Abstract
Although the staphylococcal methicillin resistance determinant, mecA, resides on a mobile genetic element, staphylococcus cassette chromosome mec (SCCmec), its distribution in nature is limited to as few as five clusters of related methicillin-resistant Staphylococcus aureus (MRSA) clones. To investigate the potential role of the host chromosome in clonal restriction of the methicillin resistance determinant, we constructed plasmid pYK20, carrying intact mecA, and introduced it into several methicillin-susceptible Staphylococcus aureus strains, five of which were naive hosts (i.e., mecA not previously resident on the host chromosome) and five of which were experienced hosts (i.e., methicillin-susceptible variants of MRSA strains from which SCCmec was excised). We next assessed the effect of the recipient background on the methicillin resistance phenotype by population analysis, by assaying the mecA expression of PBP2a by Western blot analysis, and by screening for mutations affecting mecA. Each experienced host transformed with pYK20 had a resistance phenotype and expressed PBP2a similar to that of the parent with chromosomal SCCmec, but naive hosts transformed with pYK20 selected against its expression, indicative of a host barrier. Either inducible β-lactamase regulatory genes blaR1-blaI or homologous regulatory genes mecR1-mecI, which control mecA expression, acted as compensatory elements, permitting the maintenance and expression of plasmid-carried mecA.
Staphylococcus aureus is an important human pathogen causing both community and hospital-associated infections. Penicillins and related β-lactams have dramatically reduced the morbidity and mortality of S. aureus infections, but steadily rising resistance threatens to erode their utility (5). Staphylococcal resistance to β-lactam antibiotics is mediated by either of two mechanisms: (i) production of β-lactamase and (ii) production of an altered target penicillin-binding protein (PBP), PBP2a.
β-Lactamase, encoded by blaZ, is an inducible, typically plasmid-carried, narrow-spectrum penicillinase that inactivates penicillin G and structurally related penicillins. It is regulated by two genes, blaR1-blaI (1), which are located immediately upstream and transcribed in the direction opposite that of blaZ.
PBP2a, which confers broad resistance to the entire β-lactam class (which is termed methicillin or oxacillin resistance) is a bacterial cell wall synthetic PBP that probably functions as a transpeptidase. The β-lactam antibiotics that are currently used clinically do not bind PBP2a at therapeutic concentrations and therefore lack efficacy in infections caused by methicillin-resistant staphylococci. PBP2a is encoded by mecA (8, 11, 33) and, like β-lactamase, also is inducible. Upstream from mecA are two genes, mecR1-mecI, which are homologs of blaR1-blaI (12, 18, 28, 29). MecI and BlaI are repressors of mecA as well as blaZ transcription (19, 20). MecR1 and BlaR1, which are specific for their cognate repressors and cannot substitute for each other (20), are the corresponding sensor-transducer molecules. A transmembrane signal is generated by binding of the inducer β-lactam to the extracellular sensor domain, which triggers the cleavage of the sensor transducer (19, 35) and the repressor, enabling the transcription of both blaZ and blaR1-blaI transcripts and mecA and mecR1-mecI transcripts, respectively (28).
mecA is located on a mobile element, staphylococcal cassette chromosome mec (SCCmec), which is horizontally transferable among staphylococcal species (14-16, 24). Four major SCCmec types, ranging in size from approximately 20 to over 50 kb, have been identified. Despite this potential mobility, mecA is nevertheless restricted to relatively few closely related methicillin-resistant Staphylococcus aureus (MRSA) clonal complexes, approximately five worldwide (10, 22), possibly because three of the four elements are too large to be transducible. Although type IV SCCmec is small enough to be transducible, it is found in the same clonal clusters that harbor the other types, suggesting that some genetic backgrounds are better adapted than others to whatever fitness burden might be imposed by SCCmec or mecA.
Chromosomal elements residing outside SCCmec are known to influence, sometimes dramatically, the methicillin resistance phenotype, which classically is heterogeneous, such that the vast majority of cells, although fully expressing PBP2a, can nevertheless be susceptible or express only low-level resistance. In the course of experiments to elucidate the contribution of the genome “hosting” mecA to the phenotype, we found that the host also can influence the genotype by restricting the horizontal acquisition of mecA. Differences among potential methicillin-susceptible recipient strains in the ability to tolerate mecA expression are another factor, in addition to SCCmec size and antibiotic selective pressure, that could account for the relatively limited clonal distribution of mecA in nature. Regulatory genes appear to have an important permissive role that allows an otherwise restrictive host genome to become parasitized by mecA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains are shown in Table 1. Five strains carried mecA: COLn (nafcillin MIC, 400 μg/ml); COL52 (nafcillin MIC, 3,200 μg/ml); BB270 (nafcillin MIC, 400 μg/ml); COL8a, a spontaneous methicillin-susceptible variant of COLn; and N315 (nafcillin MIC, 50 μg/ml). All MRSA strains were tetracycline susceptible and β-lactamase negative, except for N315. We constructed mecA-negative, experienced S. aureus variants (i.e., a genetic background that has harbored mecA) from the mecA-carrying strains by introducing into them plasmid pSR (provided by K. Hiramatsu [16]). pSR provides two chromosomal cassette recombinase genes, ccrA and ccrB, in trans to precisely excise SCCmec, and mutants are identified by selecting for the methicillin susceptibility phenotype as described previously (16). The suffix “ex” designates an experienced S. aureus variant from which mecA has been excised (e.g., COLnex is the SCCmec excision mutant of COLn). All S. aureus strains and transformants were grown in Trypticase soy broth (Difco Laboratories, Detroit, Mich.) or on Trypticase soy agar with aeration for 37°C unless indicated otherwise. Tetracycline and chloramphenicol (Sigma Chemical Co., St. Louis, Mo.) were used at a concentration of 10 μg/ml unless indicated otherwise.
TABLE 1.
Strains and plasmid vectors
| Strain or plasmid | Description | Relevant phenotype or genotypea | Reference(s) or source |
|---|---|---|---|
| Strains | |||
| MRSA | |||
| COL | Homogeneous methicillin-resistant strain | Mcr Tcr | 31 |
| COLn | Tcs isolated derivative of COL | Mcr | This study |
| COL52 | Antibiotic-selected mutant of COLn expressing higher level of resistance | Mcr | 6 |
| COL8a | Methicillin-susceptible spontaneous mutant of COLn; stop codon at position of amino acid residue 115 in PBP2a | Mcs | 6 |
| BB270 | Homogeneous methicillin-resistant strain; SCCmec-positive transductant of NCTC 8325 | Mcr | 2 |
| N315 | Heterogeneous methicillin-resistant strain carrying functional mecl-mecR1 regulatory genes of mecA | Mcr; β-lac | 12 |
| mecA-negative experienced hosts | |||
| COLnex | SCCmec excision, strain obtained from COLn | Mcs | This study |
| COL52ex | SCCmec excision strain obtained from COL52 | Mcs | This study |
| COL8aex | SCCmec excision strain obtained from COL8a | Mcs | This study |
| BB270ex | SCCmec excision strain obtained from BB270 | Mcs | This study |
| N315ex | SCCmec excision strain obtained from N315 | Mcs; β-lac | 15 |
| mecA-negative naive hosts | |||
| 8325-4 | Derivative of NCTC 8325 cured of all known prophages | Mcs | 23 |
| RN4220 | Restriction-deficient derivative of 8325-4 | Mcs | 17 |
| 1-63 | Wild-type clinical isolate | Mcs | 7, 8 |
| 83A | Wild-type strain ATCC 27706 | Mcs | |
| Newman | Wild-type strain ATCC 25904 | Mcs; β-lac | |
| NewmanP | Newman cured of β-lactamase plasmid | Mcs | This study |
| Plasmids | |||
| pAW8 | E. faecalis ori in pAMα1; E. coli ori in ColEI | Tcr; 5.1 kb | 34 |
| pYK20 | 2.8-kb PCR product; mecA gene cloned into pAW8 (BamHI) | Tcr; 7.9 kb | This study |
| pSR2 | ccr-2 complex cloned into temperature-sensitive pYT3 | Tcr; 12.2 kb | 16 |
| pYK644 | mecI-mecRl-mecA fragment from pGO644 cloned into pAW8 (BamHI·EcoRI) | Tcr; 9.3 kb | This study |
| pRN5542 | pSK256 with HindIII site in multiple cloning site deleted; derivative of pC194 | Cmr; 3.0 kb | 19 |
| pZRI | blaI-blaR1-blaZ complex gene cloned into pRN5542 and pBluescript-SK | Cmr Ampr; 9.2 kb | 35 |
| pZRE/AI | Contains an amino acid substitution (Glu202 to Ala) on B1aR1 in pZRI | Cmr Ampr; 9.2 kb | 35 |
| pZRIc | Termination of blaI after amino acid 101 (BlaI cleavage site) in pZRI | Cmr Ampr; 9.0 kb | This study |
Abbreviations: Mcr, methicillin resistant; Tcr, tetracycline resistant; Mcs, methicillin susceptible; Cmr, chloramphenicol resistant; Ampr, ampicillin resistant; β-lac, carrying wild-type β-lactamase plasmid.
Plasmids and DNA manipulations.
All plasmids and primers used in this study are shown in Tables 1 and 2, respectively. For transformation experiments, mecA was cloned into low-copy-number plasmid pAW8, an Enterococcus faecalis-Escherichia coli shuttle vector with a tetracycline-selectable marker (34). The mecA product, including the promoter and the first 223 nucleotides (nt) of mecR1, was obtained by PCR (35) amplification of COLn mecA with primers K34 and K38 and 1 U of Clontaq polymerase (Clontech, PaloAlto, Calif.). Plasmid pYK20 was isolated from E. coli DH5α by standard procedures. Plasmid pYK644, derived from pGO644 (provided by G. L. Archer), carries mecA complex genes (mecI-mecR1-mecA) from strain N315 (12). The BamHI-EcoRI-digested fragment from pGO644 was inserted into those sites in pAW8. Plasmid pZRI contains blaZ complex genes (blaI-blaR1-blaZ), and plasmid pZRE/AI contains an amino acid substitution (Glu202 → Ala) in BlaR1, as previously described (35). Plasmid pZRIc was constructed by PCR amplification of the 2.1-kb fragment of pCH2278 carrying blaR1 minus the first 13 nt through blaI minus the last 78 nt with primers P1-101 (Table 2) and P4 (35). The product was digested with HindIII and ligated to the HindIII site of pSK1.0 (35), restoring the bla region, with termination of blaI after amino acid residue 101, which is the BlaI cleavage site. All blaZ complex plasmids were cloned into S. aureus chloramphenicol-selectable plasmid vector pRN5542 (2). We confirmed the absence of mutations by nucleotide sequencing (performed at the UCSF Biomolecular Resources Center DNA Sequencing Facility). β-Lactamase activity was assessed with nitrocefin disks (Becton-Dickinson and Company, Sparks, Md.) (35). Naive and experienced host strains were transformed with plasmids by electroporation (16). After 48 h of incubation, transformants were tested for mecA by PCR amplification with vector-specific primers for the sequence immediately adjacent to the insert (K3 and K4).
TABLE 2.
Synthetic oligonucleotide primers
| Genetic elementa and primer designation | Sequenceb | Nucleotide positions |
|---|---|---|
| mecA | ||
| K15 | 5′-AGCACACCTTCATATGACGTCT-3′ | 32473-32494 |
| K16 | 5′-TGGATCAAAATTGGGTACAAGA-3′ | 31991-32012 |
| K17 | 5′-AGTTGTAGTTGTCGGGTTTGGT-3′ | 31425-31446 |
| K18 | 5′-TGGCAATATTAACGCACCTCACT-3′ | 33012-33034 |
| K34 | 5′-TATGCGGATCCTCGTGTCAGATACATTTCGATTCA-3′ | 31068-31091 |
| K38 | 5′-ATTTCGGATCCGTTGTAGCAGGAACACAAATGAATAAC-3′ | 33645-33619 |
| lacZ | ||
| K3 | 5′-TATGTTGTGTGGAATTGTGAGCGGA-3′ | 167-191 |
| K4 | 5′-AGGCGATTAAGTTGGGTAACGCCAG-3′ | 329-353 |
| blaR1-blaI, P1-101 | 5′-GTCGATAAGCTTTTAATTCAGCACTAAACTTTTCATG-3′ | 2205-2184 |
The primers listed are specific for mecA genes of strain NCTC 10442 (GenBank accession no. AB033763), for lacZ in pUC119 (GenBank accession no. U07649), and for blaR1-blaI in pI258 (GenBank accession no. M62650).
Underlining indicates BamHI sites introduced in K34 and K38 and HindIII site introduced in P1-101.
Construction of pYK20 or pYK644 transformants.
Plasmid pYK20 carrying intact mecA or pYK644 carrying the mecI-mecR1-mecA complex but lacking other SCCmec DNA (12, 14, 18) was introduced by electroporation into several methicillin-susceptible S. aureus strains, which were either naive (i.e., naturally free of mecA) or experienced (i.e., methicillin-susceptible variants of MRSA strains from which SCCmec had been excised, denoted by the “ex” suffix). β-Lactam antibiotic selection, which could affect phenotype, was avoided by selecting for the tetracycline resistance marker of the plasmid vector. We then assessed the effect of recipient background on the methicillin resistance phenotype by population analysis to determine the number of highly resistant CFU and assayed the mecA expression of PBP2a by Western blot analysis.
Population analysis.
Population analysis was done by the agar plate method, in which approximately 108 CFU are quantitatively inoculated onto a series of agar plates containing increasing concentrations of nafcillin (Sigma) (32).
Detection of PBP2a.
We assayed for PBP2a production by Western blotting. S. aureus membrane proteins were prepared from late-exponential-stage cultures, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred as described previously (35). PBP2a was detected by using a mouse monoclonal anti-PBP2a antibody (a gift from Denka Seiken Co., Ltd., Niigata, Japan) as the primary antibody, diluted 1:100,000, and alkaline phosphatase-conjugated anti-mouse immunoglobulin (Promega, Madison, Wis.). We detected bound antibodies by color development as directed by the manufacturer. Relative amounts of PBP2a proteins were measured by scanning densitometry.
COLnex reporter assay.
We screened for mecA mutations in pYK20 transformants by using COLnex which, when transformed with wild-type mecA in pYK20, expresses homogeneous resistance. A representative colony of the transformant yielding the predicted 2.8-kb PCR mecA amplification product was diluted 1:104 and regrown to a cell density of approximately 5 × 108 CFU/ml. Plasmid pYK20 was extracted, purified, and introduced into host strain COLnex by electroporation. After 48 h of incubation, COLnex(pYK20) transformants were selected on tetracycline-containing agar and replicated on Trypticase soy agar containing both tetracycline and nafcillin at concentrations of 0, 2.5, 10, and 100 μg/ml. After 24 h of incubation, CFU growing at each nafcillin concentration were counted, and the proportions were calculated relative to CFU growing on nafcillin-free agar. The COLn homogeneous phenotype is 100% growth of CFU at 100 μg of nafcillin/ml.
RESULTS
Phenotypes of naive and experienced host transformants.
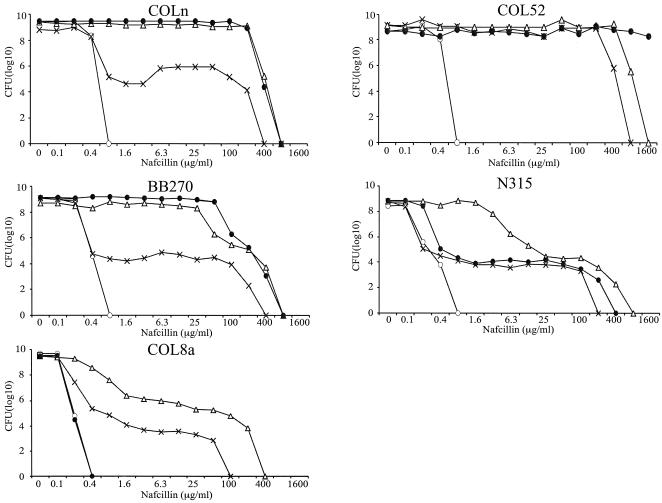
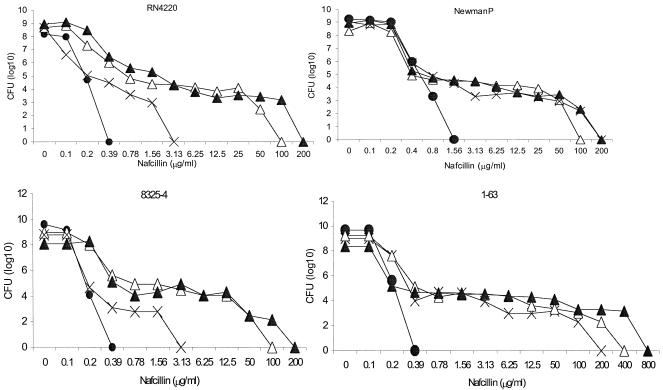
The resistance phenotype in a population analysis of experienced host transformants with pYK20 carrying unregulated mecA was in every instance identical to that of the parent (Fig. 1): COLnex and COL52ex transformants exhibited their characteristic homogeneous phenotype (i.e., >1% of plated CFU grew on agar containing 100 μg of nafcillin/ml). BB270ex, N315ex, COL8a, and COL8aex transformants had class 3 heterogeneous resistance (i.e., 1 CFU in 102 to 103 CFU at 100 μg of nafcillin/ml), as defined by Tomasz et al. (32). Resistance was extremely heterogeneous for naive host transformants carrying pYK20 (Fig. 2), with a class 2 pattern (only 1 CFU in 105 to 106 CFU at 100 μg of nafcillin/ml) for 1-63 and 8325-4 and a class 1 pattern (only 103 CFU in 108 CFU at 6.25 μg of nafcillin/ml) for RN4220.
FIG. 1.
Population analysis with nafcillin for five MRSA strains and experienced host MRSA (MRSAex) transformants. Symbols: •, wild-type MRSA with SCCmec present plus cloning vector pAW8 only; ○, MRSAex with SCCmec excised plus cloning vector only; ▵, MRSAex transformed with pYK20, carrying unregulated mecA; ×, MRSAex transformed with pYK644, carrying the intact mecI-mecR1-mecA complex. Shown on the y axis is the number of cells (per milliliter) growing on nafcillin-containing agar.
FIG. 2.
Population analysis with nafcillin for four naive host methicillin-susceptible S. aureus transformants. Symbols: •, wild-type naive host with cloning vector pAW8 only; ▵, naive host transformed with pYK20, carrying unregulated mecA; ×, naive host transformed with pYK644, carrying the intact mecI-mecR1-mecA complex; ▴, naive host cotransformed with pYK20 and pZRI, carrying the intact blaI-blaR1-blaZ complex.
The phenotype of each of the experienced hosts transformed with pYK644 carrying mecR1-mecI-regulated mecA was heterogeneous, except for strain COL52 (Fig. 1). The curve for N315ex(pYK644) was identical to that for parent strain N315, both of which expressed a more heterogeneous pattern than N315ex(pYK20), in which mecA regulatory genes are absent. Likewise, COL8a(pYK644) and BB270ex(pYK644) transformants, with intact mecR1-mecI, had a more heterogeneous pattern of resistance than their respective pYK20 transformants. pYK644 transformants of naive host 8325-4 and its derivative strain, RN4220, also had a more heterogeneous pattern of resistance than their pYK20 counterparts. In contrast, the phenotypes of naive host NewmanP(pYK644) and 1-63(pYK644) transformants and pYK20 transformants were the same (Fig. 2).
Stability of plasmid pYK20 in naive and experienced hosts.
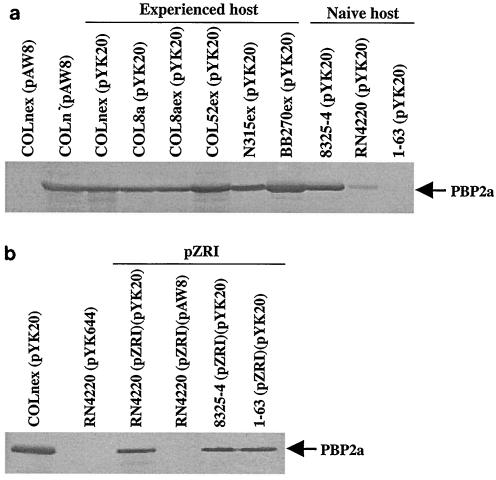
The expression of PBP2a detected in Western blots for each of the experienced host transformants carrying pYK20 was similar to that detected for the COLn(pAW8) control strain, in which an unregulated, single copy of mecA naturally resides on SCCmec (Fig. 3a). PBP2a expression varied greatly among the naive host transformants carrying pYK20: 8325-4 was similar to the COLn(pAW8) control strain, RN4220 was weakly positive, and 1-63 had no PBP2a detected.
FIG. 3.
Detection by Western blot analysis of PBP2a (78 kDa) encoded by mecA. COLn(pAW8) is the positive control; COLnex(pAW8), RN4220(pAW8), and RN4220(pZRI)(pAW8) are the negative controls. (a) Comparison of experienced host and naive host transformants carrying pYK20. (b) Representative transformants with cloning vector pAW8 only, pYK644 (carrying intact mecR1-mecI-mecA), and pYK20 (carrying unregulated mecA) and cotransformants with pZRI (carrying blaR1-blaI-blaZ). All were grown under noninducing conditions.
As regulatory elements that could repress mecA expression are absent in these constructs, the diminished amounts of PBP2a in naive host transformants suggested the presence of mutations affecting mecA expression. We screened transformants for mecA deletions by PCR. Of 36 COLnex(pYK20) transformants tested, all had the expected 2.8-kb fragment. In contrast, 24.4, 4.7, and 70%, respectively, of transformants of naive hosts RN4220, 8325-4,and 1-63 carrying pYK20 yielded PCR products of <2.8 kb, indicating the presence of a deletion mutation in mecA (Table 3).
TABLE 3.
Detection by PCR of mecA insert in plasmid pYK20
| Transformant | No. of transformants | Fragment ratioa | Relative amt of PBP2 (%)b |
|---|---|---|---|
| COLnex(pYK20) | 125 | 0 | 96 |
| RN4220(pYK20) | 100 | 24.4 | 33 |
| 8325-4(pYK20) | 53 | 4.4 | 107 |
| 1-63(pYK20) | 26 | 70 | 18 |
Ratio of unexpected <2.8-kb fragments in to total fragments screened transformants.
Percent density of PBP2a in each transformant compared to PBP2a in COLn.
We developed a reporter assay using COLnex to investigate whether smaller mecA mutations undetectable by PCR screening might also be present in apparently intact pYK20. Resistance is homogeneous upon transformation of COLnex with pYK20 carrying intact mecA (Fig. 1); therefore, COLnex transformed with pYK20 in which there was a mutation of mecA or its promoter that affected PBP2a production or activity should be detectable by a loss of resistance in COLnex transformants. As expected, 100% of the COLnex(pKY20) transformants purified from experienced host donors were resistant to 100 μg of nafcillin/ml (Table 4). A total of 96.9% of the COLnex (pYK20) transformants obtained with pYK20 isolated from naive hosts 83A(pYK20) and NewmanP(pYK20) were resistant to 100 μg of nafcillin/ml. In contrast, 3.7 to 76.8% of the COLnex(pYK20) transformants obtained with pYK20 isolated from naive hosts RN4220(pYK20), 1-63(pYK20), and 8325-4(pYK20) were resistant to 100 μg of nafcillin/ml.
TABLE 4.
Assay of phenotype of COLnex transformants with pAW8, pYK20, or pYK644 and purified from control, experienced, and naive plasmid donors
| Plasmid donor strain | No. of COLnex transformants
|
Ratioa | |||
|---|---|---|---|---|---|
| Total | Replicated on plate containing at nafcillin at (μg/ml):
|
||||
| 2.5 | 10 | 100 | |||
| Controls | |||||
| DH5α(pYK20) | 157 | 157 | 100 | ||
| COLnex(pYK20) | 21 | 21 | 100b | ||
| COLnex(pAW8) | 211 | 0 | 0 | 0 | 0b |
| RN4220(pZRI)(pAW8) | 17 | 0 | 0 | 0 | 0b |
| COLnex(pYK644) | 36 | 36 | 100 | ||
| Experienced host | |||||
| COL8aex(pYK20) | 775 | 775 | 100b | ||
| COL8a(pYK20) | 212 | 212 | 100b | ||
| N315ex(pYK20) | 195 | 195 | 100b | ||
| COL52ex(pYK20) | 543 | 543 | 100b | ||
| BB270ex(pYK20) | 45 | 45 | 100b | ||
| Naive host | |||||
| RN4220(pYK20) | 62 | 6 | 6 | 6 | 9.7b |
| 8325-4(pYK20) | 168 | 168 | 168 | 129 | 76.8b |
| 1-63(pYK20) | 54 | 12 | 2 | 2 | 3.7b |
| 83A(pYK20) | 128 | 123 | 123 | 123 | 96.9 |
| NewmanP(pYK20) | 355 | 344 | 344 | 344 | 96.9 |
| Naive host (mecI-mecR1-mecA complex in pYK644) | |||||
| RN4220(pYK644) | 48 | 48 | 100b | ||
| 8325-4(pYK644) | 51 | 51 | 100 | ||
| 1-63(pYK644) | 125 | 125 | 100 | ||
| Naive host + mecA + β-lactamase plasmid | |||||
| RN4220(pYK20)(pZR1) | 48 | 46 | 46 | 46 | 95.8b |
| RN4220(pYK20)(pRN5542)c | 712 | 23 | 18 | 18 | 2.5 |
| RN4220(pYK20)(pZRIc) | 911 | 41 | 7 | 7 | 0.8 |
| RN4220(pYK20)(pZRE/AI) | 48 | 48 | 100 | ||
| RN4220(pYK644)(pRN5542)c | 48 | 48 | 100 | ||
| RN4220(pYK644)(pZRI) | 130 | 130 | 100 | ||
| 8325-4(pYK20)(pZRI) | 354 | 354 | 100b | ||
| 1-63(pYK20)(pZRI) | 232 | 232 | 100b | ||
| Newman(pYK20) | 285 | 285 | 100 | ||
Ratio of number of transformants growing on agar containing 100 μg of nafcillin per ml to total number of transformants.
Transformant used in Western blot analysis.
pRN5542 is the vector into which the β-lactamase regulon, blaI-blaR1-blaZ, was cloned.
We passaged four RN4220(pYK20) transformants and one COLnex(pYK20) transformant for 5 days in the absence of tetracycline and repeated the reporter assay. A total of 100% of the COLnex(pYK20) transformants originating from RN4220(pYK20) were susceptible to 2.5 μg of nafcillin/ml, whereas 100% of the COLnex(pYK20) transformants obtained from COLnex(pYK20) were resistant. We picked two passaged RN4220(pYK20) transformants and determined the mecA sequence. One had a point mutation in the mecA −35 promoter sequence (TTGACA → TTGAAA), and the other had a stop codon at the position of amino acid residue 95 in PBP2a. Thus, in the absence of β-lactam-selective pressure, naive strains to various degrees selected against the presence or expression of PBP2a.
Stability of plasmid pYK644 in naive hosts.
In contrast to the genetic instability of unregulated mecA in the pYK20 construct, mecA under the strong repressor control (Fig. 3b) of mecR1-mecI in the pYK644 construct was faithfully maintained in naive hosts. A total of 100% of COLnex transformants with plasmid pYK644 donated from naive hosts grew on agar containing 100 μg of nafcillin/ml. These results suggested that regulatory genes play a role in stabilizing mecA in a new host that is otherwise intolerant of its presence.
Effect of the blaZ-blaR1-blaI complex on PBP2a expression in a naive strain.
In many, and perhaps most, clinical isolates, the expression of mecA is under the control of β-lactamase regulatory genes, blaR1-blaI, rather than mecR1-mecI genes, which either are deleted or mutated (26). The blaR1-blaI complex has significant amino acid homology to the mecR1-mecI complex (12), and the two systems function in the same way (18, 29). MecI and BlaI are repressors of both mecA transcription and blaZ transcription (19, 20), and MecR1 and BlaR1 are signal-transducing molecules which function as antirepressors for their cognate repressors and which cannot substitute for each other (20). To investigate whether the bla regulon also could stabilize mecA, we constructed double transformants of RN4220 possessing both the wild-type blaZ-blaR1-blaI-carrying plasmid pZRI (35) and pYK20. Reflecting the weaker repressor activity of bla than of mec regulatory elements, PBP2a was readily detectable in Western blots of RN4220(pZRI)(pYK20), 8325-4(pZRI)(pYK20), and 1-63(pZRI)(pYK20) (Fig. 3b). RN4220(pZRI)(pYK20), 8325-4(pZRI)(pYK20), and 1-63(pZRI)(pYK20) double transformants and Newman(pYK20) yielded 95.8 or 100% of COLnex(pYK20) transformants resistant to 100 μg of nafcillin/ml (Table 4). An RN4220 double transformant carrying the pRN5542 vector lacking bla genes yielded only 2.5% of COLnex(pYK20) transformants that were nafcillin resistant.
To determine which bla gene products were important for stabilizing mecA, we constructed RN4220 double transformants with pYK20 plus one of two β-lactamase mutant plasmids, pZRE/AI or pZRIc. pZRE/AI has a single amino acid substitution of alanine for glutamic acid in the metalloprotease motif of the BlaR1 sensory-transducer protein (35). This mutation blocks the proteolytic cleavage of BlaI and completely represses β-lactamase production (19, 35). The introduction of pYK20 from the RN4220(pZRE/AI)(pYK20) double transformant into COLnex yielded 100% of COLnex transformants that were resistant. pZRIc has a stop codon engineered into the β-lactamase repressor BlaI, so that only the inactive, cleaved 11-kDa form of the protein is expressed and β-lactamase is constitutively expressed. pYK20 from the RN4220(pZRIc)(pYK20) double transformant failed to stabilize mecA, yielding 0.8% of COLnex transformants growing on nafcillin (Table 4). These results indicate that it is the β-lactamase repressor that stabilizes mecA in a naive background. PBP2a expression was not detected in either of these RN4220 double transformants by Western blotting, but for different reasons. Mutant mecA not expressing PBP2a was selected for in the absence of an active repressor with pZRIc. Unopposed repressor activity of pZRE/AI, although it preserved mecA, as demonstrated in the COLnex reporter assay, did not allow for its expression in the RN4220 cotransformant.
DISCUSSION
These experiments demonstrate that some MRSA strains select against the expression of PBP2a, presumably due to a fitness cost posing a barrier to the stability and maintenance of mecA. The unregulated mecA plasmid construct was stable in the experienced strains previously harboring mecA. The results underscore the important contribution of the genetic background to both the phenotype and the genotype of methicillin resistance and suggest that strains harboring mecA perhaps have adapted to it, compensated for it, previously experienced it, or are otherwise properly prepared for it. This instability of mecA in some genetic backgrounds may account in part for the relatively restricted clonal clustering of the mobile SCCmec element.
Particularly interesting is the ability of mec or bla regulatory genes to stabilize mecA, a finding which suggests a previously unrecognized role of these elements in facilitating the dissemination of mecA. The repression of mecA may be an important first step in its successful engraftment by a new host. Although in principle this role could be provided by either mec or bla regulatory genes, the latter is the more likely to do so, because the mecR1-mecI genes strongly repress mecA expression, and the latter could be a survival disadvantage in the presence of a β-lactam antibiotic (18, 29).
The relatively much stronger repression of mecA expression by mec than by bla regulatory genes is evident from the Western blots of RN4220(pYK644), in which mecA is paired with mecR1-mecI, compared to RN4220(pZRI)(pYK20), in which mecA is paired with blaR1-blaI (Fig. 3b). Strong repression can translate to a significant survival disadvantage in some (e.g., RN4220 and 8325-4) but not all (e.g., NewmanP and 1-63) naive genetic backgrounds (Fig. 2). mec-regulated repression of mecA in experienced strains resulted in a very heterogeneous resistance pattern for COLn and BB270 compared to that for parents and transformants with unregulated mecA (Fig. 1). The effect of mec regulation on the phenotype of a naturally very heterogeneous strain, N315, was also quite apparent. The parent and the pYK644 transformant, in which the bla and mec regulatory genes both are present, exhibit a much more heterogeneous resistance pattern than the pYK20 transformant, in which the β-lactamase plasmid is present and the mecR1-mecI genes are absent. The important contribution of the genetic background to the resistance phenotype is again demonstrated by the behavior of strain COL52, the homogeneous phenotype of which is fundamentally unchanged by the imposition of mec regulation. On balance, β-lactamase regulatory genes seem to provide a compromise solution to the need for some control over PBP2a production to minimize the cost of maintaining mecA while also being able to express the protein in the presence of an antibiotic.
These experimental results correlate well with what others have found concerning the relationship between β-lactamase-mediated and PBP2a-mediated resistance. As would be predicted from the strong repression of mecA, mecR1-mecI elements in SCCmec typically are deleted or mutated (15, 24). The early observation that β-lactamase was a critical determinant for transduction of the methicillin resistance determinant (4, 9, 28) and reports that methicillin resistance tends to be unstable in clinical isolates when the penicillinase plasmid is absent are understandable in view of the ability of the β-lactamase repressor to stabilize mecA in some genetic backgrounds (13, 25, 27, 30). This interesting synergy between the two dissimilar resistance mechanisms can be thought of as a novel variation on the theme of compensatory mutations in drug resistance (3, 21), such that merely introducing a gene is often not enough. The genetic or biochemical basis of the barrier to mecA and or tolerance to it (aside from regulatory genes) is unknown but may have important implications for drug resistance and drug development, as the genes involved could be drug targets. We also propose that this barrier may account for the clonality that is typical of MRSA by restricting the element to certain backgrounds.
Acknowledgments
This work was supported by grant AI46610 from the National Institute of Allergy and Infectious Diseases.
We thank Keiichi Hiramatsu for plasmid pSR and Gordon L. Archer for plasmid pGO644. We also thank John Imboden and Israel Charo for constructive comments on the preparation of the manuscript.
REFERENCES
- 1.Asheshov, E. H., and K. G. Dyke. 1968. Regulation of the synthesis of penicillinase in diploids of Staphylococcus aureus. Biochem. Biophys. Res. Commun. 30:213-218. [DOI] [PubMed] [Google Scholar]
- 2.Berger-Bächi, B., and M. L. Köhler. 1983. A novel site on the chromosome of Staphylococcus aureus influencing the level methicillin resistance: genetic mapping. FEMS Microbiol. Lett. 20:305-309. [Google Scholar]
- 3.Bjorkman, J., I. Nagaev, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479-1482. [DOI] [PubMed] [Google Scholar]
- 4.Boyce, J. M., and A. A. Medeiros. 1987. Role of beta-lactamase in expression of resistance by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 31:1426-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brumfitt, W., and J. Hamilton-Miller. 1989. Methicillin-resistant Staphylococcus aureus. N. Engl. J. Med. 320:1188-1196. [DOI] [PubMed] [Google Scholar]
- 6.Chambers, H. F. 1995. In vitro and in vivo antistaphylococcal activities of L-695,256, a carbapenem with high affinity for the penicillin-binding protein PBP2a. Antimicrob. Agents Chemother. 39:462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers, H. F., and C. J. Hackbarth. 1987. Effect of NaCl and nafcillin on penicillin-binding protein 2a and heterogeneous expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 31:1982-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers, H. F., G. L. Archer, and M. Matsuhashi. 1989. Low-level methicillin resistance in strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 33:424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen, S., and H. M. Sweeney. 1973. Effect of the prophage and penicillinase plasmid of the recipient strain upon the transduction and the stability of methicillin resistance in Staphylococcus aureus. J. Bacteriol. 116:803-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartman, B. J., and A. Tomasz. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiramatsu, K., K. Asada, E. Suzuki, K. Okonogi, and T. Yokota. 1992. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 298:133-136. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu, K., E. Suzuki, H. Takayama, Y. Katayama, and T. Yokota. 1990. Role of penicillinase plasmids in the stability of the mecA gene in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 34:600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 18.Kuwahara-Arai, K., N. Kondo, S. Hori, E. Tateda-Suzuki, and K. Hiramatsu. 1996. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP2′ production. Antimicrob. Agents Chemother. 40:2680-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis, R. A., and K. G. Dyke. 2000. MecI represses synthesis from the beta-lactamase operon of Staphylococcus aureus. J. Antimicrob. Chemother. 45:139-144. [DOI] [PubMed] [Google Scholar]
- 20.McKinney, T. K., V. K. Sharma, W. A. Craig, and G. L. Archer. 2001. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate mecA and beta-lactamase regulators. J. Bacteriol. 183:6862-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagaev, I., J. Bjorkman, D. I. Andersson, and D. Hughes. 2001. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol. Microbiol. 40:433-439. [DOI] [PubMed] [Google Scholar]
- 22.Nicholas, P. J. D., E. M. Catrin, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114-116. [DOI] [PubMed] [Google Scholar]
- 23.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 24.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richmond, M. H. 1965. Dominance of the inducible state in strains of Staphylococcus aureus containing two distinct penicillinase plasmids. J. Bacteriol. 90:370-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosato, A. E., B. N. Kreiswirth, W. A. Craig, W. Eisner, M. W. Climo, and G. L. Archer. 2003. mecA-blaZ corepressors in clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 47:1460-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryffel, C., A. Strassle, F. H. Kayser, and B. Berger-Bächi. 1994. Mechanisms of heteroresistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 38:724-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryffel, C., F. H. Kayser, and B. Berger-Bächi. 1992. Correlation between regulation of mecA transcription and expression of methicillin resistance in staphylococci. Antimicrob. Agents Chemother. 36:25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma, V. K., C. J. Hackbarth, and T. M. Dickinson. 1998. Interaction of native and mutant MecI repressors with sequences that regulate mecA, the gene encoding penicillin binding protein 2a in methicillin-resistant staphylococci. J. Bacteriol. 180:2160-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart, G. C., and E. D. Rosenblum. 1980. Transduction of methicillin resistance in Staphylococcus aureus: recipient effectiveness and β-lactamase production. Antimicrob. Agents Chemother. 18:424-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomasz, A., H. B. Drugeon, H. M. de Lencastre, D. Jabes, L. McDougall, and J. Bille. 1989. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob. Agents Chemother. 33:1869-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomasz, A., S. Nachman, and H. Leaf. 1991. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 35:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Utsui, Y., and T. Yokota. 1985. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 28:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wada, A., and H. Watanabe. 1998. Penicillin-binding protein 1 of Staphylococcus aureus is essential for growth. J. Bacteriol. 180:2759-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, H. Z., C. J. Hackbarth, K. M. Chansky, and H. F. Chambers. 2001. A proteolytic transmembrane signaling pathway and resistance to β-lactams in staphylococci. Science 291:1962-1965. [DOI] [PubMed] [Google Scholar]