplotypes with C1 and B haplotypes with C2 in human genomes. The C1 ligand is a poorer inhibitor than C2, its allelic alternative [19, 21, 22]. Thus the stronger ligand (C2) is associated with the presence of more activating KIR (B haplotypes) while the weaker ligand (C1) is associated with mainly inhibitory receptors (A haplotypes). Both A and B haplotypes contain inhibitory receptors for HLA-C1 and C2. Therefore, for KIR genotypes with few activating receptors (AA genotypes), inhibition can be maintained by a weaker inhibitory ligand (C1) than for genotypes containing more activating receptors (Bx) where stronger inhibition by C2 is required to offset activating signals. That presence of C2 decreases the likelihood of aGVHD suggests that the stronger inhibition offered by C2 is able to control the alloreactions of KIR-expressing lymphocytes to a greater extent than C1. Therefore activating KIR in the donor, combined with mainly inhibitory KIR and weak HLA ligands in the recipient facilitate unhelpful alloreactions following HLA-matched transplantation from a sibling donor.
Differential association of C1 and C2 with resolution of hepatitis C infection and the incidence of pre-eclampsia in pregnancy has been described and interpreted in a similar manner [48, 49]. Cook and colleagues found that patients transplanted for myeloid leukemia had poor overall survival when they were HLA-C2 homozygotes and KIR2DS2 was present in the donor [40]. We observed a similar trend for the 18 transplants of this type in our cohort, though it did not reach statistical significance (data not shown). Such similarity encourages further evaluation in larger patient populations.
An effect of inhibitory HLA-B ligands on the incidence of aGVHD was observed. In contrast to the HLA-C effect, here it was the stronger inhibitor, Bw4, that was dominantly associated with aGVHD and not its weaker alternative, Bw6. That strong inhibition by HLA-C appears advantageous and strong inhibition by HLA-B is disadvantageous indicates a difference in the underlying mechanism. This is also suggested by the observation that presence of Bw4 does not modify the effect of the donor Bx-recipient AA genotype combination, whereas C2 does. One reasonable possibility based upon known mechanisms, is that the effect of C2 is to inhibit T cells that cause GVHD, whereas Bw4 inhibits alloreactive T cells or NK cells that suppress GVHD. In contrast to its effect on aGVHD, the presence of Bw4 and its cognate inhibitory receptor, KIR3DL1, had a dominant, beneficial effect upon non-relapse mortality. That similar effects were not seen for the combination of HLA-A and HLA-C ligands with their cognate KIR is also consistent with Bw4-mediated inhibition affecting a different population of cells.
For T cell-depleted, HLA-matched, related transplants, increased overall survival and decreased relapse was observed when the donor had an inhibitory KIR for which the recipient did not have the cognate HLA ligand [41]. In that scenario the potential for alloreactions in the graft-versus-host direction was beneficial. In contrast, our analysis of T cell replete, HLA-matched transplants showed increased non-relapse death when the recipient lacked cognate ligands for donor inhibitory KIR, especially when lacking two such HLA ligands. Similarly, study of HLA-A, B, and DR-matched, but predominantly HLA-C-mismatched, transplants revealed increased relapse, NRM and aGVHD when the recipient lacked a ligand for a donor inhibitory KIR [34]. The likely cause of these striking differences is the T-cell content of the grafts. Whereas T cell alloreactions appear dominant following transplantation with T-cell replete grafts, NK cell alloreactions can emerge following transplantation with T-cell depleted grafts. These contrasting results show how differences in transplant protocol can influence the impact of the genetic factors of KIR and HLA class I and the importance of considering them when comparing the results of different studies.
Gagne and colleagues reported increased aGVHD in patients receiving HLA-matched transplants from an unrelated donor, especially when KIR2DS3 was present in the donor [33]. As well as a general correlation of donor activating KIR with aGVHD, we found that KIR2DS3 was protective against chronic GVHD, the only significant correlation we observed with an individual KIR locus. As nothing is known of the function and ligand specificity of KIR2DS3, there is no clear interpretation for these associations.
In human populations there is a variable balance between group A and group B KIR haplotypes, that appears maintained by balancing selection for inhibitory and activating functions [28, 45]. In part this selection is mediated by the interaction of inhibitory KIR with their HLA class I ligands. As a consequence the frequency of the strong inhibitory ligand, C2, is inversely correlated with the frequency of group A haplotypes, in which inhibitory receptors predominate. Pregnancies involving AA KIR mothers and a C2 homozygous fetus are at risk for pre-eclampsia, which may arise from excessive inhibition of KIR-expressing NK cells by C2-expressing fetal trophoblast. An unexpected feature of this recognition of haploidentical fetus by maternal lymphocytes is its indifference to whether the mother also expresses C2 [48]. In its conventional immunological meaning, self was unimportant; what mattered was the extent to which HLA class I ligands could engage inhibitory KIR on maternal lymphocytes.
This type of recognition also pertains to the setting of hematopoietic cell transplantation, as indicated by the results of this and other studies that followed the work on haploidentical transplants [40, 41]. Consequently, in HLA identical transplants there is evidence for lymphocyte reactions that effect the outcome and are determined by the number of inhibitory KIR for which there are cognate ligands in the HLA genotype shared by donor and recipient. If assessment of KIR genotype is to have any impact on donor selection for hematopoietic cell transplantation then it cannot be a stringent selection, because of that already imposed by HLA matching. For this reason, and because of the biological significance of the A/B haplotype difference, our study aimed to assess whether combinations of A and B haplotype influenced clinical outcome. The results indicate that they do and point to simple choices between common, and easily distinguished, KIR genotypes that could inform donor selection, particularly for unrelated donors, and help improve transplant outcome.
Supplementary Material
Acknowledgements
We thank D. Kathryn Tierney, RN and Dr Robert S. Negrin for their help in facilitating the collection of patient samples, Drs Phil Lavori and Jerry Halpern of the Department of Health and Research Policy for advice regarding statistical methods, and Drs Carl Grumet and Debra Hiraki for providing DNA samples. We would like to acknowledge the staff of the Cellular Therapeutics and Transplantation Laboratory and the BMT Administration Office at the Stanford Medical Center for their assistance in patient sample preparation and data collection. We would also like to thank Dr Sally Arai for clinical advice.
Abbreviations
- KIR
killer immunoglobulin-like receptor
- HCT
hematopoietic cell transplantation
- GVHD
graft-versus-host disease
- aGVHD
acute GVHD
- cGVHD
chronic GVHD
- LRC
leukocyte receptor complex
- AML
acute myelogenous leukemia
- CML
chronic myelogenous leukemia
- MDS
myelodysplastic syndrome
- MPD
myeloproliferative disease
- ALL
acute lymphocytic leukemia
- CLL
chronic lymphocytic leukemia
- NHL
non-Hodgkins lymphoma
- HD
Hodgkins disease
- MM
mulitiple myeloma
- SAA
severe aplastic anemia
- PNH
paroxysmal nocturnal hemoglobinuria
- DFS
disease-free survival
- NRM
non-relapse mortality
Footnotes
This research was supported by National Institutes of Health Grant CA49605. K.L. McQueen was supported by a Cancer Research Institute Fellowship.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parham P, McQueen KL. Alloreactive killer cells: hindrance and help for haematopoietic transplants. Nat Rev Immunol. 2003;3(2):108. doi: 10.1038/nri999. [DOI] [PubMed] [Google Scholar]
- 2.Giebel S, Locatelli F, Lamparelli T, Velardi A, Davies S, Frumento G, Maccario R, Bonetti F, Wojnar J, Martinetti M, Frassoni F, Giorgiani G, Bacigalupo A, Holowiecki J. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003;102(3):814. doi: 10.1182/blood-2003-01-0091. [DOI] [PubMed] [Google Scholar]
- 3.Ruggeri L, Capanni M, Casucci M, Volpi I, Tosti A, Perruccio K, Urbani E, Negrin RS, Martelli MF, Velardi A. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999;94(1):333. [PubMed] [Google Scholar]
- 4.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, Posati S, Rogaia D, Frassoni F, Aversa F, Martelli MF, Velardi A. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295(5562):2097. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 5.Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome. 1999;10(2):154. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 6.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97(9):4778. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantoni C, Verdiani S, Falco M, Pessino A, Cilli M, Conte R, Pende D, Ponte M, Mikaelsson MS, Moretta L, Biassoni R. p49, a putative HLA class I-specific inhibitory NK receptor belonging to the immunoglobulin superfamily. Eur J Immunol. 1998;28(6):1980. doi: 10.1002/(SICI)1521-4141(199806)28:06<1980::AID-IMMU1980>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 8.Faure M, Long EO. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J Immunol. 2002;168(12):6208. doi: 10.4049/jimmunol.168.12.6208. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189(7):1093. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yusa S, Catina TL, Campbell KS. SHP-1- and phosphotyrosine-independent inhibitory signaling by a killer cell Ig-like receptor cytoplasmic domain in human NK cells. J Immunol. 2002;168(10):5047. doi: 10.4049/jimmunol.168.10.5047. [DOI] [PubMed] [Google Scholar]
- 11.Dohring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A1,2. J Immunol. 1996;156(9):3098. [PubMed] [Google Scholar]
- 12.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, Rowland-Jones S, Braud VM. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34(6):1673. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 13.D'Andrea A, Chang C, Franz-Bacon K, McClanahan T, Phillips JH, Lanier LL. Molecular cloning of NKB1. A natural killer cell receptor for HLA-B allotypes. J Immunol. 1995;155(5):2306. [PubMed] [Google Scholar]
- 14.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181(3):1133. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, Moretta L, Moretta A. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur J Immunol. 1997;27(12):3095. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 16.Vales-Gomez M, Reyburn HT, Mandelboim M, Strominger JL. Kinetics of interaction of HLA-C ligands with natural killer cell inhibitory receptors. Immunity. 1998;9(3):337. doi: 10.1016/s1074-7613(00)80616-0. [DOI] [PubMed] [Google Scholar]
- 17.Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158(9):4026. [PubMed] [Google Scholar]
- 18.Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A. 1993;90(24):12000. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268(5209):405. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 20.Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, Gauthier L, Romagne F, Ferracci G, Arosa FA, Moretta A, Sun PD, Ugolini S, Vivier E. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102(37):13224. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati MS, Vitale M, Bottino C, Moretta L, Moretta A, Long EO. Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra- and intracellular domains. Immunity. 1995;2(5):439. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 22.Wagtmann N, Rajagopalan S, Winter CC, Peruzzi M, Long EO. Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity. 1995;3(6):801. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 23.Uhrberg M, Valiante NM, Young NT, Lanier LL, Phillips JH, Parham P. The repertoire of killer cell Ig-like receptor and CD94:NKG2A receptors in T cells: clones sharing identical alpha beta TCR rearrangement express highly diverse killer cell Ig-like receptor patterns. J Immunol. 2001;166(6):3923. doi: 10.4049/jimmunol.166.6.3923. [DOI] [PubMed] [Google Scholar]
- 24.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D'Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7(6):739. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 25.Stewart CA, Van Bergen J, Trowsdale J. Different and divergent regulation of the KIR2DL4 and KIR3DL1 promoters. J Immunol. 2003;170(12):6073. doi: 10.4049/jimmunol.170.12.6073. [DOI] [PubMed] [Google Scholar]
- 26.Trompeter HI, Gomez-Lozano N, Santourlidis S, Eisermann B, Wernet P, Vilches C, Uhrberg M. Three structurally and functionally divergent kinds of promoters regulate expression of clonally distributed killer cell Ig-like receptors (KIR), of KIR2DL4, and of KIR3DL3. J Immunol. 2005;174(7):4135. doi: 10.4049/jimmunol.174.7.4135. [DOI] [PubMed] [Google Scholar]
- 27.Shilling HG, Young N, Guethlein LA, Cheng NW, Gardiner CM, Tyan D, Parham P. Genetic control of human NK cell repertoire. J Immunol. 2002;169(1):239. doi: 10.4049/jimmunol.169.1.239. [DOI] [PubMed] [Google Scholar]
- 28.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7(6):753. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 29.Shilling HG, McQueen KL, Cheng NW, Shizuru JA, Negrin RS, Parham P. Reconstitution of NK cell receptor repertoire following HLA-matched hematopoietic cell transplantation. Blood. 2003;101(9):3730. doi: 10.1182/blood-2002-08-2568. [DOI] [PubMed] [Google Scholar]
- 30.Uhrberg M. Shaping the human NK cell repertoire: an epigenetic glance at KIR gene regulation. Mol Immunol. 2005;42(4):471. doi: 10.1016/j.molimm.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 31.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5(3):201. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 32.Bishara A, De Santis D, Witt CC, Brautbar C, Christiansen FT, Or R, Nagler A, Slavin S. The beneficial role of inhibitory KIR genes of HLA class I NK epitopes in haploidentically mismatched stem cell allografts may be masked by residual donor-alloreactive T cells causing GVHD. Tissue Antigens. 2004;63(3):204. doi: 10.1111/j.0001-2815.2004.00182.x. [DOI] [PubMed] [Google Scholar]
- 33.Gagne K, Brizard G, Gueglio B, Milpied N, Herry P, Bonneville F, Cheneau ML, Schleinitz N, Cesbron A, Follea G, Harrousseau JL, Bignon JD. Relevance of KIR gene polymorphisms in bone marrow transplantation outcome. Hum Immunol. 2002;63(4):271. doi: 10.1016/s0198-8859(02)00373-7. [DOI] [PubMed] [Google Scholar]
- 34.De Santis D, Bishara A, Witt CS, Nagler A, Brautbar C, Slavin S, Christiansen FT. Natural killer cell HLA-C epitopes and killer cell immunoglobulin-like receptors both influence outcome of mismatched unrelated donor bone marrow transplants. Tissue Antigens. 2005;65(6):519. doi: 10.1111/j.1399-0039.2005.00396.x. [DOI] [PubMed] [Google Scholar]
- 35.Sun JY, Gaidulis L, Dagis A, Palmer J, Rodriguez R, Miller MM, Forman SJ, Senitzer D. Killer Ig-like receptor (KIR) compatibility plays a role in the prevalence of acute GVHD in unrelated hematopoietic cell transplants for AML. Bone Marrow Transplant. 2005;36(6):525. doi: 10.1038/sj.bmt.1705089. [DOI] [PubMed] [Google Scholar]
- 36.Verheyden S, Schots R, Duquet W, Demanet C. A defined donor activating natural killer cell receptor genotype protects against leukemic relapse after related HLA-identical hematopoietic stem cell transplantation. Leukemia. 2005;19(8):1446. doi: 10.1038/sj.leu.2403839. [DOI] [PubMed] [Google Scholar]
- 37.Gomez-Lozano N, Vilches C. Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: an update. Tissue Antigens. 2002;59(3):184. doi: 10.1034/j.1399-0039.2002.590302.x. [DOI] [PubMed] [Google Scholar]
- 38.Yawata M, Yawata N, McQueen KL, Cheng NW, Guethlein LA, Rajalingam R, Shilling HG, Parham P. Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics. 2002;54(8):543. doi: 10.1007/s00251-002-0497-x. [DOI] [PubMed] [Google Scholar]
- 39.Frohn C, Schlenke P, Ebel B, Dannenberg C, Bein G, Kirchner H. DNA typing for natural killer cell inhibiting HLA-Cw groups NK1 and NK2 by PCR-SSP. J Immunol Methods. 1998;218(12):155. doi: 10.1016/s0022-1759(98)00126-4. [DOI] [PubMed] [Google Scholar]
- 40.Cook MA, Milligan DW, Fegan CD, Darbyshire PJ, Mahendra P, Craddock CF, Moss PAH, Briggs DC. The impact of donor KIR and patient HLA-C genotypes on outcome following HLA-identical sibling haematopoietic stem cell transplantation for myeloid luekaemia. Blood. 2004;103(4):1521. doi: 10.1182/blood-2003-02-0438. [DOI] [PubMed] [Google Scholar]
- 41.Hsu KC, Keever-Taylor CA, Wilton A, Pinto C, Heller G, Arkun K, O'Reilly RJ, Horowitz MM, Dupont B. Improved outcome in HLA-identical sibling hematopoietic stem-cell transplantation for acute myelogenous leukemia predicted by KIR and HLA genotypes. Blood. 2005;105(12):4878. doi: 10.1182/blood-2004-12-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norman PJ, Stephens HA, Verity DH, Chandanayingyong D, Vaughan RW. Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics. 2001;52(34):195. doi: 10.1007/s002510000281. [DOI] [PubMed] [Google Scholar]
- 43.Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168(5):2307. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 44.Toneva M, Lepage V, Lafay G, Dulphy N, Busson M, Lester S, Vu-Trien A, Michaylova A, Naumova E, McCluskey J, Charron D. Genomic diversity of natural killer cell receptor genes in three populations. Tissue Antigens. 2001;57(4):358. doi: 10.1034/j.1399-0039.2001.057004358.x. [DOI] [PubMed] [Google Scholar]
- 45.Uhrberg M, Parham P, Wernet P. Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics. 2002;54(4):221. doi: 10.1007/s00251-002-0463-7. [DOI] [PubMed] [Google Scholar]
- 46.Bacigalupo A, Sormani MP, Lamparelli T, Gualandi F, Occhini D, Bregante S, Raiola AM, di Grazia C, Dominietto A, Tedone E, Piaggio G, Podesta M, Bruno B, Oneto R, Lombardi A, Frassoni F, Rolla D, Rollandi G, Viscoli C, Ferro C, Garbarino L, Van Lint MT. Reducing transplant-related mortality after allogeneic hematopoietic stem cell transplantation. Haematologica. 2004;89(10):1238. [PubMed] [Google Scholar]
- 47.Stem Cell Trialists' Collaborative Group Allogeneic peripheral blood stem-cell compared with bone marrow transplantation in the management of hematologic malignancies: an individual patient data meta-analysis of nine randomized trials. J Clin Oncol. 2005;23(22):5074. doi: 10.1200/JCO.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hiby SE, Walker JJ, O'Shaughnessy KM, Redman CW, Carrington M, Trowsdale J, Moffett A. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200(8):957. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O'Brien SJ, Rosenberg WM, Thomas DL, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305(5685):872. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.