Abstract
Two oxygen-responsive regulatory systems controlling numerous symbiotic genes in Bradyrhizobium japonicum were assayed in free-living cultures for their capacity to activate target genes under different oxygen conditions. NifA- and FixLJ-controlled target genes showed disparate relative expression patterns. Induction of NifA-dependent genes was observed only at oxygen concentrations below 2% in the gas phase, whereas that of FixLJ-controlled targets progressively increased when the oxygen concentration was lowered from 21 to 5, 2, or 0.5%. We propose that this reflects a response to a gradient of increasing oxygen deprivation as bacteria invade their host during root nodule development.
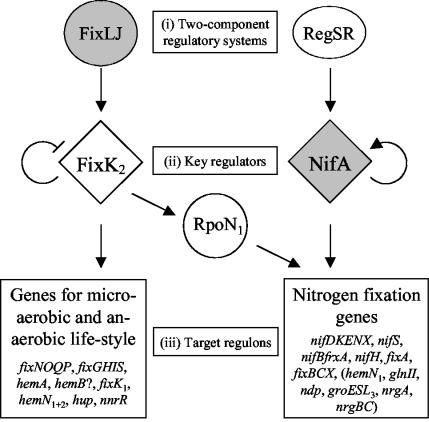
Expression of nitrogen fixation genes and symbiosis-related genes in the soybean symbiont Bradyrhizobium japonicum largely depends on two regulatory cascades (Fig. 1; see reference 10 for a review). In each cascade, three levels can be distinguished: (i) the two-component regulatory system level (FixLJ and RegSR), (ii) the key regulator level (FixK2 and NifA), and (iii) the target regulon level. The first cascade is composed of the two-component regulatory system FixLJ, the key regulator FixK2, and its regulon, comprising genes related to microaerobic and anaerobic life styles. Transcription factor FixJ is phosphorylated by the oxygen-inhibitable sensor kinase FixL and activates transcription of fixK2. FixK2 down-regulates the expression of its own gene and acts as a transcriptional activator of fixK1, the fixNOQP and fixGHIS operons for the synthesis of a high-affinity terminal oxidase (21), heme biosynthetic genes (hemA [24] and possibly hemB [6] and hemN1+2 [14]), uptake hydrogenase genes (hup [9]), the nnrR gene required for transcriptional activation of denitrification genes (20), and the rpoN1 gene encoding σ54 (RpoN1), the specialized σ factor (21).
FIG. 1.
Schematic representation of the two regulatory cascades controlling genes required for microaerobic and anaerobic lifestyles and for nitrogen fixation in B. japonicum. The latter group also includes genes that are not directly involved in nitrogen fixation (in parentheses). Oxygen-responsive regulatory components are shaded gray. For details, see text.
Activation of the second cascade is initiated by the RegSR two-component regulatory system via induction of the fixR-nifA operon (4). Transcription of this operon is controlled by two overlapping but functionally distinct promoters, fixRp1 and fixRp2, that are dependent on NifA and RpoN and on RegSR, respectively (1, 2). The precise nature of the signal that is transduced by the RegSR system to the level of fixRp2 activity is presently unknown (see below). By contrast, on the basis of indirect evidence, NifA was suggested to sense oxygen or redox conditions directly, probably via a metal cofactor (12, 13, 19). Under microoxic or anoxic conditions, active NifA enhances its own synthesis via induction of the fixRp1 promoter. In addition, it activates expression of the NifA regulon that comprises nitrogen fixation genes (e.g., the nitrogenase structural genes) and additional genes (22). NifA acts in concert with the RNA polymerase containing σ54, which, in B. japonicum, is encoded by two highly similar, functionally equivalent genes (rpoN1 and rpoN2 [18]). Thus, RpoN1 represents a link between the two regulatory cascades (Fig. 1). The FixR protein which is encoded by the promoter-proximal gene of the fixR-nifA operon belongs to the protein family of short chain dehydrogenases-reductases (Pfam database accession number PF00106 [3; http://www.sanger.ac.uk/Software/Pfam/]), yet its precise function in B. japonicum is unknown.
Although it was known from previous studies that the FixLJ- and NifA-mediated transcriptional activation is oxygen responsive, neither the kinetics of induction nor the question of whether or not the two systems differ in their oxygen responsiveness has been addressed in detail in the past. These issues are subjects of the present work.
Modulation of the transcriptional activation function of NifA and FixLJ by oxygen.
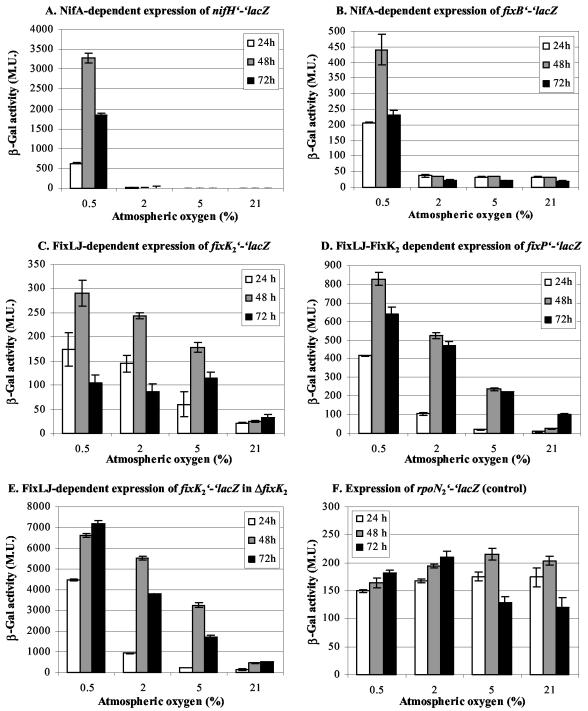
To test the activity of FixLJ and NifA in vivo, we used B. japonicum strains harboring chromosomally integrated, translational lacZ fusions to the corresponding direct or subordinate target genes fixK2 (strain 9054 [21]), fixP (strain 3604 [28]), nifH (strain 110-48 [15]), and fixB (strain 110-47 [15]). The strains were grown at different oxygen concentrations and assayed for β-galactosidase activity after 24, 48, and 72 h (Fig. 2). Monitoring over a period of three days was crucial in order to account for the differences in growth rates resulting from the different oxygen concentrations present in the gas phase of the individual cultures. B. japonicum strain 8015, harboring a chromosomal rpoN2′-′lacZ fusion (18), always served as a control for gene expression, because its expression is largely oxygen insensitive (Fig. 2F).
FIG. 2.
Expression (dependent on the oxygen-responding regulators NifA [A and B] and FixLJ [C to E]) of chromosomally integrated, translational lacZ reporter gene fusions. B. japonicum strains 110-48 (nifH′-′lacZ [A]), 110-47 (fixB′-′lacZ [B]), 9054 (fixK2′-′lacZ [C]), 3604 (fixP′-′lacZ [D]), 9054K2 (fixK2′-′lacZ in ΔfixK2 [E] [21]) and 8015 (rpoN2′-′lacZ [control] [F]) were grown at 30°C in 20 ml of PSY medium (25) supplemented with 0.1% l-arabinose in 500-ml infusion bottles (microaerobic cultures) or in 500-ml Erlenmeyer flasks (aerobic cultures). The gas atmosphere in the microaerobic cultures was replaced with an oxygen-nitrogen mixture containing the indicated concentration of oxygen and refreshed two times every 24 h. Thus, the oxygen concentrations indicated refer to maximal oxygen concentrations in the gas phase, which probably decreased slightly until the next flush due to oxygen consumption of the growing cultures. Samples were collected after 24, 48, and 72 h and assayed for β-galactosidase activity (indicated in Miller units [M. U.]) as described previously (11). The results of a typical experiment, in which two parallel cultures of each strain were grown and assayed in duplicate, are shown. Two to four independent experiments were performed with individual strains.
No expression of nifH′-′lacZ was detected under oxic conditions (21% oxygen) or in 5 and 2% oxygen (Fig. 2A). However, strong induction (>500-fold) was observed in cells grown for 48 h in 0.5% oxygen. Similarly, the fixB′-′lacZ fusion was expressed only very weakly in 21, 5, and 2% oxygen but it was strongly induced (>14-fold) after growth for 48 h in 0.5% oxygen (Fig. 2B). Thus, maximal induction of NifA-dependent gene expression required <2% oxygen to be present in the gas phase of the culture system used in these experiments. The reason for the significant difference in basal expression of the nifH and the fixB reporter fusion at oxygen concentrations of ≥2% is not obvious. The difference might have been caused by the presence of a cryptic, NifA-independent promoter associated with fixB.
Expression of the FixLJ-dependent fixK2-lacZ fusion in the wild type showed a low, basal level under oxic conditions which, however, increased at least sevenfold after 48-h growth at an oxygen concentration of 5% (Fig. 2C). The induction factor (relative to aerobic conditions) gradually increased when cultures were grown at lower oxygen concentrations for 24 or 48 h. Essentially the same expression pattern was observed for the fixP′-′lacZ reporter fusion which is indirectly controlled by FixLJ via FixK2 (Fig. 2D). Thus, unlike NifA, the FixLJ system is (partially) active at relatively high oxygen concentrations and mediates gradual activation of target genes in response to decreasing oxygen conditions. It is important to note here that microaerobic conditions per se are not sufficient for FixLJ-FixK2-mediated activation of genes involved in hydrogen uptake and denitrification (via nnrR). Maximal induction of hup genes requires the simultaneous presence of hydrogen gas and nickel (9 and references therein), whereas that of denitrification genes depends on N oxides such as nitrate or nitrite (20). In the latter case, transduction of the N oxide signal involves the product of nnrR, which is the primary target of FixLJ-FixK2-mediated control.
In previous work, Nellen-Anthamatten et al. demonstrated that fixK2 expression is negatively autoregulated by a factor of at least 10 (21). Therefore, we have also assayed fixK2-lacZ expression in a fixK2 mutant strain. Expression levels were indeed strongly elevated, but the relative expression pattern observed under the different oxygen conditions was the same as that determined in the fixK2+ background (Fig. 2E). Thus, oxygen-mediated regulation of fixK2 is solely dependent on FixLJ, whereas its autoregulation is not oxygen responsive.
Previously, Barrios et al. presented evidence indicating that expression of the fixR-nifA operon is redox controlled not only via autoregulation of the fixRp1 promoter but also via redox regulation of the fixRp2 promoter (1). However, the RegSR two-component regulatory system had not been identified at that time and thus its potential role in oxygen regulation of fixRp2 could not be tested. Given the redox responsiveness of the RegSR-homologous systems RegBA and PrrBA from Rhodobacter capsulatus and Rhodobacter sphaeroides, respectively (23, 27), it now seemed attractive to speculate that RegSR mediates redox regulation of fixRp2. Yet results from recent studies with B. japonicum mutant strains that were assumed to lack a functional fixRp1 promoter were not conclusive and partially conflicted with previous findings (H.-M. Fischer, unpublished results). Thus, the signal transduced by the RegSR two-component regulatory system of B. japonicum remains unknown and its identification requires further experimental work, ideally with a target promoter that is controlled exclusively by RegSR. In any case, RegR is essential for aerobic expression of the fixR-nifA operon (4), and it also may contribute to the fine-tuning of expression level under microoxic conditions.
Notably, the expression of the nifA gene in Sinorhizobium meliloti is indeed oxygen regulated; yet this type of control is not mediated via an RegSR-like system but rather by the FixLJ two-component regulatory system (7; for reviews, see references 10 and 17). As a consequence, the expression pattern of the S. meliloti nifA gene under different oxygen partial pressures is similar to that of fixK2 in B. japonicum (8). Oxygen regulation of NifA-dependent target genes is comparable between B. japonicum and S. meliloti, because the NifA proteins of both species respond to the cellular oxygen status in an equal manner (5, 13, 16).
Oxygen as a key regulator for the controlled expression of symbiotic genes.
The differential expression of S. meliloti symbiotic genes in disparate segments of alfalfa root nodules has provided visual evidence for the critical role of oxygen as a developmental regulator (26). Along similar lines, the disparate oxygen responsiveness of the two B. japonicum regulatory cascades might have implications for the temporally and spatially ordered expression of symbiotic genes during the infection process and the formation of nitrogen-fixing nodules. Expression of symbiotic genes is kept largely silent when B. japonicum exists as a free-living, aerobic bacterium under the oxic conditions of the soil. During root hair invasion and migration through the infection thread into the root tissue, bacteria are probably facing an increasing oxygen limitation. This would first lead to the induction of FixLJ-FixK2-dependent genes whose products, among which is the high-affinity cbb3-type oxidase, enable the bacteria to adapt their respiratory energy metabolism to the new, microoxic environment. Subsequently, the NifA-dependent nif and fix genes are to be activated when the oxygen concentration within the colonized nodule has dropped to such a low level that it is compatible not only with NifA function but also with nitrogenase activity. In parallel, fixR-nifA expression is enhanced via NifA-dependent activation of fixRp1. At this stage of the symbiosis, optimal low-oxygen conditions allowing concurrent respiration and nitrogen fixation are maintained by the plant-derived protein leghemoglobin, which acts both as an oxygen buffer and as an oxygen transport protein in support of bacteroids. In conclusion, the disparate oxygen responsiveness characteristics of the two regulatory systems appear to contribute to the synchronization of nodule development and expression of symbiotic genes in the microsymbiont.
Acknowledgments
We thank Franziska Biellmann for excellent technical assistance.
This work was supported by the Swiss Federal Institute of Technology, Zürich, Switzerland.
REFERENCES
- 1.Barrios, H., H. M. Fischer, H. Hennecke, and E. Morett. 1995. Overlapping promoters for two different RNA polymerase holoenzymes control Bradyrhizobium japonicum nifA expression. J. Bacteriol. 177:1760-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrios, H., R. Grande, L. Olvera, and E. Morett. 1998. In vivo genomic footprinting analysis reveals that the complex Bradyrhizobium japonicum fixRnifA promoter region is differently occupied by two distinct RNA polymerase holoenzymes. Proc. Natl. Acad. Sci. USA 95:1014-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, E., T. Kaspar, H. M. Fischer, and H. Hennecke. 1998. Expression of the fixR-nifA operon in Bradyrhizobium japonicum depends on a new response regulator, RegR. J. Bacteriol. 180:3853-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beynon, J. L., M. K. Williams, and F. C. Cannon. 1988. Expression and functional analysis of the Rhizobium meliloti nifA gene. EMBO J. 7:7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chauhan, S., and M. R. O'Brian. 1997. Transcriptional regulation of δ-aminolevulinic acid dehydratase synthesis by oxygen in Bradyrhizobium japonicum and evidence for developmental control of the hemB gene. J. Bacteriol. 179:3706-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David, M., M.-L. Daveran, J. Batut, A. Dedieu, O. Domergue, J. Ghai, C. Hertig, P. Boistard, and D. Kahn. 1988. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell 54:671-683. [DOI] [PubMed] [Google Scholar]
- 8.Ditta, G., E. Virts, A. Palomares, and K. Choong-Hyun. 1987. The nifA gene of Rhizobium meliloti is oxygen regulated. J. Bacteriol. 169:3217-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durmowicz, M. C., and R. J. Maier. 1998. The fixK2 gene is involved in regulation of symbiotic hydrogenase expression in Bradyrhizobium japonicum. J. Bacteriol. 180:3253-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer, H. M. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58:352-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, H. M., M. Babst, T. Kaspar, G. Acuña, F. Arigoni, and H. Hennecke. 1993. One member of a groESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 12:2901-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer, H. M., S. Fritsche, B. Herzog, and H. Hennecke. 1989. Critical spacing between two essential cysteine residues in the interdomain linker of the Bradyrhizobium japonicum NifA protein. FEBS Lett. 255:167-171. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, H. M., and H. Hennecke. 1987. Direct response of Bradyrhizobium japonicum nifA-mediated nif gene regulation to cellular oxygen status. Mol. Gen. Genet. 209:621-626. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, H. M., L. Velasco, M. J. Delgado, E. J. Bedmar, S. Schären, D. Zingg, M. Göttfert, and H. Hennecke. 2001. One of two hemN genes in Bradyrhizobium japonicum is functional during anaerobic growth and in symbiosis. J. Bacteriol. 183:1300-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gubler, M., and H. Hennecke. 1988. Regulation of the fixA gene and fixBC operon in Bradyrhizobium japonicum. J. Bacteriol. 170:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huala, E., and F. M. Ausubel. 1989. The central domain of Rhizobium meliloti NifA is sufficient to activate transcription from the R. meliloti nifH promoter. J. Bacteriol. 171:3354-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski, P. A., J. Batut, and P. Boistard. 1998. A survey of symbiotic nitrogen fixation by rhizobia, p. 431-460. In H. P. Spaink, A. Kondorosi, and P. J. J. Hooykaas (ed.), The Rhizobiaceae. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 18.Kullik, I., S. Fritsche, H. Knobel, J. Sanjuan, H. Hennecke, and H. M. Fischer. 1991. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the σ54 gene (rpoN). J. Bacteriol. 173:1125-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kullik, I., H. Hennecke, and H. M. Fischer. 1989. Inhibition of Bradyrhizobium japonicum nifA-dependent nif gene activation by oxygen occurs at the NifA protein level and is irreversible. Arch. Microbiol. 151:191-197. [Google Scholar]
- 20.Mesa, S., E. J. Bedmar, A. Chanfon, H. Hennecke, and H. M. Fischer. 2003. Bradyrhizobium japonicum NnrR, a denitrification regulator, expands the FixLJ-FixK2 regulatory cascade. J. Bacteriol. 185:3978-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nellen-Anthamatten, D., P. Rossi, O. Preisig, I. Kullik, M. Babst, H. M. Fischer, and H. Hennecke. 1998. Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for control of genes inducible by low oxygen levels. J. Bacteriol. 180:5251-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nienaber, A., A. Huber, M. Göttfert, H. Hennecke, and H. M. Fischer. 2000. Three new NifA-regulated genes in the Bradyrhizobium japonicum symbiotic gene region discovered by competitive DNA-RNA hybridization. J. Bacteriol. 182:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh, J. I., and S. Kaplan. 2000. Redox signaling: globalization of gene expression. EMBO J. 19:4237-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page, K. M., and M. L. Guerinot. 1995. Oxygen control of the Bradyrhizobium japonicum hemA gene. J. Bacteriol. 177:3979-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regensburger, B., and H. Hennecke. 1983. RNA polymerase from Rhizobium japonicum. Arch. Microbiol. 135:103-109. [DOI] [PubMed] [Google Scholar]
- 26.Soupène, E., M. Foussard, P. Boistard, G. Truchet, and J. Batut. 1995. Oxygen as a key developmental regulator of Rhizobium meliloti N2-fixation gene expression within the alfalfa root nodule. Proc. Natl. Acad. Sci. USA 92:3759-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swem, L. R., S. Elsen, T. H. Bird, D. L. Swem, H. G. Koch, H. Myllykallio, F. Daldal, and C. E. Bauer. 2001. The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J. Mol. Biol. 309:121-138. [DOI] [PubMed] [Google Scholar]
- 28.Zufferey, R., O. Preisig, H. Hennecke, and L. Thöny-Meyer. 1996. Assembly and function of the cytochrome cbb3 oxidase subunits in Bradyrhizobium japonicum. J. Biol. Chem. 271:9114-9119. [DOI] [PubMed] [Google Scholar]