Abstract
Mycobacterium strains that grow on ethene and vinyl chloride (VC) are widely distributed in the environment and are potentially useful for biocatalysis and bioremediation. The catabolic pathway of alkene assimilation in mycobacteria is not well characterized. It is clear that the initial step is a monooxygenase-mediated epoxidation that produces epoxyethane from ethene and chlorooxirane from VC, but the enzymes involved in subsequent transformation of the epoxides have not been identified. We investigated epoxyethane metabolism in Mycobacterium strain JS60 and discovered a coenzyme M (CoM)-dependent enzyme activity in extracts from VC- and ethene-grown cells. PCR amplifications using primers targeted at epoxyalkane:CoM transferase (EaCoMT) genes yielded part of the JS60 EaCoMT gene, which was used to clone an 8.4-kb genomic DNA fragment. The complete EaCoMT gene (etnE) was recovered, along with genes (etnABCD) encoding a four-component monooxygenase and two genes possibly involved in acyl-CoA ester metabolism. Reverse transcription-PCR indicated that the etnE and etnA genes were cotranscribed and inducible by ethene and VC. Heterologous expression of the etnE gene in Mycobacterium smegmatis mc2155 using the pMV261 vector gave a recombinant strain capable of transforming epoxyethane, epoxypropane, and chlorooxirane. A metabolite identified by mass spectrometry as 2-hydroxyethyl-CoM was produced from epoxyethane. The results indicate that the EaCoMT and monooxygenase enzymes encoded by a single operon (etnEABCD) catalyze the initial reactions in both the VC and ethene assimilation pathways. CoM-mediated reactions appear to be more widespread in bacteria than was previously believed.
Ethene and vinyl chloride (VC) are important industrial chemicals and are also produced naturally by abiotic (22) and biosynthetic (14, 53) reactions. Anaerobic bacteria such as Dehalococcoides spp. produce VC and ethene during dehalorespiration of the chlorinated solvents perchloroethene and trichloroethene (29, 30), thus VC and/or ethene can accumulate in anaerobic subsurface zones at contaminated sites (12, 56). Monooxygenases, such as liver cytochrome P-450 (16, 44), oxidize VC and ethene to highly reactive epoxides that pose potential health risks. Whereas uncertainty exists concerning the dangers of low-level ethene exposure (43), VC is known to be a human carcinogen (23).
Mycobacterium strains capable of aerobic growth on ethene as the sole carbon and energy source were first isolated almost 30 years ago (8). More recently, strains of Mycobacterium (7, 18), Nocardioides (7), and Pseudomonas (50, 51) capable of growth on both ethene and VC have been discovered. The VC- and ethene-assimilating bacteria may be useful for the bioremediation of sites contaminated with chlorinated solvents (38). In addition, several ethene-assimilating strains have been evaluated as biocatalysts for the production of epoxides (17, 46). Much research has focused on the kinetics of VC and ethene oxidation and on the cometabolism of related substrates (7, 24, 49), while fundamental questions concerning the catabolic pathways and enzymes involved have been somewhat neglected. The biochemical traits that distinguish bacteria that grow on both substrates from those that grow on ethene alone are unknown, as are most of the metabolic intermediates in both pathways.
In bacteria, the initial enzymatic attack on VC and ethene is similar to the reactions observed in mammalian systems, i.e., a monooxygenase catalyzes the formation of epoxyethane (ethylene oxide) from ethene and the formation of chlorooxirane (VC epoxide) from VC (18, 50). The ethene monooxygenase from Mycobacterium strain E3 has been examined in some detail (19) and is a multicomponent enzyme, most likely with a binuclear iron active site similar to those of methane and propene monooxygenases (13, 39). The reductase component of the monooxygenase from strain E3 has been purified (55), but the other monooxygenase components have not been characterized. None of the genes encoding VC or ethene monooxygenases have been cloned or sequenced.
Epoxide metabolism in the VC and ethene catabolic pathways has been investigated in a few cases, but the results are inconclusive. Early work with the ethene-assimilating Mycobacterium strain E20 (10) suggested the involvement of a coenzyme A (CoA)- and NAD+-dependent enzyme that converted epoxyethane into acetyl-CoA, but the specific activities reported were very low (approximately 2 nmol/min/mg of protein). This “epoxyethane dehydrogenase” activity was also detected in the VC-assimilating Mycobacterium strain L1 (18), but further investigations in that case were hampered by the instability of the activity in cell extracts and because of practical difficulties in working with chlorooxirane.
Various other bacterial enzymes can transform epoxides, and such alternative reactions must also be considered with respect to epoxyethane and chlorooxirane metabolism. Epoxide hydrolases play a role in many biodegradative pathways, including those involving chlorinated aliphatic compounds. The epichlorohydrin hydrolase from Agrobacterium strain AD1 is the best-studied example (35). Glutathione S-transferase (GST) enzymes are also widely distributed among bacteria. Notably, the GST from the isoprene degrader Rhodococcus strain AD45 can transform cis-dichloroepoxyethane (48), a substrate similar to the epoxides of VC and ethene. In the propene-assimilating bacteria Xanthobacter strain Py2 and Rhodococcus strain B-276, an epoxide carboxylase enzyme complex catalyzes the conversion of epoxypropane into acetoacetate. This unusual system consists of epoxyalkane:CoM transferase (EaCoMT), two stereoselective dehydrogenases, and an oxidoreductase-carboxylase (11).
The aim of the present study was to investigate the initial reactions of the VC and ethene assimilation pathways in Mycobacterium strain JS60, a bacterium previously isolated from groundwater (7). In particular, our focus was on determining the mechanism of epoxide metabolism.
MATERIALS AND METHODS
Chemicals, enzymes, and media.
Most chemicals and media were described previously (7). CoM (98%), CoA (95%), NAD (98%), glutathione (98%), epoxyethane (99.5%), and epoxypropane (99%) were from Sigma-Aldrich. All molecular biology enzymes were from Roche except for DNase-free RNase, which was from Qiagen. MSM minimal medium (7) or 1/10-strength trypticase-soy medium with 1% glucose (7) was used for growing Mycobacterium strain JS60. MSM medium was modified by increasing the phosphate concentration to 40 mM and decreasing the pH to 6.5. Luria-Bertani (LB) medium (37) was used for both Escherichia coli strain JM109 and Mycobacterium strain mc2155. Transformants of JM109 were plated on LB containing kanamycin (Km; 50 μg/ml) or ampicillin (Ap; 100 μg/ml). 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and isopropyl-β-d-thiogalactopyranoside (IPTG) (37) were added where required. Transformants of mc2155 were plated on LB with 20 μg of Km/ml or grown in LB broths containing 10 μg of Km/ml and 0.05% Tween 80. Strain mc2155(pMV-CoM) was grown in LB-Tween-Km medium containing 0.1 mM ZnSO4.
Plasmids, bacterial strains, and incubation conditions.
Plasmids and bacterial strains used in this study are described in Table 1. Strain JS60 was grown in 50 or 700 ml of MSM medium in crimp-sealed flasks (7). VC was added at 2% (vol/vol) of the total flask volume and resupplied as necessary. Ethene was added once at 10% (vol/vol), and potassium acetate was added at 20 mM. Mycobacterium strains JS60 and mc2155 were grown at 30°C; E. coli strain JM109 was grown at 37°C. All cultures were incubated aerobically with shaking at 200 rpm.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides
| Material | Relevant characteristics | Reference or Source |
|---|---|---|
| Strains | ||
| Mycobacterium rhodesiae JS60 | Grows on VC and ethene as sole carbon sources | 7 |
| Mycobacterium smegmatis mc2155 | Highly electrotransformable derivative of ATCC 607 | 40 |
| Escherichia coli JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′[traD36 proAB+laclqlacZ ΔM15] | 58 |
| Plasmids | ||
| pK18 | Kmr, 2.7 kb, general purpose cloning vector | 34 |
| pGEM-T Easy | Apr, 3.0 kb, T/A cloning vector | Promega |
| pMV261 | Kmr, 4.5 kb, E. coli-Mycobacterium shuttle vector | 41 |
| pMV-CoM | Kmr, 5.8 kb, pMV261 carrying JS60 EaCoMT gene (etnE) | This study |
| Oligonucleotides (5′-3′) | ||
| CoM-F1 | ATGGTGGGGAACTACCCGAATCC | This study |
| CoM-R1 | ATGAGGCCGCAGTCGGTGGACA | This study |
| CoM-F1L | AACTACCCSAAYCCSCGCTGGTACGAC | This study |
| CoM-R2 | TCGTCGGCAGTTTCGGTGATCGTGCTCTT | This study |
| 60M-F2 | GGGGATATCAGTGATCGCCATACAGCAATTAAGGAG | This study |
| 60M-R2 | GGGAAGCTTTTGTGTCCGGACAGGCCGATAGTC | This study |
| RTM-F1 | AAGGAGTGCATCATGAAGGTTGGGG | This study |
| RTβ-R1 | TTGGCTGATACCGGGTCGAGGTTGAG | This study |
General analytical methods.
VC, ethene, epoxyethane, and epoxypropane were analyzed in headspace samples (250 μl) by gas chromatography (HP 5890 series II) on a megabore capillary column (GSQ; Agilent) with flame-ionization detection. The growth of cultures was followed by measurement of the optical density at 600 nm (OD600). Protein concentrations in cell extracts were measured by a UV absorbance assay, as described previously (7) but omitting the lysis and neutralization steps. Chloride was analyzed as described previously (6).
Preparation of cell extracts.
Cultures of JS60 cells were grown to early exponential phase (OD600 = 0.2 to 0.3) in MSM with VC, ethene, or acetate as carbon sources. Tween 80 was added to 0.05%, and the cells were harvested by centrifugation, washed in buffer (20 mM K2HPO4, 0.05% Tween 80; pH 7.0), and suspended in 2 ml of morpholinepropanesulfonic acid (MOPS)-glycerol-dithiothreitol (DTT) buffer (3). The cells were broken by three passages through a chilled French pressure cell (130,000 kPa), and the lysate was centrifuged (16,000 × g for 15 min). The supernatant was retained and diluted to 2.0 mg of protein/ml in the same buffer.
EaCoMT assay.
Serum bottles (25 ml) containing 900 μl of Tris-HCl (50 mM; pH 8.0) and 50 μl of CoM (200 mM) were crimp sealed. Epoxyethane (5 μmol) was added, and the bottles were incubated at 30°C with shaking at 300 rpm. After 15 min, cell extract (50 μl) was added, and after a further 5 min of equilibration headspace samples were analyzed at intervals to quantify epoxyethane. Specific activity was calculated as nanomoles of epoxyethane consumed per minute per milligram of protein.
DNA extraction, PCR, and T/A cloning.
Extraction of genomic DNA from strain JS60 was performed essentially as described previously (7), except that ethene-grown cultures (700 ml) were used and ampicillin (200 μg/ml) was included with glycine in the overnight incubation. Plasmid extraction from E. coli strains was done by alkaline lysis (37). A Qiaquick kit (Qiagen) was used for purification of plasmid DNA and PCR products, while the Qiaex II kit (Qiagen) was used for the purification of genomic DNA fragments. PCR mixtures (25 μl) contained 1.5 mM Mg2+, 50 pmol of each primer, 2.5 U of Taq polymerase, and 5 to 50 ng of template DNA. Primers used in this work are listed in Table 1. Unless indicated otherwise, the thermocycling protocol was 95°C for 2 min, then 30 cycles of 94°C (30 s), 60°C (30 s), and 72°C (1 min), followed by a final extension cycle (72°C, 10 min). In initial PCR experiments with JS60, the primers CoM-F1 and CoM-R1 were used. A band of the expected size (981 bp) was excised from the gel, purified, ligated to the pGEM-T-Easy vector (Promega), and introduced into strain JM109 by electroporation. Recombinant clones were screened by restriction digestion, and several representatives containing inserts of the expected size were sequenced (Roswell Park Cancer Institute Biopolymer Facility; PE-ABI model 373A Stretch sequencer).
Southern blotting and construction of clone library.
Restriction digests (80 μl) containing 10 to 15 μg of JS60 genomic DNA and 60 to 80 U of restriction enzyme were incubated for 16 h at 37°C. After gel electrophoresis, Southern blotting was done according to standard methods (37), using the ECL kit (Pharmacia) for detection. The probe consisted of an 893-bp fragment of the EaCoMT gene of strain JS60, amplified by PCR using the primers CoM-F1L and CoM-R2. For clone library construction, NheI restriction digests were repeated at a fivefold-larger scale and DNA fragments (7 to 10 kb) were excised, purified, and ligated to XbaI-cut, phosphatase-treated pK18 vector. The ligation mixture was electroporated into strain JM109, which was plated on LB-Km-X-Gal-IPTG medium.
Screening of clone library.
Recombinant JM109(pK18) clones (480 white colonies) were transferred to five microtiter plates containing LB-Km broth (100 μl), and the plates were incubated with shaking overnight. Cultures (30 μl) from eight wells of a microtiter plate were pooled and centrifuged (16,000 × g, 1 min), and the cells were suspended in 200 μl of Tris-HCl buffer (5 mM; pH 8.0), yielding 60 clone pools. Each cell suspension was heat treated (95°C, 5 min) and centrifuged (16,000 × g, 2 min), and then 1 μl of the supernatant was used in PCR amplifications with the CoM-F1L and CoM-R2 primers, as described above. Individual cultures from pools containing positive clones were subsequently screened by a similar method, except that the template was 1 μl of culture from each well. Several positive clones were analyzed by restriction digestion, and the insert DNA (8.4 kb) from one representative was sequenced on both strands.
RNA extraction and RT-PCR.
Cultures of strain JS60 were grown on VC, ethene, or acetate to mid-exponential phase. The cells were washed in TE buffer (37), suspended in the same buffer (OD600 = 30), and frozen in 500-μl aliquots at −80°C. The RNeasy kit (Qiagen) was used for RNA extraction, with modifications as follows. Cells from one frozen aliquot were thawed, pelleted, suspended in 500 μl of buffer RLT, and added to a 2-ml screw-cap tube containing 1.5 ml of buffer RLT-saturated zirconia-silica beads (0.1-mm diameter). The mixture was subjected to beadbeating (Mini-Beadbeater; BioSpec Products) for 1 min at high speed. The supernatant (350 μl) was column purified according to the RNeasy protocol for bacteria, and the eluate was treated with DNase (30 U; 15 min; 20°C) and then repurified using the RNeasy protocol for RNA cleanup. The final RNA solutions were diluted to 5 ng/μl in water, and 2 μl of each was used for reverse transcription-PCR (RT-PCR) with the Titan One-Tube kit (Roche). Thermocycling was done with the RTM-F1 and RTβ-R1 primers, with annealing at 60°C (30 s), extension at 68°C (2 min), and other parameters according to the kit instructions. Negative controls (no reverse transcriptase) were prepared with Expand DNA polymerase mixture, and a positive control contained JS60 DNA (1 ng) instead of RNA.
Construction of Mycobacterium strain mc2155(pMV-CoM).
The JS60 EaCoMT gene etnE was amplified using the primers 60 M-F2 and 60 M-R2. The product (1.3 kb) was digested with EcoRV and HindIII, ligated into PvuII-HindIII-digested pMV261, and electroporated into strain JM109. The recombinant plasmid was designated pMV-CoM. Electrocompetent cells of Mycobacterium strain mc2155 were prepared essentially as described previously (21). Competent mc2155 cells (100 μl) were electroporated with plasmid pMV-CoM, recovered for 4 h with shaking in 1 ml of LB-Tween broth, and plated on LB-Km medium. Clones were screened by PCR with the 60 M-F2 and 60 M-R2 primers, using as the template 1 μl of cell lysate obtained from beadbeating a loopful of cells in 500 μl of Tris-HCl (5 mM; pH 8.0). The size of the plasmid in one PCR-positive clone was checked by electroporating 1 μl of beadbeater lysate into JM109, followed by plasmid extraction and restriction digestion. One clone, designated mc2155(pMV-CoM), was used for subsequent experiments. A transformant of mc2155 containing the pMV261 vector without insert DNA was prepared by similar methods for use as a negative control.
Expression of JS60 EaCoMT in strain mc2155(pMV-CoM).
Mycobacterium strain mc2155(pMV-CoM) and mc2155(pMV261) cultures were grown in LB-Tween-Km-Zn medium to an OD600 of 1.0 to 1.5, and EaCoMT expression was induced by heat shock (45°C for 30 min). Cell lysis and EaCoMT assays were as described above for JS60, except that 0.5% Tween was added to the lysis buffer. The protein concentration in the extracts was standardized to 3.0 to 3.5 mg/ml unless indicated otherwise. The activities of mc2155(pMV-CoM) and mc2155(pMV261) extracts with epoxypropane were assayed by similar methods.
Chlorooxirane transformation by cell extracts.
Chlorooxirane was synthesized by photochlorination of epoxyethane with tert-butyl hypochlorite (16, 20, 32, 52). Liquid epoxyethane (20 ml) and tert-butyl hypochlorite (1 ml) were reacted under N2 at 0°C for 45 min, illuminated by a 150 W incandescent bulb. Epoxyethane was removed by sparging with N2 at room temperature for 10 min. The resultant mixture of chlorooxirane and tert-butyl alcohol was diluted in anhydrous tetrahydrofuran (THF) and used immediately. Reactions were set up in crimp-sealed 10-ml bottles containing 755 μl of Tris-acetate buffer (50 mM; pH 8), 200 μl of cell extract [13 mg of protein/ml, from either mc2155(pMV-CoM) or mc2155(pMV261)], 20 μl of CoM (0.2 M), and 25 μl of chlorooxirane (12 mM in THF), with magnetic stirring. A control containing only Tris-acetate buffer (775 μl), MOPS-glycerol-DTT buffer (200 μl), and chlorooxirane (25 μl) was also included. The assays were conducted at 20°C to slow the rate of reactions and allow reasonable sampling times. At appropriate intervals, samples of the reaction mixtures (100 μl) were added to 4-(p-nitrobenzyl)pyridine reagent (500 μl) (16), and after 5 min triethylamine reagent (400 μl) (16) was added. The absorbance at 550 nm was measured, and the chlorooxirane concentration was calculated using the molar extinction coefficient (ɛ = 14,300 M/cm at 550 nm [5]).
Analysis of metabolites by MS.
Cell extract (12 mg of protein in 2 ml) from strain mc2155(pMV-CoM) was added to dialysis tubing (15-kDa cutoff) and dialyzed overnight at 4°C in 500 ml of MOPS-glycerol-DTT buffer. Dialyzed extract (50 μl) was added to crimp-sealed 25-ml serum bottles containing ammonium acetate (900 μl; 50 mM; pH 8.0) and CoM (50 μl; 200 mM). Epoxyethane (360 μl; 15 μmol) was added, and the bottles were incubated at 30°C with shaking at 300 rpm. At intervals, reaction mixtures were added to chilled prerinsed filters (5-kDa cutoff; Ultrafree-MC; Sigma) and centrifuged (5,000 × g; 20 min; 0°C). The eluates were injected directly into an LCQ Advantage mass spectrometer (Thermo Finnigan) operated in negative ionization electrospray injection mode. Tandem mass spectrometry (MS-MS) analysis was performed on the m/z 141 and 185 parent ions to determine their fragmentation patterns. Metabolites produced by extracts of mc2155(pMV261) cultures and in abiotic controls (no cell extract) were analyzed by similar methods.
RESULTS
Growth of Mycobacterium strain JS60 on VC and ethene.
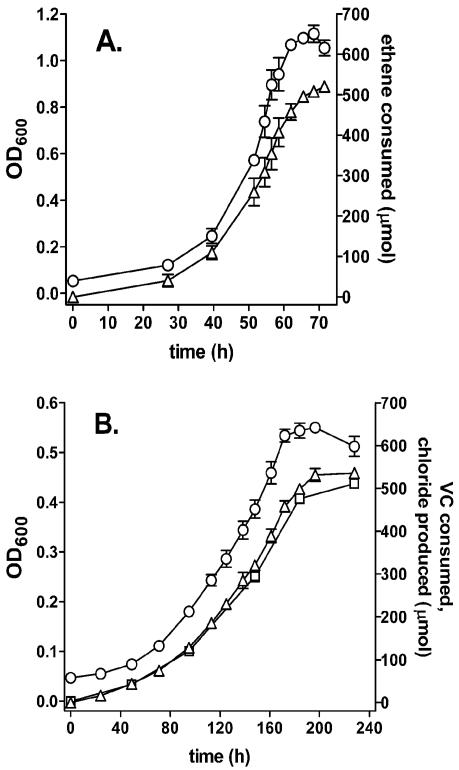
Near-stoichiometric production of inorganic chloride from VC (0.95 mol/mol) occurred during growth of strain JS60 on VC as the sole carbon source (Fig. 1B). Ethene was clearly a better substrate than VC in terms of both growth rate and growth yield (Fig. 1A). The maximum specific substrate utilization rates of whole cells calculated from the growth rates (Fig. 1) and growth yields (7) were 118 nmol/min/mg of protein (ethene) and 44 nmol/min/mg of protein (VC). The increases in growth rates and substrate utilization rates compared to those reported previously (7) are most likely due to optimization of culture conditions (the temperature was increased from 20 to 30°C, and the pH was decreased from 7.0 to 6.5).
FIG. 1.
Growth of Mycobacterium strain JS60 on ethene (A) and VC (B) as sole carbon and energy sources. ○, biomass measured as OD600; Δ, cumulative amount of substrate consumed; □, cumulative amount of chloride produced. Growth rates (0.080 h−1 with ethene, 0.017 h−1 with VC) were calculated by plotting an exponential curve through a subset of the OD600 data (results not shown). The inoculum for both experiments was a frozen stock of washed, VC-grown cells. Data are averages of three replicates, and error bars are the standard deviations.
Epoxyethane metabolism in cell extracts.
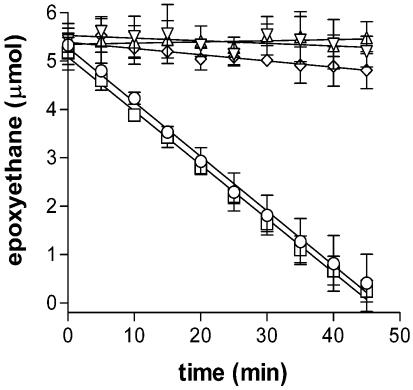
Various cofactors were tested for the ability to stimulate epoxyethane metabolism in extracts of ethene-grown Mycobacterium strain JS60. There was no significant loss of epoxyethane in the absence of cofactors (Fig. 2), and neither glutathione (48) nor a combination of NAD+ and CoA (10) stimulated activity under the assay conditions used (data not shown). Epoxyethane metabolism was rapid when CoM (2-mercaptoethanesulfonate) was added to cell extracts (Fig. 2), suggesting the presence of an EaCoMT enzyme (4). The specific activities of EaCoMT in extracts from VC-grown, ethene-grown, and acetate-grown cells were calculated to be 980 ± 50, 990 ± 60, and 0 ± 50 nmol/min/mg of protein (errors from 95% confidence intervals of linear regressions in Fig. 2) after the abiotic rate of epoxyethane loss was subtracted (13 ± 2 nmol/min in Tris buffer with CoM [data not shown]). The EaCoMT activity in extracts was an order of magnitude higher than the alkene oxidation rates calculated for whole cells (above), and it is more than sufficient to account for the measured growth rates on VC and ethene. The high EaCoMT activity in cell extracts is probably due in part to the high concentrations of reduced CoM and epoxyethane available to the enzyme under the assay conditions.
FIG. 2.
Effect of CoM on epoxyethane metabolism in JS60 cell extracts. ○, ethene-grown cell extract with CoM; □, VC-grown cell extract with CoM; ⋄, acetate-grown cell extract with CoM; ▿, ethene-grown cell extract with no cofactor; Δ, VC-grown cell extract with no cofactor. Data are the averages of three independent experiments, and error bars are the standard deviations.
Cloning of the EaCoMT gene from strain JS60.
PCR primers (CoM-F1 and CoM-R1) based on conserved regions of the EaCoMT genes of Xanthobacter strain Py2 (X79863) and Rhodococcus strain B-276 (AF426826) yielded several products in PCR amplifications with JS60 DNA. A major product at the expected size (981 bp) had very high sequence identity to the EaCoMT gene from strain B-276. Improved primers (CoM-F1L and CoM-R2) yielded a single strong EaCoMT PCR product (893 bp), which was subsequently used as a probe in Southern blotting experiments. Genomic DNA from JS60 digested with enzymes that did not cut within the 893-bp sequence (PstI, KpnI, NsiI, SphI, and NheI) gave one hybridizing band in all cases (data not shown), indicating that a single EaCoMT allele was present. The NheI fragment (8.4 kb) was cloned into E. coli strain JM109 using the pK18 vector, positive clones were detected by PCR screening, and the insert DNA from one representative clone (E4H) was sequenced.
Sequence analysis of the JS60 EaCoMT gene and flanking DNA.
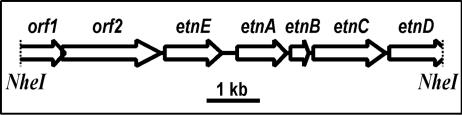
The NheI fragment of JS60 DNA (8,364 bp) (GenBank accession number AY243034) had an overall GC content of 59%, which is slightly lower than typical for Mycobacterium (54). Seven open reading frames (ORFs), the first and last of which were incomplete, were identified on one strand of the DNA (Fig. 3). Five of the ORFs started with ATG, one started with TTG, and all were preceded by plausible ribosome binding sites. Two genes possibly involved in acyl-CoA ester metabolism (ORF1 and ORF2) were located upstream of the EaCoMT gene, while genes likely to encode a four-component monooxygenase were located downstream (Table 2).
FIG. 3.
Schematic diagram of genes on the 8,364-bp NheI restriction fragment cloned from Mycobacterium strain JS60. orf1 and orf2 are likely to be involved in acyl-CoA ester metabolism, etnE encodes an EaCoMT, and the etnABCD genes encode a putative four-component alkene monooxygenase.
TABLE 2.
Gene products of the etn locus: predicted functions and database similarities
| ORF | Position in sequence (nucleotides) | Predicted molecular mass (Da) | Predicted function | Similar protein | Organism | Amino acid identity (%) | Protein accession no. |
|---|---|---|---|---|---|---|---|
| ORF1a | 0-795 | 28,767 | CoA transferase | GctB | Acidaminococcus fermentans | 29.9 | Q59112 |
| CatJ | Pseudomonas strain B13 | 27.8 | AAL02406 | ||||
| ORF2 | 783-2771 | 70,192 | Acyl-CoA synthetase | CAA10043 | Pseudomonas mendocina strain 35 | 30.4 | CAA10043 |
| FerA | Sphingomonas strain SYK6 | 28.3 | BAB86294 | ||||
| etnE | 2887-3996 | 40,928 | EaCoMT | AAL28081 | Rhodococcus strain B-276 | 71.3 | AAL28081 |
| XecA | Xanthobacter strain Py2 | 47.3 | CAA56241 | ||||
| etnA | 4315-5367 | 39,732 | Monooxygenase β-subunit | AmoA | Rhodococcus strain B-276 | 39.1 | D37875 |
| ThmB | Pseudonocardia strain K1 | 31.6 | CAC10509 | ||||
| etnB | 5391-5720 | 12,334 | Monooxygenase coupling-effector protein | AmoB | Rhodococcus strain B-276 | 50.8 | D37875 |
| MmoB | Methylocystis strain WI14 | 28.3 | AAF01270 | ||||
| etnC | 5767-7269 | 56,926 | Monooxygenase α-subunit | AmoC | Rhodococcus strain B-276 | 59.4 | BAA07114 |
| MmoX | Methylosinus strain OB3b | 35.1 | P27353 | ||||
| etnDa | 7389-8364 | 35,149 | Monooxygenase reductase | AmoD | Rhodococcus strain B-276 | 41.8 | BAA07115 |
| DmpP | Pseudomonas strain CF600 | 40.4 | P19734 |
Incomplete ORF.
The monooxygenase genes were similar in sequence and identical in arrangement to the propene monooxygenase genes (amoABCD) of Rhodococcus strain B-276. The sequence similarity, inducibility by VC and ethene (see below), and proximity to a gene involved in epoxyethane and chlorooxirane metabolism (see below) provide strong evidence that the four genes encode the VC-ethene monooxygenase of JS60. Due to the likelihood that ethene rather than VC was the original substrate of the putative monooxygenase, we designated the genes etnABCD. Because the JS60 EaCoMT gene was functional in epoxide metabolism and cotranscribed with at least etnA (see below), it was designated etnE.
Sequence alignment (data not shown) of the EtnC gene product with α-subunit components from methane (MmoX), propene (AmoC, XamoA), butane (BmoX), isoprene (IsoA), THF (ThmA), benzene (BmoA), phenol (DmpN), and toluene (TomA3) monooxygenases confirmed the presence of four conserved glutamate residues and two conserved histidine residues involved in binding nonheme iron atoms at the active site (15, 36, 39, 59). The sequence comparison therefore indicates that the JS60 ethene-VC monooxygenase is a binuclear iron enzyme. The National Center for Biotechnology Information conserved domain database (CDD) detected the presence of binding sites for FAD, NAD, and a 2Fe-2S iron-sulfur cluster in the EtnD gene product, as would be expected for a monooxygenase reductase (28, 55).
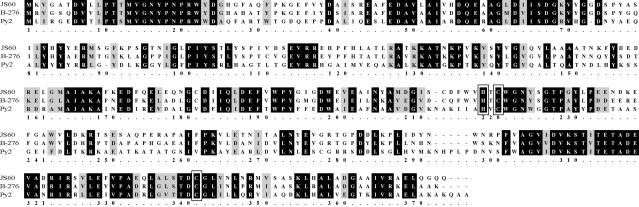
A sequence alignment of the EaCoMTs from JS60, Py2, and B-276 (Fig. 4) emphasized the high similarity of these enzymes, particularly those from JS60 and B-276. The His-X-Cys-Xn-Cys motif involved in zinc binding in the Py2 EaCoMT enzyme (26) and in the more distantly related methionine synthases and methanogenic CoM transferases (60) is conserved in the EaCoMT from strain JS60, which suggests that this enzyme also possesses a zinc cofactor.
FIG. 4.
Sequence alignment of EaCoMT proteins from Mycobacterium strain JS60, Rhodococcus strain B-276, and Xanthobacter strain Py2. Identical residues are shaded black, while similar residues are shaded gray. The conserved histidine (220) and cysteine (222, 343) residues likely to be involved in zinc binding are boxed.
RT-PCR of etn locus genes.
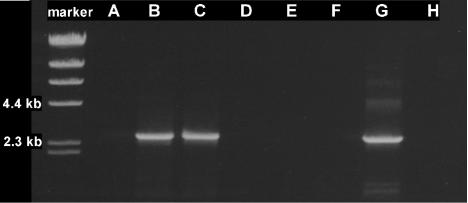
Primers (RTM-F1 and RTβ-R1) were designed to amplify a 2.5-kb region of the etn locus from the start of the EaCoMT gene etnE to the end of the putative monooxygenase β-subunit gene etnA. A product of the expected size was seen in RT-PCRs with RNA extracted from VC- or ethene-grown JS60 cultures, whereas no products were seen in reactions using RNA from acetate-grown JS60 cultures (Fig. 5). Negative controls lacking reverse transcriptase gave no detectable products, indicating that the amplicons observed in RT-PCRs were derived from RNA and not DNA. The results indicate that the etnE and etnA genes are cotranscribed and are inducible by ethene and VC.
FIG. 5.
RT-PCR analysis of etnEA expression in strain JS60. Lanes: A, acetate-grown cells; B, VC-grown cells; C, ethene-grown cells; D, acetate-grown cells (no RT); E, VC-grown cells (no RT); F, ethene-grown cells (no RT); G, DNA template; H, no template.
Heterologous expression of the JS60 EaCoMT gene.
Based on the work of Krum and Ensign with the EaCoMT from Xanthobacter strain Py2 (25), we initially attempted to express the JS60 EaCoMT gene etnE in E. coli by using a T7-based expression system [pET-21a vector, C43(DE3) host], but the experiments were unsuccessful (data not shown). We hypothesized that heterologous expression in a more closely related host strain might be more successful and, therefore, we cloned the etnE gene into Mycobacterium smegmatis mc2155 (40) under the control of the hsp60 promoter in the pMV261 vector (41). Good EaCoMT activity was obtained in cell extracts of the recombinant strain, designated mc2155(pMV-CoM).
In contrast to previously published work (41), we observed better expression of the cloned gene (increase in specific activity of approximately 30% [data not shown]) in extracts from mc2155(pMV-CoM) cultures that were heat shocked (45°C, 30 min) before harvest, and therefore cell extracts from heat-shocked cultures were used for all subsequent work. Our results indicate that the hsp promoter in the pMV261 vector is partially inducible above a basal constitutive expression level. The EaCoMT activity in mc2155(pMV-CoM) cell extracts with epoxyethane as a substrate (Table 3) was 62% of the activity in cell extracts of JS60 (Fig. 2). Epoxypropane was also transformed by mc2155(pMV-CoM) cell extracts, although the specific activity was only 23% of that seen with epoxyethane. Apart from the slow abiotic reaction between epoxide and CoM, no significant transformation of epoxyethane or epoxypropane was seen in cell extracts of strain mc2155(pMV261), confirming that the EaCoMT activities were due to expression of the cloned JS60 etnE gene. EaCoMT-like genes were not found in BLAST searches of the in-progress genome sequence of M. smegmatis strain mc2155.
TABLE 3.
EaCoMT activity in cell extracts of strains mc2155(pMV-CoM) and mc2155(pMV261)
| Substrate | EaCoMT activity (nmol/min/mg of protein)a,b (n)
|
|
|---|---|---|
| mc2155(pMV-CoM) | mc2155(pMV261) | |
| Epoxyethane | 615 ± 98 (6) | 12 ± 20 (3) |
| Epoxypropane | 143 ± 23 (4) | 1 ± 3 (4) |
Averages and standard deviations derived from several independent experiments.
Activities calculated from substrate depletion rate, after subtraction of abiotic rates of substrate loss in controls without cell extract. The abiotic rates of epoxyethane and epoxypropane loss were 13 ± 2 and 12 ± 2 nmol/min, respectively.
Chlorooxirane metabolism in cell extracts.
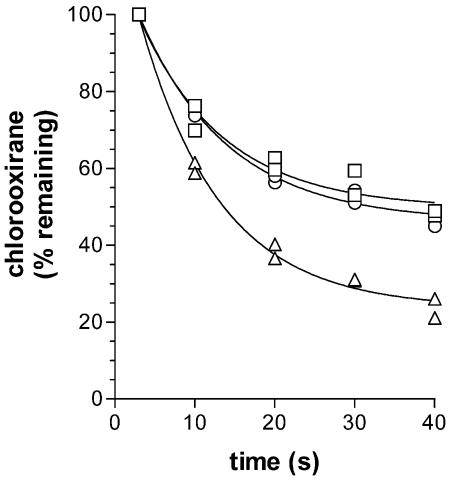
Transformation of chlorooxirane was more extensive, and the initial rate more rapid, in reactions containing strain mc2155(pMV-CoM) extracts than in reactions containing strain mc2155(pMV261) extracts or buffer alone (Fig. 6). Based on the data between 3 and 20 s, the enzyme-catalyzed rate of reaction was calculated to be 57 nmol/min/mg of protein at 20°C. This specific activity should be considered an approximation, due to both the nonlinearity of the data and the very high rate of abiotic chlorooxirane decomposition. Although much lower than the activity with epoxyethane (Fig. 2), the EaCoMT activity with chlorooxirane is sufficient to account for VC oxidation (10 nmol/min/mg of protein in whole cells at 20°C [7]), which would be essential to avoid any accumulation of the highly reactive chlorooxirane in the cells. The reason for the incomplete metabolism of chlorooxirane in the assay is unknown. CoM was present in 10-fold excess and, thus, is not expected to be limiting. Either a high Km or inactivation of the EaCoMT enzyme by chlorooxirane could be responsible. Respiking of chlorooxirane or desalting of the mixture and repeating the assay would distinguish between these possibilities. In combination with the results from JS60 cell extracts (Fig. 2) and RT-PCR (Fig. 5), the results indicate that the JS60 EaCoMT enzyme, EtnE, can catalyze the transformation of chlorooxirane in addition to epoxyethane and thus plays a role in both the ethene and VC catabolic pathways.
FIG. 6.
Chlorooxirane transformation in cell extract reactions. ○, buffer alone; □, buffer, CoM, and cell extract from strain mc2155(pMV261); Δ, buffer, CoM, and cell extract from strain mc2155(pMV-CoM). Data from two experiments are shown. Chlorooxirane was quantified by the absorbance (550 nm) of its 4-(p-nitrobenzyl)pyridine adduct. Initial concentrations of chlorooxirane in the assays ranged from 0.2 to 0.3 mM.
Analysis of metabolites produced from epoxyethane.
Reactions between CoM and epoxyethane were analyzed by MS (Fig. 7), using dialyzed cell extracts from strain mc2155(pMV-CoM) assayed in ammonium acetate buffer. Under these conditions, the EaCoMT activity was approximately 75% of that seen with undialyzed extracts in Tris buffer (data not shown). A strong peak at m/z 141 that yielded a fragment ion at m/z 81 was present at the start of the reactions. The spectrum matched that of a CoM standard, and thus m/z 141 and m/z 81 are probably HS-CH2-CH2-SO3− and HSO3−, respectively. After 1 h, the m/z 141 peak was almost undetectable and a peak at m/z 185 was dominant. Based on analogy with the CoM adduct in the propene assimilation pathway (4), the m/z 185 ion is consistent with 2-hydroxyethyl-CoM (HO-CH2-CH2-S-CH2-CH2-SO3−). The proposed structure is supported by the fragmentation pattern, assuming that m/z 77 is HO-CH2-CH2-S−, m/z 81 is HSO3−, and m/z 107 is CH2=CH-SO3−. Several minor peaks also appeared during the course of the EaCoMT reaction. The ion at m/z 208 could be derived from MOPS buffer. The peaks at m/z 233 and 277 are possibly glycerol adducts (sulfonate esters) of m/z 141 and 185, respectively.
FIG. 7.
MS analysis of metabolites formed from epoxyethane during EaCoMT reaction. (A) Zero time sample; (B) 30-min sample; (C) 1-h sample.
The m/z 185 ion did not appear in reactions when either CoM or epoxyethane was omitted, but it was seen in reactions containing cell extract from strain mc2155(pMV261) and also in abiotic reactions containing only buffer, CoM, and epoxyethane (data not shown). In these latter two cases, the intensity of the m/z 185 ion after 1 h of incubation was approximately 10% of that observed with cell extract from strain mc2155(pMV-CoM). The data suggest that the JS60 EaCoMT enzyme increases the rate of a reaction that also occurs spontaneously between epoxyethane and CoM.
DISCUSSION
We have conclusively determined the mechanism responsible for bacterial epoxyethane metabolism, and we obtained strong circumstantial evidence that the same mechanism is also involved in chlorooxirane metabolism. Three lines of investigation support the latter conclusion: the JS60 etnE gene was expressed in response to both VC and ethene; there were equal levels of EaCoMT activity in extracts from VC- and ethene-grown JS60 cells; and chlorooxirane was transformed more rapidly by cell extracts of strain mc2155(pMV-CoM) than of strain mc2155(pMV261). Our finding of EaCoMT in strain JS60 adds Mycobacterium to the small group of eubacterial genera (Xanthobacter, Rhodococcus) thus far known to use CoM in catabolic reactions (25). The fact that Mycobacterium strain JS60 contains an active EaCoMT enzyme and grows on alkenes in minimal medium indicates that the strain can biosynthesize CoM, although we did not obtain direct evidence of this.
Our results suggest answers to questions raised by earlier studies on epoxyethane metabolism in other Mycobacterium strains. It is likely that CoM is the unknown heat-stable, dialyzable cofactor that was required for epoxyethane metabolism in cell extracts of Mycobacterium strain E20 (10). The authors of that study noted that only extracts from ethene-grown cells contained the stimulatory cofactor, which agrees with the more recent finding of inducible CoM biosynthesis in Xanthobacter strain Py2 and Rhodococcus strain B-276 (25). It is also possible that EaCoMT is responsible for epoxyethane metabolism in the dibromoethane-degrading Mycobacterium strain GP1 (33), which is phylogenetically similar to strain JS60. In strain GP1, epoxyethane is produced from 2-bromoethanol through the action of a haloalcohol dehalogenase, but the mechanism of subsequent epoxyethane transformation was not elucidated.
Early experiments with ethene- and VC-assimilating mycobacteria indicated that epoxyethane metabolism was dependent on NAD and CoA (10, 18). Although we saw no evidence of a requirement for such cofactors, our reaction conditions were very different in that they contained an exogenous supply of CoM, 10- to 50-fold less protein, and 10- to 50-fold more epoxyethane. Different strains of alkene-utilizing mycobacteria could possess different mechanisms of epoxide metabolism, but based on preliminary results from several other strains (unpublished data) we believe this is unlikely. NAD and CoA could play a role in the downstream reactions of the ethene pathway, e.g., in regenerating the reduced form of CoM. In our reactions that contained an excess of CoM, recycling of the cofactor would not be expected to affect the initial reaction rate, unlike in earlier studies where CoM would have been extremely rate limiting.
The JS60 EaCoMT accepted epoxypropane as a substrate, although the strain does not grow on propene (data not shown). The activities of the EaCoMTs from the propene-oxidizing strains with epoxyethane have not been reported, but such activity might be expected at least in strain Py2, which can grow (albeit slowly) on ethene as a carbon source (45). The amino acid identity (71.3%) between the EaCoMTs of strains JS60 and B-276 was much higher than that between the Py2 and B-276 enzymes (49.2%). This is unusual in light of the fact that the enzymes from Py2 and B-276 are both part of a propene assimilation pathway, whereas the JS60 enzyme is part of an ethene and VC assimilation pathway. It appears that, in this case, the similarity of the catabolic enzymes was better predicted by the phylogeny of the strains than by the substrates of the pathways. The correspondence between JS60 and B-276 held also for the alkene monooxygenase genes in terms of gene sequence and gene organization. The alkene monooxygenases of strains JS60 and B-276 are both more similar to each other than to anything else currently in GenBank.
Strong evidence that we have identified the VC-ethene monooxygenase genes comes from the sequence and operon similarity of etnABCD to the strain B-276 propene monooxygenase (amoABCD), the expression of etnA in response to growth on ethene and VC, and the cotranscription of etnA with the EaCoMT gene etnE. We attempted to express the etnABC genes with the pMV261/mc2155 cloning system, but we did not detect any activity against ethene (data not shown). This failure could be due to the need for a monooxygenase reductase in addition to the alpha, beta, and coupling protein components (etnD was incomplete in the DNA fragment we cloned). It is conceivable that additional monooxygenase genes are found downstream of etnABCD, but based on the high sequence similarity and identical gene organization of the JS60 and B-276 systems we believe that the JS60 monooxygenase is a four-component enzyme.
The genetic organization of the etn locus in strain JS60 was considerably different from that of the alkene catabolic genes of Xanthobacter strain Py2 (27). In JS60, the EaCoMT gene etnE is located upstream of the alkene monooxygenase genes etnABCD, and it is cotranscribed with at least etnA. In contrast, the Py2 EaCoMT gene xecA is in an operon with the other three components of the epoxide carboxylase complex, and it is not immediately adjacent to the alkene monooxygenase genes. The organization of the etn locus in JS60 is more reminiscent of the isoprene gene cluster of Rhodococcus strain AD45 (47), where the GST gene isoI responsible for epoxide metabolism is just upstream of the alkene monooxygenase genes isoABCDEF. However, in strain AD45 a transcriptional terminator is present between the GST and monooxygenase genes, unlike in JS60, where transcription of the alkene monooxygenase and EaCoMT genes appears to be coupled.
We found no evidence for epoxide carboxylase genes (1, 2) near the etnE gene of strain JS60, but we did not attempt to find such genes elsewhere in the genome or assay for the corresponding enzyme activities. Based on the propene assimilation pathway determined by Ensign (11), the most likely product of a carboxylase reaction with 2-hydroxyethyl-CoM would be malonate semialdehyde, which can readily support bacterial growth (31, 57). Downstream metabolism in the VC and ethene pathways most likely occurs via acetyl-CoA, based on the results of previous work with Mycobacterium strain E20 (9, 10). Similar fluoroacetate inhibition effects to those reported in strain E20 were also seen in strain JS60 (i.e., epoxide accumulation in resting cell assays [data not shown]). Clues to the metabolic intermediates between 2-hydroxyethyl-CoM and acetyl-CoA may be found in the genes (orf1, orf2) upstream of the JS60 EaCoMT gene. The products of these two genes are similar to CoA transferase and acyl-CoA synthetase, respectively, and thus it is plausible that ORF1 and ORF2 transfer the alkene-derived two-carbon unit from CoM to CoA.
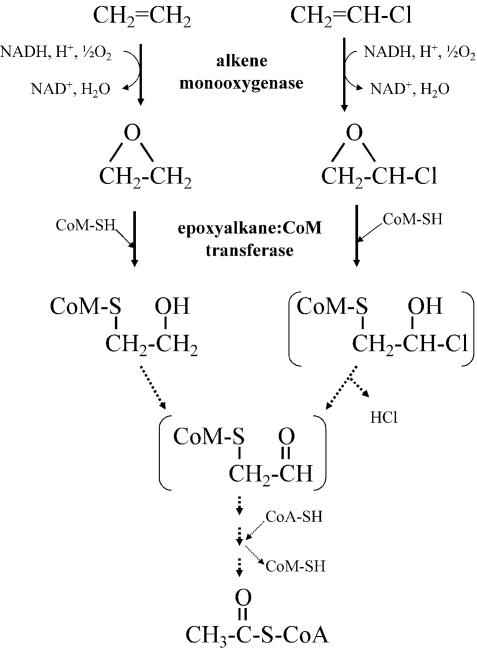
Our enzyme assays and RT-PCR experiments indicate that the alkene monooxygenase and EaCoMT enzymes of strain JS60 are active in both the VC and ethene assimilation pathways. The point at which the two pathways converge is unclear at present: of key importance is the mechanism and timing of chloride release in the VC pathway. Several different metabolites could result from the nucleophilic attack of CoM on chlorooxirane. Consideration of metabolic economy leads us to believe that 2-chloro-2-hydroxyethyl-CoM is most likely, because it could spontaneously eliminate HCl (42) to give an aldehyde that could readily feed into the ethene assimilation pathway. VC and ethene catabolic pathways consistent with our results are shown in Fig. 8.
FIG. 8.
Proposed pathways of VC and ethene assimilation in Mycobacterium strains. Intermediates that have not been identified are in brackets, and hypothetical reactions are indicated by dotted lines.
The cloning and sequencing of EaCoMT and alkene monooxygenase genes reported in the present study provide a foundation for the development of DNA-based methods of detecting VC- and ethene-assimilating mycobacteria in the environment. Such methods would be invaluable for monitoring natural attenuation and bioaugmentation processes at chlorinated-ethene-contaminated sites. The alkene monooxygenase and EaCoMT of Mycobacterium strain JS60 could also have applications in biocatalysis, for example in the stereoselective synthesis of epoxides.
Acknowledgments
We thank Torin Weisbrod and Bill Jacobs for supplying pMV261 and mc2155. F. P. Guengerich advised on chlorooxirane synthesis. Heather Luckarift and Lloyd Nadeau assisted with MS analyses. George Paoli and Glenn Johnson are thanked for stimulating discussions on molecular biology.
This work was funded by the U.S. Strategic Environmental Research and Development Program. N.V.C. was supported by a postdoctoral fellowship from the Oak Ridge Institute for Science and Education (U.S. Department of Energy).
REFERENCES
- 1.Allen, J. R., and S. A. Ensign. 1997. Characterization of three protein components required for functional reconstitution of the epoxide carboxylase multienzyme complex from Xanthobacter strain Py2. J. Bacteriol. 179:3110-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, J. R., and S. A. Ensign. 1998. Identification and characterization of epoxide carboxylase activity in cell extracts of Nocardia corallina B276. J. Bacteriol. 180:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, J. R., and S. A. Ensign. 1997. Purification to homogeneity and reconstitution of the individual components of the epoxide carboxylase multiprotein enzyme complex from Xanthobacter strain Py2. J. Biol. Chem. 272:32121-32128. [DOI] [PubMed] [Google Scholar]
- 4.Allen, J. R., D. D. Clark, J. G. Krum, and S. A. Ensign. 1999. A role for coenzyme M (2-mercaptoethanesulfonic acid) in a bacterial pathway of aliphatic epoxide carboxylation. Proc. Natl. Acad. Sci. USA 96:8432-8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbin, A., J.-C. Béréziat, A. Croisy, I. K. O'Neill, and H. Bartsch. 1990. Nucleophilic selectivity and reaction kinetics of chloroethylene oxide assessed by the 4-(p-nitrobenzyl)pyridine assay and proton nuclear magnetic resonance spectroscopy. Chem. Biol. Interact. 73:261-277. [DOI] [PubMed] [Google Scholar]
- 6.Coleman, N. V., T. E. Mattes, J. M. Gossett, and J. C. Spain. 2002. Biodegradation of cis-dichloroethene as the sole carbon source by a beta-proteobacterium. Appl. Environ. Microbiol. 68:2726-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman, N. V., T. E. Mattes, J. M. Gossett, and J. C. Spain. 2002. Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl. Environ. Microbiol. 68:6162-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Bont, J. A. M. 1976. Oxidation of ethylene by soil bacteria. Antonie Leeuwenhoek 42:59-71. [DOI] [PubMed] [Google Scholar]
- 9.de Bont, J. A. M., and R. A. J. M. Albers. 1976. Microbial metabolism of ethylene. Antonie Leeuwenhoek 42:73-80. [DOI] [PubMed] [Google Scholar]
- 10.de Bont, J. A. M., and W. Harder. 1978. Metabolism of ethylene by Mycobacterium E 20. FEMS Microbiol. Lett. 3:89-93. [Google Scholar]
- 11.Ensign, S. A. 2001. Microbial metabolism of aliphatic alkenes. Biochemistry 40:5845-5853. [DOI] [PubMed] [Google Scholar]
- 12.Fennell, D. E., A. B. Carroll, J. M. Gossett, and S. H. Zinder. 2001. Assessment of indigenous reductive dechlorinating potential at a TCE-contaminated site using microcosms, polymerase chain reaction analysis, and site data. Environ. Sci. Technol. 35:1830-1839. [DOI] [PubMed] [Google Scholar]
- 13.Fox, B. G., W. A. Froland, J. E. Dege, and J. D. Lipscomb. 1989. Methane monooxygenase from Methylosinus trichosporium OB3b. Purification and properties of a three-component system with high specific activity from a type II methanotroph. J. Biol. Chem. 264:10023-10033. [PubMed] [Google Scholar]
- 14.Fukuda, H., T. Ogawa, and S. Tanase. 1993. Ethylene production by micro-organisms. Adv. Microb. Physiol. 35:275-306. [DOI] [PubMed] [Google Scholar]
- 15.Gallagher, S. C., R. Cammack, and H. Dalton. 1997. Alkene monooxygenase from Nocardia corallina B-276 is a member of the class of dinuclear iron proteins capable of stereospecific epoxygenation reactions. Eur. J. Biochem. 247:635-641. [DOI] [PubMed] [Google Scholar]
- 16.Guengerich, F. P., W. M. Crawford, Jr., and P. G. Watanabe. 1979. Activation of vinyl chloride to covalently bound metabolites: roles of 2-chloroethylene oxide and 2-chloroacetaldeyhde. Biochemistry 18:5177-5182. [DOI] [PubMed] [Google Scholar]
- 17.Habets-Crützen, A. Q. H., L. E. S. Brink, C. G. van Ginkel, J. A. M. de Bont, and J. Tramper. 1984. Production of epoxides from gaseous alkenes by resting-cell suspensions and immobilized cells of alkene-utilizing bacteria. Appl. Microbiol. Biotechnol. 20:245-250. [Google Scholar]
- 18.Hartmans, S., and J. A. M. de Bont. 1992. Aerobic vinyl chloride metabolism in Mycobacterium aurum L1. Appl. Environ. Microbiol. 58:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmans, S., F. J. Weber, D. P. M. Somhorst, and J. A. M. de Bont. 1991. Alkene monooxygenase from Mycobacterium: a multicomponent enzyme. J. Gen. Microbiol. 137:2555-2560. [DOI] [PubMed] [Google Scholar]
- 20.Holt, S., T. Y. Yen, R. Sangaiah, and J. A. Swenberg. 1998. Detection of 1,N6-ethenoadenine in rat urine after chloroethylene oxide exposure. Carcinogenesis 19:1763-1769. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs, W. R., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 22.Keppler, F., R. Borchars, J. Pracht, S. Rheinberger, and H. Scholer. 2002. Natural formation of vinyl chloride in the terrestrial environment. Environ. Sci. Technol. 36:2479-2483. [DOI] [PubMed] [Google Scholar]
- 23.Kielhorn, J., C. Melber, U. Wahnschaffe, A. Aitio, and I. Mangelsdorf. 2000. Vinyl chloride: still a cause for concern. Environ. Health Perspect. 108:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koziollek, P., D. Bryniok, and H.-J. Knackmuss. 1999. Ethene as an auxiliary substrate for the cooxidation of cis-dichloroethene and vinyl chloride. Arch. Microbiol. 172:240-246. [DOI] [PubMed] [Google Scholar]
- 25.Krum, J. G., and S. A. Ensign. 2000. Heterologous expression of bacterial epoxyalkane:coenzyme M transferase and inducible coenzyme M biosynthesis in Xanthobacter strain Py2 and Rhodococcus rhodochrous B276. J. Bacteriol. 182:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krum, J. G., H. Ellsworth, R. R. Sargeant, G. Rich, and S. A. Ensign. 2002. Kinetic and microcalorimetric analysis of substrate and cofactor interactions in epoxyalkane:CoM transferase, a zinc-dependent epoxidase. Biochemistry 41:5005-5014. [DOI] [PubMed] [Google Scholar]
- 27.Krum, J. G., and S. A. Ensign. 2001. Evidence that a linear megaplasmid encodes enzymes of aliphatic alkene and epoxide metabolism and coenzyme M (2-mercaptoethanesulfonate) biosynthesis in Xanthobacter strain Py2. J. Bacteriol. 183:2172-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lund, J., M. P. Woodland, and H. Dalton. 1985. Electron transfer reactions in the soluble methane monooxygenase of Methylococcus capsulatus (Bath). Eur. J. Biochem. 147:297-305. [DOI] [PubMed] [Google Scholar]
- 29.Maymó-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1, 2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maymó-Gatell, X., Y.-T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 31.Menendez, C., Z. Bauer, H. Huber, N. Gad'on, K. O. Stetter, and G. Fuchs. 1999. Presence of acetyl coenzyme A (CoA) carboxylase and propionyl-CoA carboxylase in autotrophic Crenarchaeota and indication for operation of a 3-hydroxypropionate cycle in autotrophic carbon fixation. J. Bacteriol. 181:1088-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mintz, M. J., and C. Walling. 1969. tert-Butyl hypochlorite. Organic Syntheses 49:9-12. [Google Scholar]
- 33.Poelarends, G. J., J. E. T van Hylckama Vlieg, J. R. Marchesi, L. M. Freitas dos Santos, and D. B. Janssen. 1999. Degradation of 1, 2-dibromoethane by Mycobacterium sp. strain GP1. J. Bacteriol. 181:2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pridmore, R. D. 1987. New and versatile cloning vectors with kanamycin-resistance marker. Gene 56:309-312. [DOI] [PubMed] [Google Scholar]
- 35.Rink, R., M. Fennema, M. Smids, U. Dehmel, and D. B. Janssen. 1997. Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J. Biol. Chem. 272:14650-14657. [DOI] [PubMed] [Google Scholar]
- 36.Rosenzweig, A. C., C. A. Frederick, S. J. Lippard, and P. Nordlund. 1993. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature 366:537-543. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Semprini, L. 1995. In situ bioremediation of chlorinated solvents. Environ. Health Perspect. 103:101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Small, F. J., and S. A. Ensign. 1997. Alkene monooxygenase from Xanthobacter strain Py2. J. Biol. Chem. 272:24913-24920. [DOI] [PubMed] [Google Scholar]
- 40.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 41.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. Jacobs, and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 42.Stucki, G., U. Krebser, and T. Leisinger. 1983. Bacterial growth on 1,2-dichloroethane. Experientia 39:1271-1273. [DOI] [PubMed] [Google Scholar]
- 43.Thier, R., and H. M. Bolt. 2000. Carcinogenicity and genotoxicity of ethylene oxide: new aspects and recent advances. Crit. Rev. Toxicol. 30:595-608. [DOI] [PubMed] [Google Scholar]
- 44.Tornqvist, M. 1994. Is ambient ethene a cancer risk factor? Environ. Health Perspect. 102:157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Ginkel, C. G., and J. A. M. de Bont. 1986. Isolation and characterization of alkene-utilizing Xanthobacter spp. Arch. Microbiol. 145:403-407. [Google Scholar]
- 46.van Ginkel, C. G., H. G. J. Welten, and J. A. M. de Bont. 1987. Oxidation of gaseous and volatile hydrocarbons by selected alkene-utilizing bacteria. Appl. Environ. Microbiol. 53:2903-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Hylckama Vlieg, J. E., H. Leemhuis, J. H. L. Spelberg, and D. B. Janssen. 2000. Characterization of the gene cluster involved in isoprene metabolism in Rhodococcus sp. strain AD45. J. Bacteriol. 182:1956-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Hylckama Vlieg, J. E. T., J. Kingma, A. J. van den Wijngaard, and D. B. Janssen. 1998. A glutathione S-transferase with activity towards cis-dichloroepoxyethane is involved in isoprene utilization by Rhodococcus sp. strain AD45. Appl. Environ. Microbiol. 64:2800-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verce, M. F., C. K. Gunsch, A. S. Danko, and D. L. Freedman. 2002. Cometabolism of cis-1,2-dichloroethene by aerobic cultures grown on vinyl chloride as the primary substrate. Environ. Sci. Technol. 36:2171-2177. [DOI] [PubMed] [Google Scholar]
- 50.Verce, M. F., R. L. Ulrich, and D. L. Freedman. 2000. Characterization of an isolate that uses vinyl chloride as a growth substrate under aerobic conditions. Appl. Environ. Microbiol. 66:3535-3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verce, M. F., R. L. Ulrich, and D. L. Freedman. 2001. Transition from cometabolic to growth-linked biodegradation of vinyl chloride by a Pseudomonas sp. isolated on ethene. Environ. Sci. Technol. 35:4242-4251. [DOI] [PubMed] [Google Scholar]
- 52.Walling, C., and P. S. Fredericks. 1962. Positive halogen compounds. IV. Radical reactions of chlorine and t-butyl hypochlorite with some small ring compounds. J. Am. Chem. Soc. 84:3326-3331. [Google Scholar]
- 53.Wang, K. L., H. Li, and J. R. Ecker. 2002. Ethylene biosynthesis and signaling networks. Plant Cell 14:131-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wayne, L. G., and G. P. Kubica. 1986. Genus Mycobacterium, p. 1436-1457. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md.
- 55.Weber, F. J., W. J. H. van Berkel, S. Hartmans, and J. A. M. de Bont. 1992. Purification and properties of the NADH reductase component of alkene monooxygenase from Mycobacterium strain E3. J. Bacteriol. 174:3275-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Witt, M. E., G. M. Klecka, E. J. Lutz, T. A. Ei, N. R. Grosso, and F. H. Chapelle. 2002. Natural attenuation of chlorinated solvents at Area 6, Dover Air Force Base: groundwater biogeochemistry. J. Contam. Hydrol. 57:61-80. [DOI] [PubMed] [Google Scholar]
- 57.Yamada, E. W., and W. B. Jakoby. 1960. Aldehyde oxidation. V. Direct conversion of malonic semialdehyde to acetyl-coenzyme A. J. Biol. Chem. 235:589-594. [PubMed] [Google Scholar]
- 58.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 59.Zhou, N. Y., A. Jenkins, C. K. Chan Kwo Chion, and D. J. Leak. 1998. The alkene monooxygenase from Xanthobacter Py2 is a binuclear non-haem iron protein closely related to toluene 4-monooxygenase. FEBS Lett. 430:181-185. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, Z. S., K. Peariso, J. E. Penner-Hahn, and R. G. Matthews. 1999. Identification of the zinc ligands in cobalamin-independent methionine synthase (MetE) from Escherichia coli. Biochemistry 38:15915-15926. [DOI] [PubMed] [Google Scholar]