Abstract
The furA-katG region of Mycobacterium tuberculosis, encoding a Fur-like protein and the catalase-peroxidase, is highly conserved among mycobacteria. Both genes are induced upon oxidative stress. In this work we analyzed the M. tuberculosis furA promoter region. DNA fragments were cloned upstream of the luciferase reporter gene, and promoter activity in Mycobacterium smegmatis was measured in both the presence and absence of oxidative stress. The shortest fragment containing an inducible promoter extends 45 bp upstream of furA. In this region, −35 and −10 promoter consensus sequences can be identified, as well as a 23-bp AT-rich sequence that is conserved in the nonpathogenic but closely related M. smegmatis. M. tuberculosis FurA was purified and found to bind upstream of furA by gel shift analysis. A ca. 30-bp DNA sequence, centered on the AT-rich region, was essential for FurA binding and protected by FurA in footprinting analysis. Peroxide treatment of FurA abolished DNA binding. Three different AT-rich sequences mutagenized by site-directed mutagenesis were constructed. In each mutant, both M. tuberculosis FurA binding in vitro and pfurA regulation upon oxidative-stress in M. smegmatis were abolished. Thus, pfurA is an oxidative stress-responsive promoter controlled by the FurA protein.
Fur-like proteins are widespread in both gram-negative and gram-positive bacteria. Their major role in the regulation of iron uptake systems in response to environmental iron was first described for Escherichia coli (for a review, see reference 14). Under iron-replete conditions, the Fur protein, using Fe2+ as a corepressor, binds to a 19-bp operator sequence located upstream of the Fur-regulated genes (2, 10, 11, 13).
Fur and Fur-like proteins were found to be involved in regulation of functions as varied as the acid shock response (17), detoxification of oxygen radicals (12, 18), production of toxins and virulence factors (20, 31), and metabolic pathways (24, 29). These observations led Fur-like proteins to be considered not only transcriptional repressors but also global regulators.
The E. coli Fur protein (17 kDa) contains a nonclassical helix-turn-helix DNA binding domain and is active as a homodimer (7). In vivo assays showed that the N-terminal domain is involved in DNA binding and that the C-terminal domain is involved in dimerization (29). Biochemical analysis indicated that repressor activation occurs upon a conformational change due to metal binding to the C-terminal domain (8, 15, 30). The Fur protein contains two metal ion binding sites: the iron binding regulatory site and a tightly binding zinc site that plays a role in stability of the protein (1, 19).
Several redox-sensing proteins of the Fur family have been identified in various bacteria. Their role in the oxidative stress response might be mediated via metal-catalyzed oxidation of the protein or a change in the oxidation state of the bound metal ion (4, 16). This is the case for CatR in Streptomyces coelicolor, which regulates its own gene and the catalase CatA by binding to operator sequences upstream of their promoters under reducing conditions (16). Similarly, in Streptomyces reticuli, FurS in the thiol-reduced form specifically binds to a motif upstream of the furS gene (25).
The furA gene has been identified in Mycobacterium tuberculosis as well as other mycobacterial species (6, 23, 26). S. reticuli FurS and M. tuberculosis FurA show 53% identity, and the predicted secondary structures of the two proteins are similar. Moreover, typical motifs involved in metal binding and several cysteines that may sense the oxidative stress are conserved (25, 32). Thus, the two proteins might play similar roles in responding to oxidative stress.
The presence of an oxidative stress-inducible promoter, immediately upstream of the furA gene, has been reported previously for M. tuberculosis, Mycobacterium smegmatis, and Mycobacterium bovis BCG (21, 23, 27). In M. bovis BCG, the 5′ end of the furA transcript was mapped in correspondence to the start codon of the furA gene (27). A 23-bp AT-rich sequence, overlapping the −35 region of pfurA, was identified and found to be conserved among mycobacteria.
In this study, we define more precisely the sequence of the pfurA inducible promoter of M. tuberculosis and demonstrate that FurA negatively controls its own transcription by binding to this region in a redox-dependent manner.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli XL1-Blue (5) was grown in LD medium (28) supplemented, when necessary, with kanamycin (50 μg/ml). M. smegmatis mc2155 was grown in LD medium containing 0.2% (vol/vol) glycerol and 0.05% (vol/vol) Tween 80 and supplemented when necessary with kanamycin (20 μg/ml). The complete M. tuberculosis genome sequence is reported under GenBank accession no. AL123456.
Construction of plasmids.
DNA fragments from M. tuberculosis were amplified by PCR with specific primers and cloned in pSG10ter (23) upstream of the luxAB reporter gene. The sequences of the primers will be provided upon request. All of the inserted fragments were sequenced.
Mutagenesis.
Complementary oligonucleotides carrying each of the 6-bp substitutions indicated in Fig. 6A were used in combination with external primers to amplify the M. tuberculosis −154/+33 region. The amplified fragments were either end labeled, purified, and used in gel retardation assays (see below) or cloned in pSG10ter upstream of luxAB.
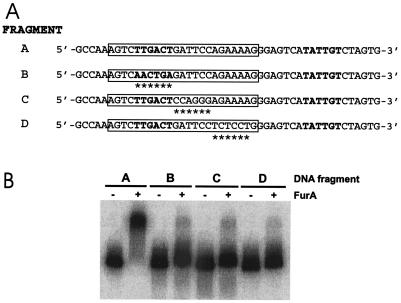
FIG. 6.
Effect of mutations in the furA operator sequence on FurA binding. (A) Sequences of the mutations. Fragment A, wild type. Fragment B, C, and D carry different 6-bp substitutions within the AT-rich region (boxed). The mutant bases are indicated by asterisks. The −10 and −35 sequences are in boldface. (B) Gel shift of the mutant fragments caused by FurA. FurA binding was performed as described in the legend to Fig. 4.
Luciferase assay.
Independent cultures of M. smegmatis mc2155 carrying the indicated plasmids were grown in LD medium at 37°C, the turbidity of the cultures at 600 nm was adjusted to 0.03, and the cultures were sampled and either mock treated or treated for 1 h at 37°C with 0.062 mM H2O2. One hundred microliters of 1% (vol/vol) decyl aldehyde solution (Sigma-Aldrich) in ethanol was then added to 1 ml of a 1:10 dilution of the culture, and luminescence was measured over 10 s, using a Stratec luminometer.
Cloning and purification of FurA.
The M. tuberculosis furA gene was amplified from cosmid DNA. The primers used contained restriction sites for BamHI and EcoRI. The PCR fragment was digested with the indicated restriction enzymes and cloned into the pGEX6P1 expression vector (Pharmacia). Recombinant plasmids were transformed into E. coli XL1-Blue, and colonies were isolated on Luria-Bertani plates containing 100 μg of ampicillin per ml. Clones were tested for the presence of the correctly inserted furA gene and sequenced.
Recombinant protein expression was obtained by growing cells for 16 h at 26°C in LB medium supplemented with ampicillin (100 μg/ml) and IPTG (isopropyl-β-d-thiogalactopyranoside) (50 μM). The culture was then harvested by centrifugation and stored at −20°C. The cell pellet from 250 ml of culture was resuspended in 5 ml of 1× phosphate-buffered saline (8 g of NaCl per liter, 0.2 g of KCl per liter, 1.44 g of Na2HPO4 per liter, 0.24 g of KH2PO4 per liter) and sonicated on ice. The lysate was centrifuged for 30 min at 13,000 rpm, and the supernatant fraction was applied to 0.5 ml of GS4B resin (Pharmacia) that was previously equilibrated with 10 volumes of 1× phosphate-buffered saline. All of the subsequent purification steps were performed according to manufacturer's instructions. Protein elution was done after digestion with Prescission protease (Pharmacia) for 16 h at 4°C; both the glutathione S-transferase (GST) tag and protease remain tightly bound to the chromatographic matrix, allowing their separation from the protein.
The protein concentration was evaluated by the Bradford assay with bovine serum albumin as a standard.
Gel retardation assay.
DNA probes for gel shift experiments were amplified by PCR with specific primers containing a BamHI site at one 5′ end, digested with BamHI, labeled with the Klenow enzyme and [α-32P]dATP, and purified from polyacrylamide gels. Binding reaction mixtures in 10 μl of binding buffer [20 mM Tris HCl (pH 8), 1 mM dithiothreitol (DTT), 50 mM KCl, 5 mM MgCl2, 0.05 mg of poly(dI-dC) per ml, 0.05 mg of bovine serum albumin per ml, 10% glycerol, and 200 μM NiSO4] containing about 7.4 fmol of the probe (40,000 cpm) were incubated with purified FurA protein (12 μM) for 20 min at room temperature. Reaction mixtures were loaded on an 8% polyacrylamide gel containing 40 mM Tris acetate (pH 8) and 200 μM NiSO4. Gels were run at 120 V at 4°C, dried, and exposed to PhosphorImager (Molecular Dynamics) detection.
DNase I footprinting.
The pMYT101 plasmid was digested with either BamHI or EcoRI, labeled with the Klenow enzyme and [α-32P]dATP, and digested with EcoRI or BamHI, respectively. The probes were extracted from a 5% polyacrylamide gel and eluted overnight in 10 mM Tris HCl (pH 7.4). Binding reactions were performed as described for the gel retardation assay, with 60,000 cpm of the probe in a final volume of 100 μl. DNase I digestion was carried out by treatment with 1 μl of DNase I (1.5 ng/μl) and 1 mM CaCl2 for 1 min at room temperature. Reactions were stopped by addition of 50 μl of Stop buffer (0.1 M EDTA, 0.8% sodium dodecyl sulfate [SDS], 1.6 M NH4-acetate, 300 mg of herring sperm DNA per ml). DNA was precipitated with 350 μl of ethanol, dried, and resuspended in 7 M urea-20 mM Tris HCl (pH 8) with tracking dyes. Samples were loaded on a 7 M urea-6% polyacrylamide sequencing gel. Maxam-Gilbert A+G sequencing reactions were performed as described previously (22).
RESULTS
Mapping of an oxidative stress-inducible promoter upstream of furA in M. tuberculosis.
We previously reported the presence of an oxidative stress-inducible promoter upstream of the furA genes in both M. smegmatis and M. tuberculosis (23). We mapped the M. tuberculosis promoter by cloning several DNA fragments upstream of the reporter luxAB gene and measuring luciferase activity expressed in M. smegmatis mc2155 (Fig. 1).
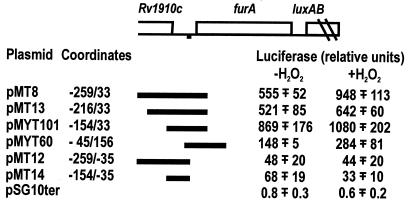
FIG. 1.
Oxidative stress induction of luciferase by plasmids carrying M. tuberculosis DNA regions. (Top) Schematic representation of the M. tuberculosis furA region. The AT-rich sequence upstream of furA is indicated by a black square. The DNA fragments cloned in pSG10ter are indicated, and the coordinates of the fragments are reported. The M. tuberculosis coordinates are arbitrary, with coordinate +1 corresponding to the first nucleotide of the furA gene. (Bottom) Luciferase activity expressed in M. smegmatis mc2155 transformed by the plasmids was measured, as described in Materials and Methods, both in the absence of (−H2O2) and after (+H2O2) oxidative stress. The mean values ± standard deviations for two to four independent clones tested are reported.
Luciferase activity was measured both before and after peroxide treatment of the cells, as described in Materials and Methods, and the results are reported in Fig. 1. Plasmids pMT8, pMT13, and pMYT101 expressed high luciferase activity that increased upon oxidative stress. In contrast, pMT12 and pMT14 did not express luciferase. The latter plasmids lack the −35/+33 region that appears to be essential for promoter activity. Deletion of the region upstream of position −45 (pMYT101) did not abolish luciferase-inducible expression, although the activity was reduced to about 20 to 25%, suggesting that the region upstream of position −45 contributes to promoter efficiency.
Upstream of the furA gene, −35 and −10 promoter consensus sequences and a 23-bp AT-rich region overlapping the −35 sequence were observed (Fig. 2A). This latter sequence is highly conserved in M. bovis, M. smegmatis, and Mycobacterium fortuitum upstream of furA (Fig. 2B), thus indicating that a regulatory protein might bind to this region.
FIG. 2.
Sequence of the M. tuberculosis DNA fragment containing the oxidative stress-inducible promoter. (A) Sequence of the −50/+10 M. tuberculosis region. The coordinates are arbitrary, with coordinate +1 corresponding to the first nucleotide of the furA gene. The −35 and −10 sequences are in boldface. The 23-bp AT-rich region is boxed. The ends of the fragments cloned in MT12 and MT14 are indicated. The region protected by FurA is indicated by a line above the sequence. (B) Alignment of the AT-rich DNA sequences upstream of furA in M. tuberculosis, M. bovis, M. smegmatis, and M. fortuitum and the sequence upstream of furS in S. reticuli. The conserved bases are indicated by asterisks.
It was found previously that FurS of S. reticuli is able to bind to a region that shares some homology with the mycobacterial 23-bp AT-rich sequence (25) (Fig. 2B). This suggested that mycobacterial FurA might bind upstream of its own promoter. Thus, we decided to express and purify M. tuberculosis FurA and test the ability of the protein to bind this DNA region.
Purification of M. tuberculosis FurA.
The M. tuberculosis furA gene was amplified by PCR and cloned in pGEX6P1, creating a fusion with the GST protein (see Materials and Methods). Expression of the GST-FurA fusion protein was induced with IPTG in E. coli XL1-Blue cultures, and the protein was recovered by using glutathione-Sepharose. After cleavage with Prescission, the partially purified FurA protein was separated by SDS-polyacrylamide gel electrophoresis (Fig. 3A). One major and three minor bands were visualized, which might represent different conformations of the protein. The major band has an apparent molecular mass of about 20 kDa, as expected for FurA.
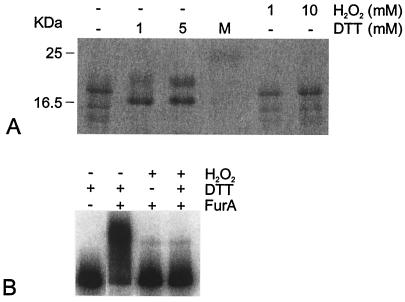
FIG. 3.
Electrophoretic mobility of the FurA protein and binding of FurA to DNA under different redox conditions. (A) Purified M. tuberculosis FurA (1.7 μg) expressed in E. coli was run in the first lane of an SDS-15% polyacrylamide gel. Equivalent samples were mixed with either DTT or H2O2 at the concentrations indicated above the lanes before being loaded on the gel. Lane M, molecular mass markers, with sizes indicated on the left. (B) Gel shift caused by FurA. The DNA probe covers the −154/+33 M. tuberculosis region. FurA (10 μM) was added, as indicated above the lanes, to 32P-labeled DNA probe in the presence of 200 μM Ni2+, and complexes were resolved on a Tris-acetate-8% polyacrylamide gel containing 200 μM Ni2+. DTT (1 mM) and H2O2 (10 mM) were added to the incubation buffer as indicated.
Reducing treatment by addition of 1 or 5 mM DTT changed the electrophoretic mobilities of all FurA bands (Fig. 3A). In contrast, addition of peroxide did not alter the electrophoretic pattern. This indicates that the purified protein is in a (partially) oxidized state. Moreover, the change in electrophoretic mobility suggests that FurA might be a redox-sensing protein.
Binding of FurA to the region upstream of M. tuberculosis furA.
The ability of FurA to bind to the region upstream of the furA gene was tested by gel shift experiments. A preliminary test, using a DNA fragment that covers the −154/+33 M. tuberculosis region, indicated that FurA was able to bind under reducing conditions (1 mM DTT), whereas peroxide treatment prevented FurA binding (Fig. 3B). A change in FurA protein conformation upon oxidative treatment was likely to abolish DNA binding. Gel shift analysis was thus performed with the addition of 1 mM DTT to the binding reaction mixture.
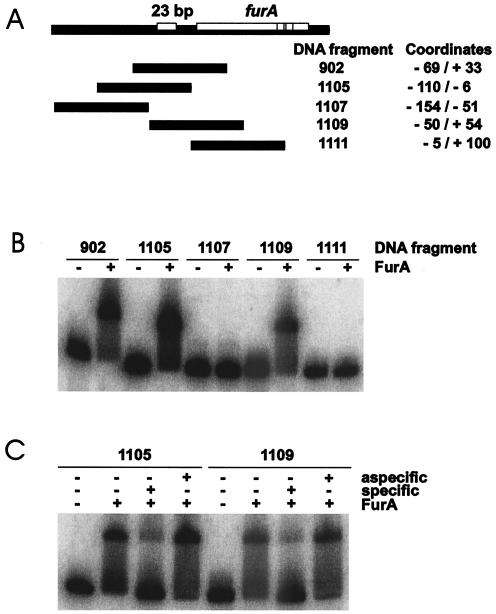
In order to map the binding region, several DNA fragments overlapping the 5′ region of M. tuberculosis furA were amplified by PCR, end labeled, and used in gel shift experiments with the purified FurA protein. The fragments tested and the results of the gel shift assay with FurA are reported in Fig. 4. Three fragments (fragments 902, 1105, and 1109) were shifted in the presence of FurA protein (Fig. 4B). All of them contain the 23-bp AT-rich region identified upstream of furA. On the other hand, two fragments lacking this sequence (fragments 1107 and 1111) were not shifted in the presence of FurA. Addition of 3,000 M specific unlabeled DNA efficiently competed FurA binding to both the 1105 and 1109 fragments, whereas no effect was observed upon addition of a similar amount of unspecific DNA (Fig. 4C). Thus, we concluded that a specific region located between positions −50 and −6 was sufficient for FurA binding.
FIG. 4.
FurA binds DNA fragments covering the M. tuberculosis furA upstream region. (A) Map of the DNA fragments. (B) Gel shift caused by FurA. FurA binding was performed as described in the legend to Fig. 3, except that 12 μM FurA was added. (C) Specificity of FurA binding. Specific (same DNA fragment) and nonspecific (1111 DNA fragment) competitor DNA was added to each binding reaction mixture at 3,000 M.
Identification of the FurA binding site in M. tuberculosis.
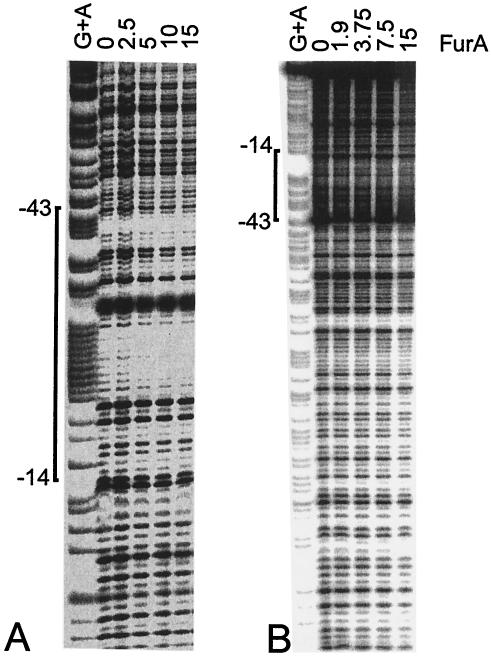
Footprinting analysis was performed in order to identify the FurA binding site upstream of furA. The 187-bp EcoRI-BamHI M. tuberculosis region carried by MYT101 was digested, end labeled at position +33 and used in footprinting experiments in the presence of increasing concentrations of FurA and 1 mM DTT (Fig. 5A). A weakly protected region extending from position −14 to −43 and centered on the AT-rich region was identified (Fig. 2A). The protected region overlaps the −35 promoter sequence, thus suggesting that FurA binding might control transcription of the furA gene. The same DNA fragment was labeled at the opposite end (position −154), and footprinting was repeated (Fig. 5B). The presence of the −14/−43 protected region, immediately upstream of furA, was confirmed. Moreover, at high FurA protein concentrations we observed an extension of the protection that suggests cooperative binding of FurA proteins on the DNA.
FIG. 5.
Footprint analysis of the FurA binding site upstream of M. tuberculosis furA. The purified −154/+33 DNA region, labeled at either the +33 end (A) or the −154 end (B), was incubated in the presence of the amount of FurA protein (in micrograms) indicated above the lanes for 20 min at room temperature and digested with DNase I as described in Materials and Methods. Maxam-Gilbert A+G sequences of the same fragments were loaded in the first lanes.
Mutagenesis of the FurA operator sequence.
Direct mutagenesis of the furA operator sequence was performed by PCR amplification with mutagenized primers (see Materials and Methods). Three different mutants were obtained, each differing from the wild-type sequence for a 6-bp stretch within the AT-rich sequence. The mutations are reported in Fig. 6A. Gel shift analysis with FurA indicated that each mutation almost completely prevented FurA binding (Fig. 6B), thus suggesting that the mutant sequence is directly involved in FurA operator recognition.
In order to test the effect of the mutations in the operator sequence on promoter activity and inducibility by oxidative stress, we cloned the fragments upstream of the luciferase gene and measured luciferase activity in M. smegmatis mc2155 (Table 1). pMYT103 and pMYT104 expressed high luciferase activity (about 9,500,000 relative light units [RLU]), compared to about 10 times less in the wild-type pMYT101. Both pMYT103 and pMYT104 were not induced upon oxidative stress. Thus, the promoter in the mutants appears to be fully functional, whereas repression by FurA was lost.
TABLE 1.
Luciferase activity expressed by mutant DNA fragments.
| Plasmida | Luciferase RLU (103)b
|
|
|---|---|---|
| Without H2O2 | With H2O2 | |
| pMYT101 | 869 ± 176 | 1,080 ± 202 |
| pMYT102 | 1,682 ± 370 | 1,638 ± 332 |
| pMYT103 | 9,528 ± 1,054 | 8,549 ± 1,314 |
| pMYT104 | 9,632 ± 2,566 | 9,616 ± 2,844 |
The plasmids were derived from pSG10ter by insertion of the M. tuberculosis −154/+33 fragments upstream of the luxAB reporter gene. pMYT101 carries the wild-type sequence, and pMYT102, pMYT103, and pMYT104 carry the B, C, and D mutant sequences indicated in Fig. 6, respectively.
Luciferase activity was measured as described in the legend to Fig. 1. The cultures were treated with 0.062 mM H2O2 for 1 h. The mean values ± standard deviations for four independent clones tested are reported.
Expression of luciferase activity by pMYT102, in which the −35 sequence has been mutated, was about 1,600,000 RLU. In this case, the mutation likely affected promoter activity. However, also in this case, no induction upon oxidative stress could be observed.
All three mutagenized promoters, in which FurA binding was abolished, were no longer controlled upon oxidative stress.
DISCUSSION
In M. tuberculosis, the region encompassing the −45/+33 sequence relative to the furA gene start codon was found to be sufficient to promote transcription and to respond to oxidative stress induction. In this region of M. bovis BCG, the 5′ end of pfurA and canonical −35 and −10 promoter sequences have been previously identified (23). Moreover, M. tuberculosis DNA fragments covering this region were found to express a downstream reporter gene (21, 23, 27). Extension of the cloned region upstream of position −45 increased promoter efficiency about fourfold, suggesting the presence of sequences that might be involved in regulation of transcription from pfurA.
In this work, we demonstrate that pfurA is negatively controlled by the mycobacterial FurA protein, which binds upstream of the furA gene. Thus, FurA autoregulates its own expression.
Band shift experiments mapped the Fur box in the 50-bp region upstream of the furA gene. Binding appears to be specific, since it was not competed by addition of a 3,000 M concentration of nonspecific DNA. By footprinting analysis a protected region covering the −14/−43 sequence upstream of the initiation codon was identified. This box overlaps the −35 consensus sequence of the pfurA promoter, suggesting that FurA competes with RNA polymerase for binding to the operator-promoter region. Moreover, the Fur box is centered on a 23-bp AT-rich sequence, which is highly conserved in mycobacteria and in S. reticuli (Fig. 2B). In S. reticuli FurS was found to bind to this sequence, thus repressing transcription of the downstream operon. Addition of metal ions enlarged the protected region, whereas oxidative conditions abolished binding (25). It might be hypothesized that FurA binding occurs by a similar mechanism. In our experiments, Ni2+ ions were always present in the binding buffer. In their absence binding appeared to be weak or absent (data not shown). Moreover, reducing conditions were necessary, since in the presence of peroxide FurA binding did not occur.
A gel shift of the DNA fragments was observed only in the presence of FurA at a concentration higher than 3 μM (data not shown), with a molar ratio of FurA to DNA of about 4 × 10−4. The large amount of protein required to observe the band shift may suggest that most of our protein preparation was inactive. Since different FurA isoforms were observed in SDS-polyacrylamide gel electrophoresis, we hypothesize that only one of them may be active.
It should be noted that in our footprinting experiments protection by FurA was not complete, since most of the bands were reduced but still visible. Only a slight increase of the protection could be observed by increasing the FurA concentration. This might indicate that not all of the DNA molecules were bound by FurA. Whether this is a consequence of an internal equilibrium or of the in vitro conditions used was not established. However, the band shift experiments are in good agreement with the protected region identified by footprinting analysis, and this result was confirmed by footprinting on the opposite DNA strand.
At high FurA concentrations, we observed an enlargement of the protected region, which extended to most of the DNA fragment used. Thus, FurA binds to the Fur box, and from this nucleoprotein complex several FurA molecules may be added cooperatively, covering the DNA fragment. A similar mechanism was proposed for Fur autoregulation in Helicobacter pylori (9).
The 23-bp AT-rich region is unique in the M. tuberculosis genome, since no homologous sequences could be found by the FINDPATTERNS program of the Genetics Computer Group package unless at least five mismatches were allowed. This might indicate either that M. tuberculosis FurA is not a global transcription regulator or that different DNA regions are recognized and bound by FurA. The 23-bp AT-rich region is almost completely conserved in M. smegmatis, and our data indicate that M. smegmatis FurA recognizes and binds to the M. tuberculosis sequence cloned upstream of the reporter gene. Thus, FurA autoregulation appears to be common to both mycobacterial species.
The addition of DTT changes FurA electrophoretic mobility, indicating that the protein senses the redox state. This property is likely to have a central role in the oxidative stress response: furA transcription is repressed by FurA binding, and, upon oxidative stress, the oxidized FurA protein loses its DNA affinity and transcription from pfurA can take place.
Three different mutations of a 6-bp stretch within the AT-rich sequence prevented FurA binding, thus confirming that the sequence is specifically recognized by the FurA protein. The effect of the mutations was to eliminate the control of furA expression. It is interesting that all of the mutations introduced in M. smegmatis caused a complete loss of the response to oxidative stress. Even more interesting is the fact that two mutations (pMYT103 and pMYT104) exhibited a very high luciferase activity when cloned upstream of the reporter luxAB, i.e., about 10 times higher than wild-type activity. Apparently, both 6-bp substitutions did not alter any sequence essential for promoter activity. This suggests that pfurA is a very strong promoter and that it is maintained in an almost completely repressed state by FurA itself. Upon oxidative stress several FurA proteins detach from the DNA, and pfurA is partially activated. This result also indicates that both M. smegmatis and M. tuberculosis FurA, which share high homology, are able to interact with pfurA promoter sequences.
The third mutant (pMYT102) carries a 6-bp substitution in the −35 sequence. This mutation had a double effect: it completely abolished FurA binding, thus preventing the oxidative stress response, and reduced, but did not abolish, pfurA activity. In this case the mutation altered a sequence relevant for promoter activity. Indeed, the presence of an intact −10 sequence enabled RNA polymerase to initiate transcription, although at a reduced rate. Promoters that lack a canonical −35 sequence but are still functional have been reported for M. tuberculosis (3).
Acknowledgments
We thank Gianni Dehò for useful discussions and for reading the manuscript. We are also grateful to Enzo Scarlato and Giovanni Bertoni for discussing some results. We thank Chiara Camisaschi and Diego Covarello for performing some experiments.
This work was supported by grant COFIN2001 n. 2001053855_003 from the Ministero dell'Istruzione, dell'Università e della Ricerca, Rome, Italy.
REFERENCES
- 1.Althaus, E. W., C. E. Outten, K. E. Olson, H. Cao, and T. V. Halloran. 1999. The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry 38:6559-6569. [DOI] [PubMed] [Google Scholar]
- 2.Bagg, A., and J. B. Neilands. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471-5477. [DOI] [PubMed] [Google Scholar]
- 3.Bashyam, M. D., D. Kaushal, S. K. Dasgupta, and A. K. Tyagi. 1996. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J. Bacteriol. 178:4847-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 5.Bullock, W., J. Fernandez, and J. Short. 1987. XLI-Blue: a high efficiency plasmid transforming recA E. coli strain with β-galactosidase selection. BioTechniques 5:376-379. [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Coy, M. 1995. The interaction of the ferric uptake regulation protein with DNA. Biochem. Biophys. Res. Commun. 212:784-792. [DOI] [PubMed] [Google Scholar]
- 8.Coy, M., and J. B. Neilands. 1991. Structural dynamics and functional domains of the Fur protein. Biochemistry 30:8201-8210. [DOI] [PubMed] [Google Scholar]
- 9.Delany, I., G. Spohn, A. Pacheco, R. Ieva, C. Alaimo, R. Rappuoli, and V. Scarlato. 2002. Autoregulation of Helicobacter pylori Fur revealed by functional analysis of the iron-binding site. Mol. Microbiol. 46:1107-1122. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., F. Giovannini, M. Herrero, and J. B. Neilands. 1988. Metal ion regulation of gene expression. Fur repressor-operator interaction at the promoter region of the aerobactin system of pColV-K30. J. Mol. Biol. 203:875-884. [DOI] [PubMed] [Google Scholar]
- 11.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (Fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubrac, S., and D. Touati. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escolar, L., V. de Lorenzo, and M. Perez. 1997. Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol. Microbiol. 26:799-808. [DOI] [PubMed] [Google Scholar]
- 14.Escolar, L., M. Perez, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez, D., P. Saint, J. M. Latour, S. Michaud, and E. Forest. 2001. Conformational changes of the ferric uptake regulation protein upon metal activation and DNA binding; first evidence of structural homologies with the diphtheria toxin repressor. J. Mol. Biol. 310:83-91. [DOI] [PubMed] [Google Scholar]
- 16.Hahn, J.-S., S.-Y. Oh, K. F. Chater, Y.-H. Cho, and J.-H. Roe. 2000. H2O2-sensisitive Fur-like repressor CatR regulating the major catalase gene in Streptomyces coelicolor. J. Biol. Chem. 275:38254-38260. [DOI] [PubMed] [Google Scholar]
- 17.Hall, H. K., and J. W. Foster. 1996. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J. Bacteriol. 178:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassett, D. J., M. L. Howell, U. A. Ochsner, M. L. Vasil, Z. Johnson, and G. E. Dean. 1997. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J. Bacteriol. 179:1452-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacquamet, L., D. Aberdam, A. Adrait, J. L. Hazemann, J. M. Latourand, and S. Michaud. 1998. X-ray absorption spectroscopy of a new zinc site in the Fur protein from Escherichia coli. Biochemistry 37:2564-2571. [DOI] [PubMed] [Google Scholar]
- 20.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Master, S., T. C. Zahrt, J. Song, and V. Deretic. 2001. Mapping of Mycobacterium tuberculosis katG promoters and their differential expression in infected macrophages. J. Bacteriol. 183:4033-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milano, A., F. Forti, C. Sala, G. Riccardi, and D. Ghisotti. 2001. Transcriptional regulation of furA and katG upon oxidative stress in Mycobacterium smegmatis. J. Bacteriol. 183:6801-6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochsner, U. A., and M. L. Vasil. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. USA 93:4409-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortiz de Orué Lucana, D., and H. Schrempf. 2000. The DNA-binding characteristics of the Streptomyces reticuli regulator FurS depend on the redox state of its cysteine residues. Mol. Gen. Genet. 264:341-353. [DOI] [PubMed] [Google Scholar]
- 26.Pagàn-Ramos, E., J. Song, M. McFalone, M. H. Mudd, and V. Deretic. 1998. Oxidative stress response and characterization of the oxyR-ahpC and furA-katG loci in Mycobacterium marinum. J. Bacteriol. 180:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pym, A. S., P. Domenech, N. Honoré, J. Song, V. Deretic, and S. T. Cole. 2001. Regulation of catalase-peroxidase (KatG) expression, isoniazid sensitivity and virulence by furA of Mycobacterium tuberculosis. Mol. Microbiol. 40:879-889. [DOI] [PubMed] [Google Scholar]
- 28.Sabbattini, P., F. Forti, D. Ghisotti, and G. Deho. 1995. Control of transcription termination by an RNA factor in bacteriophage P4 immunity: identification of the target sites. J. Bacteriol. 177:1425-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stojiljkovic, I., and K. Hantke. 1995. Functional domains of the Escherichia coli ferric uptake regulator protein (Fur). Mol. Gen. Genet. 247:199-205. [DOI] [PubMed] [Google Scholar]
- 30.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 31.Vasil, M. L., and U. A. Ochsner. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34:399-413. [DOI] [PubMed] [Google Scholar]
- 32.Zou, P., I. Borovok, D. Ortiz de Orué Lucana, D. Muller, and H. Schrempf. 1999. The mycelium-associated Streptomyces reticuli catalase-peroxidase, its gene and regulation by FurS. Microbiology 145:549-559. [DOI] [PubMed] [Google Scholar]