Abstract
The cell wall polysaccharide of Streptococcus gordonii 38 functions as a coaggregation receptor for surface adhesins on other members of the oral biofilm community. The structure of this receptor polysaccharide (RPS) is defined by a heptasaccharide repeat that includes a GalNAcβ1→3Gal-containing recognition motif. The same RPS has now been identified from S. gordonii AT, a partially sequenced strain. PCR primers designed from sequences in the genomic database of strain AT were used to identify and partially characterize the S. gordonii 38 RPS gene cluster. This cluster includes genes for seven putative glycosyltransferases, a polysaccharide polymerase (Wzy), an oligosaccharide repeating unit transporter (Wzx), and a galactofuranose mutase, the enzyme that promotes synthesis of UDP-Galf, one of five predicted RPS precursors. Genes outside this region were identified for the other four nucleotide-linked sugar precursors of RPS biosynthesis, namely, those for formation of UDP-Glc, UDP-Gal, UDP-GalNAc, and dTDP-Rha. Two genes for putative galactose 4-epimerases were identified. The first, designated galE1, was identified as a pseudogene in the galactose operon, and the second, designated galE2, was transcribed with three of the four genes for dTDP-Rha biosynthesis (i.e., rmlA, rmlC, and rmlB). Insertional inactivation of galE2 abolished (i) RPS production, (ii) growth on galactose, and (iii) both UDP-Gal and UDP-GalNAc 4-epimerase activities in cell extracts. Repair of the galE1 pseudogene in this galE2 mutant restored growth on galactose but not RPS production. Cell extracts containing functional GalE1 but not GalE2 contained UDP-Gal 4-epimerase but not UDP-GalNAc 4-epimerase activity. Thus, provision of both UDP-Gal and UDP-GalNAc for RPS production by S. gordonii 38 depends on the dual specificity of the epimerase encoded by galE2.
Specific interactions between different bacteria play an important role in development of the simple biofilm community that forms during primary colonization of the human tooth surface (36-38). The lactose-sensitive coaggregations between type 2 fimbriated strains of Actinomyces naeslundii and receptor-bearing strains of Streptococcus sanguinis, Streptococcus gordonii, Streptococcus oralis, and Streptococcus mitis are well-studied examples of such interactions (10, 19). Structural characterization of the cell wall polysaccharides isolated from over 20 receptor-bearing streptococcal strains has resulted in the identification of six different receptor polysaccharides (RPS) (1-4, 12, 30, 39), which are designated types 1Gn, 2Gn, 2G, 3G, 4Gn, and 5Gn RPS to reflect the structural relationships that exist between the different phosphodiester-linked, hexa- or heptasaccharide repeating units of these molecules (12). Each RPS repeating unit contains a host-like recognition motif consisting of Galf linked β1→6 to either Galβ1→3GalNAc (G) or GalNAcβ1→3Gal (Gn). These features are recognized as receptors by Gal- and GalNAc-binding surface adhesins on other bacteria, such as A. naeslundii (11), while other features of the individual repeating units are more closely associated with the antigenicity of these polysaccharides (12, 31). Consequently, certain types of RPS, such as 1Gn RPS of S. oralis 34 and 4Gn RPS of S. oralis C104, function as the same coaggregation receptor but react as different antigens. In contrast, other types of RPS, such as types 2Gn RPS of S. gordonii 38 and 2G RPS of S. mitis J22, function as different receptors but react as similar antigens. The functional design of these microbial receptors points to their coevolution with the host oral environment.
The overall structural similarity that exists between the RPS of viridans group streptococci and the capsular polysaccharides (CPS) of Streptococcus pneumoniae suggests a common biosynthetic pathway for these molecules. The synthesis of each CPS serotype depends on a large operon consisting of four common regulatory genes followed by a serotype-specific region (17, 20, 33, 34). The latter region includes genes for the glycosyltransferases that synthesize the lipid-linked repeating unit of the polysaccharide, a flipase (Wzx) that transports the oligosaccharide moiety to the outer surface of the membrane, and a polymerase (Wzy) that links repeating units to form a linear polysaccharide molecule. Additional genes in this region may control the synthesis of nucleotide-linked sugars that are serotype-specific polysaccharide precursors, such as dTDP-Rha, while genes that reside outside CPS clusters control the synthesis of precursors with physiological roles that extend beyond CPS biosynthesis. Examples include UDP-Glc and UDP-Gal, which in addition to being CPS precursors (20, 32) are essential for bacterial utilization of galactose by the Leloir pathway (16).
RPS production, although common among strains of S. oralis, is variable among strains of the other oral streptococci mentioned above (12). Indeed, the three streptococcal strains that are presently being sequenced, namely, S. sanguinis SK36, S. gordonii Challis, and S. mitis NCTC 12261, do not participate in lactose-sensitive coaggregations with A. naeslundii (19). In addition to the genomic databases from these streptococcal strains, a partial genomic database exists for an uncharacterized isolate of S. gordonii that was identified as a contaminant in one stock of S. pneumoniae TIGR4 (48). We now show that this isolate of S. gordonii produces type 2Gn RPS, providing a genomic approach for identification of the loci for RPS biosynthesis in S. gordonii 38, which makes this type of RPS (12, 39).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. gordonii 38 has been described previously (10-12, 19, 39). We have designated the other isolate of S. gordonii that was used in the present investigation strain AT. This strain was initially identified as a contaminant in one bacterial stock of S. pneumoniae TIGR4 (48). It was received from Brian A. Dougherty and was subsequently classified as atypical S. gordonii on the basis of its atypical hemolytic activity on blood agar and its failure to ferment amygdalin (M. Kilian, personal communication). Streptococci were routinely grown at 37°C in Todd-Hewitt Broth (Difco Laboratories, Detroit, Mich.) or on anaerobically incubated plates of brain heart infusion agar (Difco Laboratories). Streptococci were also grown in FMC chemically defined medium (47) containing 1% glucose or 1% galactose. The growth media of ermAM insertional mutant strains contained erythromycin at a final concentration of 5 or 10 μg/ml.
Chemically competent or electrocompetent Escherichia coli (strain DH5α from BRL or TOP 10 from Invitrogen [Carlsbad, Calif.]) were used for gene cloning and for the preparation of recombinant plasmids. These bacteria were grown aerobically at 37°C in Luria-Bertani broth or agar (Difco) in the presence of either 50 or 100 μg of ampicillin/ml or 200 μg of erythromycin/ml as needed.
DNA isolation, sequencing, and analysis.
Molecular techniques were performed by using standard methods (41) or by following instructions provided with various commercially available reagents and kits, including a genomic DNA isolation kit (Promega, Madison, Wis.), which was used to isolate genomic DNA from streptococci (9). DNA sequencing was performed with a BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) and an ABI model 3100 automated sequencer. Sequencing templates included PCR products of up to 2.5 kb. These were amplified from strain 38 genomic DNA by using Platinum PCR Supermix (Invitrogen), purified by using a QIAquick PCR purification kit (Qiagen, Valencia, Calif.) and added to sequencing reaction mixtures at a concentration of approximately 20 ng per μl. The primers used for these amplifications were designed from sequences identified in the available genomic database of strain AT. Access to this database, which includes 4,596 sequences totaling 4.4 Mbp of DNA sequence, was generously provided by The Institute for Genomic Research, Rockville, Md. Vectorette (Sigma Genosys, The Woodlands, Tex.) libraries were prepared from strain 38 genomic DNA according to instructions provided by the manufacturer and were used to PCR amplify uncharacterized chromosomal regions adjacent to known sequences. The sequence of the repetitive region of orfO was determined from the corresponding cloned DNA inserts in two plasmids by using a DNA Kilo deletion sequencing kit (PanVera Corp., Madison, Wis.). The 1,679-bp fragment of cloned DNA in these plasmids corresponded to the region from bases 17,952 to 19,630 in the sequence available under GenBank accession number AY147914. This region was PCR amplified from S. gordonii 38 genomic DNA by using Pfu Turbo Hotstart DNA polymerase (Stratagene, La Jolla, Calif.) and appropriate primers and was cloned into pCR4Blunt-TOPO (Invitrogen). The region containing rmlA, rmlC, and a portion of rmlB (i.e., from bases 3,253 to 5,249 of the sequence available under GenBank accession number AY147913) was sequenced from plasmids that were identified in a library of S. gordonii 38 genomic DNA by hybridization with a probe for cps19fM of S. pneumoniae (33). The library used in these studies was prepared in pDL278 (25) by cloning a 1.8-kb fraction of strain 38 genomic DNA that was prepared by sucrose density gradient untracentrifugation of a Sau3A partial digest.
DNA sequences of PCR products and cloned DNA fragments were assembled by using ContigExpress (InforMax, Inc., Bethesda, Md.). The resulting sequence was annotated by using other software modules of Vector NTI Suite 7. Nucleotide and predicted amino acid sequence homologies with genes and proteins in the database were identified by BLAST (6). Putative promoters and transcriptional terminators were identified by using MacVector 6.5 (Accelrys, Burlington, Mass.).
Northern blotting.
RNA was extracted from S. gordonii 38 harvested during the exponential phase of growth by using Trizol (Invitrogen) and was purified as previously described (28). Northern blotting was performed with a Northern Max-Gly kit (Ambion, Austin, Tex.) following the instructions of the manufacturer. Hybridization probes were prepared by random priming with [α-32P]dCTP, a RadPrime DNA Labeling system (Invitrogen), and primers designed from the sequence available under GenBank accession number AY147913. The forward primers that were used to prepare the rmlA, galE2, and rmlD probes corresponded to the sequences between bases 3,460 and 3,478, 6,072 and 6,090, and 14,922 and 14,940, respectively, of this GenBank sequence, and the corresponding reverse primers for each gene probe were complementary to the sequences between bases 3,840 and 3,860, 6,731 and 6,750, and 15,541 and 15,560, respectively. Hybridization of labeled probes with RNA on nylon membranes (Hybond-N+; Millipore Corp., Bedford, Mass.) was for 16 h at 42°C in ULTRAhyb (Ambion). Membranes were washed at 42°C in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% sodium dodecyl sulfate prior to autoradiography.
Insertional mutagenesis.
The nonreplicative plasmids (i.e., suicide plasmids) used to prepare S. gordonii mutant strains XC1, XC2, XC3, XC4, XC5, and XC6 contained the cloned ermAM cassette flanked by targeting sequences for the gene of interest (i.e., wzg, wchA, wefE, orfO, orfP, and galU, respectively). Amplification of the ermAM cassette from pKSerm2 (29) and of each 0.5- to 1-kb targeting sequence from S. gordonii 38 genomic DNA was performed by using High Fidelity Taq (Invitrogen) and PCR primers that contained complementary 5′ linkers. The linker sequence CTGCGGTC was added to the 5′ ends of the reverse primers that were used to amplify the upstream targeting sequence and the ermAM cassette. The complementary sequence, GACCGCAG, was added to the 5′ ends of the forward primers that were used to amplify the ermAM cassette and the downstream targeting sequence. The sequences of the three resulting PCR products were linked by mixed-template PCR (18) and were cloned in pCR-TOPO (Invitrogen) for transformation into E. coli. Transformants were grown on plates that contained 200 μg of erythromycin/ml and were screened by colony PCR to identify plasmids containing the expected DNA fragment (i.e., upstream targeting sequence-ermAM cassette-downstream targeting sequence).
To prepare mutant XC7, a region containing galE1 and portions of each flanking gene (bases 3,050 to 4,959 in the sequence available under GenBank accession number AY147910) was amplified by using pfu Turbo Hotstart DNA polymerase (Stratagene) and was cloned into pCR4Blunt-TOPO (Invitrogen). The resulting plasmid was digested with BamH1 and ClaI to remove a 652-bp fragment from galE1. The ermAM cassette was inserted via complementary restriction sites that were added during PCR amplification of the cassette from pKSerm2.
The suicide vectors used to prepare mutant strains XC8 and XC9 were prepared from pSKerm2 as previously described (29). Each targeting sequence was PCR amplified from strain 38 genomic DNA by using primers that contained appropriate restriction sites for insertion of the resulting PCR product into the multiple cloning site of pKSerm2, on either side of the ermAM cassette. The vector used to disrupt galE2 in mutant XC8 contained a 0.6-kb targeting sequence on the 5′ side and a 2.1-kb targeting sequence on the 3′ side of the ermAM cassette. The vector that was used to disrupt orf9 contained a 0.7-kb targeting sequence on the 5′ side and a 0.5-kb targeting sequence on the 3′ side of the ermAM cassette.
Transformation of S. gordonii 38 with each linearized suicide plasmid was performed as previously described (29). Transformants of S. gordonii 38 were grown on plates of brain heart infusion agar containing 10 μg of erythromycin/ml and were characterized by colony PCR. The location of the ermAM cassette in each insertional mutant (Table 1) was verified by the amplification of specific PCR products across the upstream and downstream boundaries of the ermAM insertion, using primers for upstream and downstream chromosomal sequences that were extraneous to those present in suicide vector constructs.
TABLE 1.
Location of the ermAM insertion in S. gordonii 38 mutant strains
| Mutant strain | Disrupted gene | Location of ermAM cassette in sequence
|
|
|---|---|---|---|
| GenBank accession no. | Between base pairs | ||
| XC1 | wzg | AY147914 | 1,838 and 1,863 |
| XC2 | wchA | AY147914 | 5,355 and 5,540 |
| XC3 | wefE | AY147914 | 15,721 and 15,817 |
| XC4 | orfO | AY147914 | 17,324 and 19,611 |
| XC5 | orfP | AY147914 | 21,346 and 21,467 |
| XC6 | galU | AY147912 | 850 and 1,149 |
| XC7 | galE1 | AY147910 | 3,680 and 4,331 |
| XC8 | galE2 | AY147913 | 6,640 and 6,644 |
| XC9 | orf9 | AY147913 | 7,813 and 7,984 |
Repair of the galE1 pseudogene in strain XC8.
The transforming DNA that was used to repair the galE1 pseudogene in mutant strain XC8 was prepared by mixed-template PCR (26) performed with a high-fidelity, proofreading DNA polymerase (Pfu Turbo Hotstart DNA polymerase; Stratagene). Initially, two adjacent sequences were amplified from strain 38 genomic DNA by using primers designed from the sequence available under GenBank accession number AY147910. One sequence was amplified by using primer F1 (bases 2,981 to 3,001) and R1 (complementary to bases 3,530 to 3,551 with thymine added to the 5′ end to restore the deleted adenine in the galE1 sequence). The adjacent sequence was amplified by using primer F2 (bases 3,552 to 3,576) and R2 (complementary to bases 4,042 to 4,061). The two sequences were then linked by 30 cycles of PCR performed in a total volume of 50 μl containing 2.5 U of Pfu Turbo Hotstart DNA polymerase, 0.5 ng of each purified PCR product as mixed template, 0.3 μM each of primers F1 and R2, and a 0.0023 μM concentration of the linking primer (complementary to bases 3,529 to 3,575 with thymine [T] inserted between bases 3,551 and 3,552). The presence of added adenine (A) at the expected position of the resulting 1,082-bp PCR product was confirmed by complete sequencing of the purified fragment. This PCR product was used to transform strain XC8, and strain XC8R(gal+) was isolated following the growth of transformants in FMC medium containing 1% galactose. Gal+ bacteria did not appear in controls that did not receive transforming DNA. The presence of complete galE1 in strain XC8R was confirmed by sequencing of a PCR product amplified from genomic DNA of this strain.
Detection, identification, and quantification of RPS.
Coaggregation between S. gordonii strain AT or 38 and A. naeslundii WVU45 or S. gordonii Challis was assessed by a visual assay (10, 19). RPS was purified by DEAE anion exchange column chromatography of a mutanolysin digest of S. gordonii AT cell walls and was identified by immunodiffusion performed with rabbit antisera and purified polysaccharide antigens as previously described (12).
Wild-type and insertional mutant strains were compared as inhibitors of an RPS-specific enzyme-linked immunosorbent assay (ELISA). The primary antibody used in this assay was affinity purified from rabbit antiserum R49 against S. mitis J22 (12) by a 4 M MgCl2 elution from a small column of Affi-Gel Hz (Bio-Rad Laboratories, Hercules, Calif.) containing coupled RPS. Prior to coupling, the RPS (10 mg/ml) was incubated with sodium periodate (0.86 mM) in 10 mM NaHCO3 buffer (pH 8.2) for 1 h at 4°C in the dark to partially oxidize adjacent hydroxyl groups. The calculated molar ratio of adjacent hydroxyl groups to periodate in the reaction mixture was approximately 70:1. RPS-coated ELISA plates were prepared by overnight incubation of biotinylated strain 38 RPS in wells of avidin-coated Immunolon 1B Flat Bottom Microtiter plates (Thermo Labsystems, Franklin, Mass.). The polysaccharide of this strain was biotinylated (presumably via amino groups of residual cell wall peptidoglycan fragments) by incubating 5 mg of RPS with 43 μmol of biotin-N-hydroxysuccinimide ester (BNHS) (Bio-Rad Laboratories) in 1 ml of 0.1 M NaHCO3 buffer (pH 8.2) for 2 h at room temperature followed by dialysis to remove free BNHS. Plates were avidin coated by incubation with 2 μg of ImmunoPure Avidin (Pierce, Rockford, Ill.)/ml in 0.05 M sodium carbonate buffer (pH 9.5) at 4°C for time periods that ranged from 1 day to 1 week. The plates were washed with 0.02 M phosphate-buffered saline (pH 7.2) containing 0.05% Tween 20 (PBS-Tween) to remove free avidin, incubated overnight at 4°C with biotinylated strain 38 RPS (30 ng/ml) in PBS-Tween, and washed with PBS-Tween immediately prior to use in ELISA.
The inhibition of ELISA was set up by incubating a constant amount of affinity-purified anti-RPS antibody (30 ng/ml) with twofold serial dilutions of washed bacterial cell suspensions or strain 38 RPS standards for 1 h at room temperature. The bacteria used in these assays were harvested from stationary-phase cultures and were adjusted by turbidity to 2 × 109 bacteria per ml (12). Positive controls containing no inhibiting antigen and negative controls containing no primary antibody were included. Reaction mixtures (100 μl) were transferred to wells of RPS-coated micro-ELISA plates and were incubated for 1 h at room temperature to allow binding of primary antibody to solid-phase antigen. ELISA plates were washed with PBS-Tween, incubated for 2 h with peroxidase-conjugated, affinity-purified goat anti-rabbit immunoglobulin G (Bio-Rad Laboratories), and developed with a tetramethylbenzidine peroxidase enzyme immunoassay substrate kit (Bio-Rad Laboratories). Absorbance was read at 450 nm in a Microplate Reader (Molecular Devices Corp., Sunnyvale, Calif.) and was used to calculate concentrations of bacteria and soluble RPS standard required for 50% inhibition of ELISA. RPS production by each bacterial strain was averaged from the results of three independent experiments.
Gal and GlcNAc 4-epimerase assays.
Suspensions of bacteria (25% weight per volume), washed in 25 mM Tris-HCl buffer (pH 7.5) containing 1 mM MgCl2, were disrupted in a Branson model 350 sonifier at 0°C. Disrupted bacteria were subjected to high-speed centrifugation (180,000 × g for 2 h) at 5°C to obtain cell extracts. Protein concentrations of extracts were determined by the bicinchoninic acid procedure (Pierce) with bovine serum albumin as the standard.
The assay for UDP-galactose 4-epimerase (EC 5.1.3.2) was performed as previously described (7) by adding 20 to 50 μl of cell extract (approximately 100 to 250 μg of protein) to 1 ml of a solution containing 50 mM Tris-HCl buffer (pH 8.0), 5 mM MgCl2, 1 mM NAD+, and 0.03 U of NAD+-dependent uridine 5′-diphosphoglucose dehydrogenase (Sigma). The assay was begun by addition of 0.5 mM UDP-Gal, and the increase in A340 was followed in a Beckman DU 640 spectrophotometer. Initial rates of NADH formation were determined by using the kinetics program installed in the instrument. A molar extinction (ɛ340) of 6,220 M−1 cm−1 was assumed in all calculations.
The assay for UDP-N-acetylglucosamine 4-epimerase (EC 5.1.3.7) was performed as previously described (15) but incorporated recent modifications (14, 53). In this procedure, the conversion of UDP-GalNAc to UDP-GlcNAc is measured after acid hydrolysis by the 3.6-fold increase in the color (A585) of free GlcNAc over GalNAc in the Morgan-Elson reaction. The reactions were carried out by adding 20 μl of cell extract to a volume containing 0.5 ml of 10 mM glycine, 1 mM MgCl2, 0.1 mM EDTA, and 0.1 mM UDP-GalNAc. Enzyme activity was halted after 5 and 10 min of incubation at 37°C by the addition of 0.8 μl of concentrated HCl. Following hydrolysis and completion of the Morgan-Elson reaction, color development was measured at 585 nm. Control assays with extract alone and with substrate only were run simultaneously. All assays were performed in triplicate. Product formation (i.e., GlcNAc formed by hydrolysis of UDP-GlcNAc) was measured from standard plots prepared by subjecting UDP-GlcNAc, UDP-GalNAc, GlcNAc, and GalNAc (Sigma-Aldrich, St. Louis, Mo.) to the same procedures.
Nucleotide sequence accession numbers.
The DNA sequences determined in this study are available under GenBank accession numbers AY147910, AY147911, AY147912, AY147913, and AY147914.
RESULTS
The coaggregation receptor of S. gordonii AT.
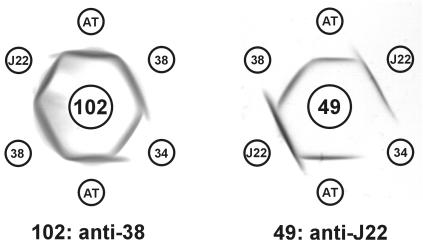
The presence of RPS on S. gordonii AT was initially detected by this strain's lactose-inhibitable coaggregation with A. naeslundii and its GalNAc-inhibitable coaggregation with S. gordonii Challis. Whereas the type 2 fimbriae of A. naeslundii recognize both Gn and G types of RPS (11), the GalNAc-binding adhesin of S. gordonii Challis (46) is specific for Gn types of RPS (12, 19). To further identify the coaggregation receptor of strain AT, a mutanolysin digest of cell walls was prepared from this strain and was fractionated by DEAE Sephacel anion exchange column chromatography. The fractions obtained from gradient elution of the column were assayed for the presence of carbohydrate and specific antigens. A peak of carbohydrate was detected at the position expected of RPS, in fractions containing from 115 to 130 mM NaCl (12). Antigenic identity was seen between the nondialyzable material in these fractions and the type 2Gn RPS of S. gordonii 38 in immunodiffusion performed with antisera against S. gordonii 38 (anti-2Gn antibody) and S. mitis J22 (anti-2G antibody) (Fig. 1). In contrast, nonidentity was seen between the strain AT polysaccharide and either type 1Gn RPS of S. oralis 34 or type 2G RPS of S. mitis. Therefore, S. gordonii AT synthesizes type 2Gn RPS.
FIG. 1.
Identification of type 2Gn RPS of S. gordonii AT by immunodiffusion. The RPS added to the outer wells was that of S. gordonii AT (AT), type 1Gn of S. oralis 34 (34), type 2Gn of S. gordonii 38 (38), or type 2G of S. mitis J22 (J22). The antiserum added to the center wells was prepared against S. gordonii 38 (102) or S. mitis J22 (49).
Identification of the RPS gene cluster of S. gordonii 38.
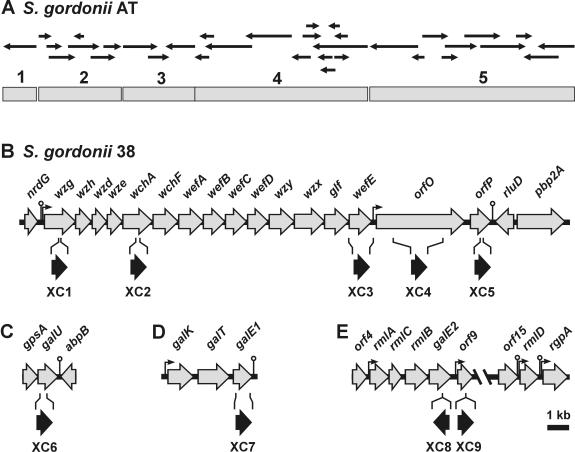
The partial genomic database of strain AT was searched for sequences encoding proteins that could be involved in polysaccharide or nucleotide-linked monosaccharide biosynthesis. Approximately 50 contigs, ranging in size from 0.5 to 2.5 kb, were found. Following the removal of associated vector sequences, 23 sequences were assembled into three larger sequences (Fig. 2A, shaded bars designated 2, 3, and 4). These were judged to be adjacent on the basis of the amplification of overlapping PCR products from strain AT genomic DNA (results not shown). BLASTX analysis of these sequences revealed a gene cluster for polysaccharide biosynthesis, but a number of sequence ambiguities prevented the clear definition of individual genes. Fourteen contiguous genes were, however, identified (Fig. 2B) following PCR amplification and complete sequencing of the corresponding region from S. gordonii 38 genomic DNA by using primers designed from strain AT sequences.
FIG. 2.
Genetic loci associated with 2Gn RPS biosynthesis in S. gordonii. (A) Sequences (small arrows) identified in the partial genomic database of S. gordonii AT were assembled into five larger contiguous sequences (shaded bars). (B) The corresponding 25.5-kb genomic sequence of S. gordonii 38 contained the gene cluster for type 2Gn RPS biosynthesis, extending from wzg to wefE, and flanking regions. Comparison (Best Fit program; GCG) of the corresponding strain 38 and AT sequences (i.e., from 1 to 5) revealed nucleotide sequence identities of 99.9, 97.0, 98.4, 99.6, and 95.1%, respectively. Other genetic loci that were identified in S. gordonii 38 included: galU flanked by gpsA and abpB (C); the galactose operon containing the galE1 pseudogene (D); and two regions of a continuous 17.3-kb sequence containing the rml-galE2 operon and rmlD (E). The positions of putative promoters (|→) and rho-independent transcriptional terminators (○) are indicated. The position of the ermAM cassette (black arrow) in each mutant strain (XC1, XC2, XC3, XC4, XC5, XC6, XC7, XC8, or XC9) is indicated below the disrupted ORF of strain 38.
The first four genes in the S. gordonii 38 cluster were homologues of the four regulatory genes found at the 5′ ends of different S. pneumoniae CPS and Streptococcus thermophilus exopolysaccharide gene clusters (Table 2). Homologous genes are also found at the 5′ end of the Streptococcus suis Cps2 cluster but are arranged in a different order than that seen in the other three streptococcal species (42). Weak homology at the nucleotide level was noted between three of the first four S. gordonii 38 genes (i.e., the first, second, and fourth) and the corresponding genes of the other streptococcal species. The similarities noted involve short sequences, generally less than 150 bp, that are approximately 80% identical. The predicted proteins encoded by these four genes of S. gordonii are, however, from 50 to 70% identical over their entire lengths to the corresponding proteins of S. pneumoniae and the other streptococci. Based on this homology, the first four genes in the S. gordonii cluster were designated wzg, wzh, wzd, and wze, respectively, the same names that were previously assigned to the corresponding four genes in the CPS clusters of S. pneumoniae (20).
TABLE 2.
S. gordonii genes and selected homologues of other bacteria
| Gene | Protein size (aa) | Selected homologue | Accession no. | % Identity (aa) | Proposed function of S. gordonii protein |
|---|---|---|---|---|---|
| nrdG | 199 | S. pneumoniae (4) NrdG | AE007335 | 92 (196) | Anaerobic ribonucleotide-triphosphate reductase-activating protein |
| wzg | 486 | S. pneumoniae (18C) Wzg | AAK20707 | 61 (447) | Transcriptional regulator |
| S. suis Cps2A | AAD24447 | 57 (482) | |||
| S. thermophilus Eps6A | AAN63713 | 52 (447) | |||
| wzh | 243 | S. pneumoniae (18C) Wzh | AAK20708 | 70 (243) | Tyrosine phosphatase |
| S. thermophilus Eps6B | AAN63714 | 64 (243) | |||
| S. suis Cps2D | AAD24450 | 57 (241) | |||
| wzd | 230 | S. pneumoniae (18C) Wxd | AAK20709 | 58 (230) | Chain length regulator |
| S. thermophilus Eps6C | AAN63715 | 52 (231) | |||
| S. suis Cps2B | AAD24448 | 52 (229) | |||
| wze | 233 | S. pneumoniae (18C) Wze | AF316642 | 62 (214) | Tyrosine kinase |
| S. suis 2C | AAD24449 | 59 (214) | |||
| S. thermophilus Eps6D | AAN63716 | 55 (214) | |||
| wchA | 458 | S. pneumoniae (18C) WchA | AAK20711 | 58 (446) | Glucosyl-1-phosphate transferase |
| S. pneumoniae Cps14E | CAA59777 | 59 (446) | |||
| S. thermophilus Eps6E | AAN63749 | 58 (456) | |||
| S. suis Cps2E | AAD24451 | 55 (454) | |||
| wchF | 390 | S. pneumoniae (18C) WchF | AAK20712 | 80 (389) | Rhamnosyltransferase (retaining) |
| S. thermophilus Eps6F | AAN63718 | 80 (390) | |||
| S. pneumoniae Cps23fF | AAC69529 | 77 (388) | |||
| S. suis Cps2F | AAD24452 | 69 (388) | |||
| wefA | 383 | S. thermophilus Eps6G | AAN63719 | 73 (379) | Glycosyltransferase (retaining) |
| S. suis Cps2G | AAD24453 | 70 (383) | |||
| wefB | 333 | S. thermophilus Eps6H | AAN63720 | 67 (209) | Glycosyltransferase |
| wefC | 333 | S. thermophilus Eps10H | AAN63768 | 78 (163) | Glycosyltransferase |
| N. meningitidis (A) SacB | AAC38286 | 27 (344) | |||
| wefD | 320 | S. pneumoniae Cap33fJ | CAA07403 | 43 (316) | Glycosyltransferase (inverting) |
| S. pneumoniae Cps23fH | AAC69531 | 36 (248) | |||
| S. thermophilus Eps6I | AAN63721 | 34 (304) | |||
| S. pneumoniae Cps14J | T50039 | 35 (223) | |||
| wzy | 388 | S. thermophilus Eps6K | AAN63723 | 22 (328) | Polysaccharide polymerase |
| S. pneumoniae Cap8K | AJ239004 | 19 (369) | |||
| wzx | 470 | S. pneumoniae Cap33fL | AJ006986 | 66 (470) | Repeat unit transporter |
| S. thermophilus Eps6L | AAN63724 | 66 (470) | |||
| glf | 367 | S. pneumoniae Cap33fN | CAA07407 | 86 (365) | Galactofuranose mutase |
| S. thermophilus Eps6M | AAN63725 | 85 (363) | |||
| E. coli Glf | AAB88403 | 60 (357) | |||
| wefE | 351 | S. thermophilus Eps6N | AAN63726 | 62 (349) | Galactofuranose transferase |
| E. coli WbbI | AAB88405 | 32 (361) | |||
| orfO | 1,366 | S. mutans SMU.689 | NP_721116 | 31 (593) | Lysozyme-like protein |
| S. pneumoniae LytC | CAA08765 | 25 (201) | |||
| orfP | 323 | S. gordonii transmembrane protein | CAB40551 | 92 (248) | Transmembrane protein |
| S. pneumoniae (18C) WciX | AAK20718 | 38 (267) | |||
| rluD | 290 | S. pneumoniae SP2011 | NP_346438 | 66 (289) | Ribosomal large subunit pseudouridine synthase D |
| pbp2A | 740 | S. pneumoniae SP2010 | NP_346437 | 65 (738) | Penicillin-binding protein 2A |
| gpsA | NDa | S. pneumoniae GpsA | NP_346510 | 89a (235) | Glycerol-3-phosphate dehydrogenase (NAD(P)+) |
| galU | 313 | S. pneumoniae GalU | CAA06172 | 87 (289) | UTP-glucose-1-phosphate uridylyl transferase |
| abpB | NDa | S. gordonii AbpB | AAK52749 | 99a (223) | Amylase-binding protein B |
| galK | 392 | S. salivarius GalK | AAL67289 | 81 (384) | Galactokinase |
| galT | 493 | S. pneumoniae GalT | NP_359259 | 70 (488) | Galactose-1-phosphate uridylyltransferase |
| galE1 | 299b | S. salivarius GalE | AAL67291 | 83 (298) | Inactive UDP-glucose 4-epimerasec |
| orf4 | 222 | Listeria monocytogenes Lmo0414 | NP_463943 | 47 (222) | Unknown |
| rmlA | 289 | S. pneumoniae (6B) RmlA | AAK20691 | 93 (288) | Glucose-1-phosphate thymidylyltransferase |
| rmlC | 197 | S. pneumoniae (6B) RmlC | AAK20692 | 91 (197) | dTDP-4-keto-6-deoxyglucose-3,5-epimerase |
| rmlB | 348 | S. pneumoniae Cps19AN | AAD19915 | 99 (348) | dTDP-glucose-4,6-dehydratase |
| galE2 | 339 | S. pneumoniae GalE | NP_346051 | 79 (339) | UDP-glucose/UDP-N-acetylglucosamine 4-epimerase |
| orf9 | 212 | S. pyogenes SPy0794 | NP_269011 | 62 (207) | Glycosyltransferase |
| orf15 | 308 | S. pneumoniae GtrB | NP_359052 | 67 (307) | Glycosyltransferase |
| rmlD | 283 | S. pneumoniae Cps2O | NP_357917 | 91 (281) | dTDP-Rhamnose synthase |
| rgpA | 382 | S. mutans RgpAc | T00086 | 68 (359) | Rhamnosyltransferase |
ND, not determined; % identity is based on the portion of the gene that was sequenced.
Amino acid residues predicted from the ORF in the galE1 pseudogene.
Complete GalE1, containing 333 amino acid residues, is a functional UDP-glucose 4-epimerase.
Of the 10 remaining S. gordonii genes that were initially identified, 7 encode putative glycosyltransferases (Table 2), the number required for synthesis of the RPS heptasaccharide repeating unit. The first two of these genes were designated wchA and wchF, based on their homology with the genes found at equivalent positions in the CPS clusters of specific S. pneumoniae serotypes (Table 2). wchA and wchF of S. pneumoniae and S. gordonii encode proteins that are approximately 60 and 80% identical, respectively. In addition, the corresponding proteins in these species are predicted to have the same glycosyltransferase activity (see Discussion). The remaining five S. gordonii genes for putative glycosyltransferases were assigned new gene names, from wefA to wefE, following recommendations of the bacterial polysaccharide gene nomenclature system (40). Two of these genes, wefA and wefE, are, however, similar at the level of predicted protein sequence to the genes identified at equivalent positions in the EPS6 cluster of S. thermophilus or the CPS2 cluster of S. suis (Table 2). Three other S. gordonii genes were identified between wefD and wefE (Fig. 2B) by their homology with genes in the database (Table 2). These included wzy for a putative polysaccharide polymerase, wzx for a repeat unit transporter, and glf for a galactofuranose mutase, the enzyme that converts UDP-Galp to UDP-Galf, an expected precursor of RPS biosynthesis.
Sequences upstream of wzg and downstream of wefE were then PCR amplified from a Vectorette library of strain 38 genomic DNA and were queried against the available genomic database, resulting in the identification of additional overlapping strain AT sequences (Fig. 2A, shaded bars designated 1 and 5). Further sequencing of the corresponding regions in strain 38 resulted in the identification of nrdG, the gene for anaerobic ribonucleotide-triphosphate reductase activating protein, immediately upstream of wzg (Table 2). A transcriptional terminator and the putative promoter of the polysaccharide gene cluster separate these two genes (Fig. 2B). A putative promoter was also found after wefE, the fourteenth gene in the cluster, followed by two genes of unknown function and a predicted transcriptional terminator (Fig. 2B and Table 2). The 1,367-amino-acid sequence encoded by orfO, the first unknown gene, includes an N-terminal signal sequence, a putative lysozyme-like domain (from amino acids 123 to 354) like those in a number of bacterial and phage muramidases, including LytC of S. pneumoniae (Table 2) and a repetitive region (from amino acids 505 to 989) consisting of five tandem repeats, each approximately 100 amino acids in length. The next gene, orfP, encodes a putative transmembrane protein that is similar to one previously identified from another strain of S. gordonii (Table 2). The predicted sequence of OrfP also resembles that of the putative transmembrane protein encoded by wciX, a gene of unknown function in the CPS18C gene cluster of S. pneumoniae (20). The two genes further downstream, rluD for ribosomal large subunit pseudouridine synthase D and pbp2A for penicillin-binding protein 2A (Table 2), are transcribed in opposite directions (Fig. 2B).
Two of the genes identified in the 5′ region of the polysaccharide gene cluster, wzg and wchA, and three downstream genes, wefE, orfO, and orfP, were insertionally disrupted (Fig. 2B) to assess their involvement in RPS production (Table 3). Insertion of the ermAM cassette into wzg, the first gene in the S. gordonii cluster, had little affect on RPS production. The resulting mutant, strain XC1, and wild-type strain 38 did not differ dramatically as inhibitors of an RPS-specific ELISA, and both strains coaggregated strongly with type 2 fimbriated A. naeslundii (Table 3). RPS production was, however, abolished by insertion of the ermAM cassette into wchA or wefE, the first and last genes, respectively, for putative glycosyltransferases. The resulting mutants (strains XC2 and XC3, respectively) gave no inhibition of ELISA and failed to coaggregate with A. naeslundii (Table 3). In contrast, RPS production was not affected by insertion of the ermAM cassette into either orfO or orfP. The surface phenotypes of the resulting mutants (strains XC4 and XC5, respectively) were indistinguishable from that of the wild type (Table 3), indicating that wefE is the last essential gene in the RPS cluster of S. gordonii 38.
TABLE 3.
RPS production by wild-type S. gordonii 38 and ermAM insertional mutant strains
| Strain | Mutant | RPS production (μg of RPS/109 bacteriaa) | Coaggregation scoreb |
|---|---|---|---|
| 38 | Wild type | 4.5 ± 1.2 | 4 |
| XC1 | wzg | 2.5 ± 0.9 | 4 |
| XC2 | wchA | <0.004 | 0 |
| XC3 | wefE | <0.004 | 0 |
| XC4 | orfO | 3.1 ± 1.5 | 4 |
| XC5 | orfP | 5.4 ± 2.5 | 4 |
RPS production was determined by inhibition of ELISA with S. gordonii 38 RPS as a standard.
Coaggregation score (0 to 4) with A. naeslundii 12104.
Identification and role of galU in RPS production and growth on galactose.
The genes identified within the RPS cluster (Fig. 2B) do not account for the synthesis of four nucleotide-linked sugars that are predicted precursors of type 2Gn RPS biosynthesis, namely, UDP-Glc, UDP-Gal, UDP-GalNAc, and dTDP-Rha. The formation of UDP-Glc from UTP and glucose-1-phosphate depends on the enzyme UTP-glucose-1-phosphate uridylyl transferase (GalU). The gene for this enzyme was identified in the genomic database of S. gordonii AT by its similarity to the S. pneumoniae homologue (Table 2) and was sequenced in strain 38 (Fig. 2C). galU of S. gordonii, like the gene in S. pneumoniae (32) or S. mutans (55), is located downstream of gpsA, which codes for an NADP-dependent glycerol-3-phosphate dehydrogenase. Cotranscription of gpsA and galU was suggested by virtue of a 10-bp overlap in the coding sequences of these genes and by the absence of a potential transcriptional terminator or promoter immediately upstream of galU. A putative Rho-independent transcriptional terminator was identified downstream of galU between this gene and abpB (Fig. 2C). The latter gene, which codes for α-amylase-binding protein B of S. gordonii (27), is transcribed in the opposite direction from that of galU. Thus, it is probable that galU is the last gene of a transcriptional unit.
Insertional mutagenesis of galU in S. gordonii 38 yielded mutant strain XC6 (Fig. 2C), whose production of RPS was less than 1% compared to that of parent strain 38 during growth in FMC chemically defined medium containing glucose as the energy source (Table 4). Likewise, the type 2 fimbriae-mediated coaggregation of A. naeslundii 12104 with mutant XC6 was much weaker than that seen with parent strain 38. Mutant strain XC6 also failed to grow in galactose-containing FMC medium, a finding consistent with the essential role of galU in metabolism of this sugar by the Leloir pathway (16).
TABLE 4.
RPS production and coaggregation of wild-type S. gordonii 38 and mutant strains grown in glucose- or galactose-containing medium
| Strain | Mutant | Carbon sourcea | RPS production (μg of RPS/109 bacteriab) | Coaggregation scorec |
|---|---|---|---|---|
| 38 | Wild type | Glucose | 8.7 ± 0.8 | 4 |
| Galactose | 4.9 ± 0.8 | 4 | ||
| XC6 | galU | Glucose | 0.068 ± 0.020 | 1 |
| Galactose | No growth | No growth | ||
| XC7 | galE1 | Glucose | 8.3 ± 0.8 | 4 |
| Galactose | 3.6 ± 0.8 | 4 | ||
| XC8 | galE2 | Glucose | <0.004 | 0 |
| Galactose | No growth | No growth | ||
| XC9 | orf9 | Glucose | 8.3 ± 0.8 | 4 |
| Galactose | 4.2 ± 0.6 | 4 | ||
| XC8Rd | galE1 (complete) | Glucose | <0.004 | 0 |
| Galactose | <0.004 | 0 |
Each strain was grown in FMC medium containing 1% glucose or 1% galactose.
Determined by inhibition of ELISA with S. gordonii 38 RPS as a standard.
Coaggregation score (0 to 4) with A. naeslundii 12104.
Strain XC8R contains disrupted galE2 and complete galE1.
Identification and role of different genes (galE1 and galE2) for galactose epimerase in RPS production and growth on galactose.
The presence of galactose in S. gordonii 38 RPS (39) implicates galactose 4-epimerase in the biosynthesis of this polysaccharide. One gene for this enzyme, designated galE1, was identified in the galactose operon downstream of galK and galT, the genes for galactokinase and galactose-1-phosphate uridylyl transferase, respectively (Fig. 2D, Table 2). Additional genes occur in the galactose operons of Streptococcus salivarius and S. thermophilus (51) and S. mutans (5), including galR (galactose repressor) and galM (galactose mutarotase). These genes were not found in the galactose operon of S. gordonii. The amino acid sequences encoded by S. gordonii 38 galE1 and S. salivarius galE, although 83% identical (Table 2), differ in length by 34 amino acid residues, the S. gordonii sequence being shorter. Significantly, the amino-terminal valine of S. gordonii 38 GalE1 aligned with valine-35 of S. salivarius GalE, raising the possibility that the galE1 open reading frame (ORF) of strain 38 is not a complete gene. Insertion of ermAM into this ORF yielded mutant strain XC7 (Fig. 2D), whose production of RPS was comparable to that of wild-type strain 38 as revealed by results of ELISA and coaggregation assays (Table 4). Furthermore, mutant XC7 grew on galactose, indicating complementation of disrupted galE1 by another gene.
The gene designated galE2 for a second galactose 4-epimerase, similar to one in S. pneumoniae (Table 2), was identified within a 17.3-kb sequence of DNA, regions of which are shown in Fig. 2E. galE2 was identified downstream of rmlA, rmlC, and rmlB, the first three genes for dTDP-Rha biosynthesis, and upstream of orf9, which codes for a putative glycosyltransferase (Table 2). rmlD, the last gene of the dTDP-Rha biosynthetic pathway, was identified further downstream between two genes, orf15 and rgpA, that encode two other putative glycosyltransferases. The organization of the four genes for dTDP-Rha biosynthesis in S. gordonii resembles that seen in S. mutans (49, 50). Moreover, as in S. mutans (56), rmlD of S. gordonii 38 is followed by rgpA and additional genes of a putative rhamnose-glucose polysaccharide (results not shown). To our knowledge, the production of such a polysaccharide by S. gordonii has not yet been demonstrated.
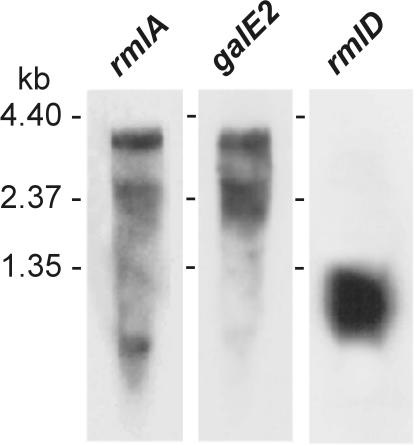
Putative promoters were identified upstream of rmlA, orf9, rmlD, and rgpA, and Rho-independent transcriptional terminators were found downstream of orf15 and rmlD (Fig. 1E). Northern blotting with probes for rmlA and galE2 revealed a transcript of the size (∼3.7 kb) expected of polycistronic rmlA-galE2 mRNA (Fig. 3). The diffuse band below this region may represent degraded transcript. In contrast, a probe for rmlD hybridized with a smaller fragment (∼1 kb) indicative of a monocistronic rmlD transcript. Thus, galE2 appears to be transcribed with the first three genes of the dTDP-Rha pathway while rmlD is transcribed independently.
FIG. 3.
Autoradiographs of Northern blots of total S. gordonii 38 RNA showing hybridization of 32P-labeled probes for rmlA, galE2, and rmlD. The positions of RNA standards on different blots are indicated.
Insertional disruption of galE2 yielded mutant strain XC8 (Fig. 2E), which grew in glucose-containing medium but failed to make detectable RPS (Table 4). Mutant XC8 also failed to grow in galactose-containing medium, providing evidence for the essential role of galE2 in the metabolism of this sugar by the Leloir pathway. To control for a possible polar effect of the mutation in strain XC8, orf9, the gene immediately downstream of galE2, was insertionally disrupted to generate mutant strain XC9 (Fig. 2E). Mutant strain XC9, unlike XC8, produced RPS and also grew on galactose (Table 4).
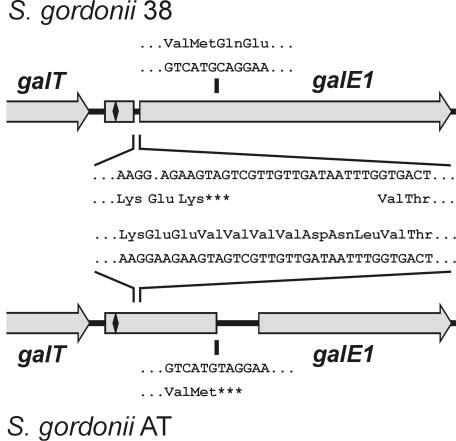
Identification of different galE1 pseudogenes in S. gordonii strains 38 and AT.
The involvement of galE2 in growth on galactose suggested that galE1 was either incomplete or that the protein formed was catalytically inactive. Closer inspection of the 299-amino-acid sequence encoded by the galE1 ORF indicated that the predicted protein lacked a G(X)G(X)(X)G NAD+ binding motif (54) that is a characteristic feature in the N-terminal region of galactose 4-epimerases. Instead, the coding sequence for this motif was found in the intragenic region between the 3′ end of galT and the putative start codon (GTG) of the galE1 ORF (Fig. 4). To examine the status of the galE1 coding region in another strain, this region was sequenced from S. gordonii AT (GenBank accession number AY147911). Interestingly, the galE1 sequence of strain AT was also interrupted but at a site different from that noted for strain 38. The different natural mutations in these S. gordonii strains were further defined by aligning the galE1 sequences of these bacteria (Fig. 4). The mutation in strain 38 involved the deletion of adenine (A) at nucleotide 76 of the complete gene, giving rise to a TAG terminator 6 bp further downstream. In contrast, the apparent mutation in strain AT involved the transition of a single base (C to T) at position 318 of the complete gene in this strain, which effectively changed a CAG (Gln) codon in strain 38 to a TAG terminator. In addition to these differences, the nucleotide sequences of the galE1 pseudogenes from the two strains differed at 42 other positions, of which only 4 were associated with differences in predicted amino acid sequence.
FIG. 4.
ORF diagrams of galE1 pseudogenes identified downstream of galT in S. gordonii strains 38 and AT. The comparison of nucleotide and predicted protein sequences reveals a deleted adenine in the strain 38 galE1 sequence with respect to strain AT (comparable sequences are shown between the ORF diagrams). Similar comparison reveals the transition of a single base (C to T) in the strain AT galE1 sequence with respect to strain 38 (comparable sequences are shown above and below the ORF diagrams). The location of the coding sequence for the G(X)G(X)(X)G NAD+ binding motif of encoded GalE is indicated by the symbol (♦) at the 5′ end of each pseudogene.
Complete galE1 permits growth of S. gordonii on galactose but not RPS production.
From consideration of the previous results, we suspected that the galE2 mutant of strain 38 (i.e., insertional mutant XC8) would grow on galactose following repair of the galE1 pseudogene in this strain. This was accomplished by selection for gal+ revertants following transformation of strain XC8 with a PCR product containing the complete galE1 sequence (i.e., the sequence of the PCR product contained the deleted adenine identified in Fig. 4 by comparison of the strain 38 and AT sequences). Transformant XC8R, containing complete galE1 and disrupted galE2, grew on galactose but failed to produce RPS during growth on either glucose or galactose (Table 4). To understand the enzymatic basis of these results, cell extracts prepared from wild-type and selected mutant strains were assayed for UDP-galactose 4-epimerase and UDP-N-acetylgalactosamine 4-epimerase activities. Both enzymatic activities were readily detected in extracts of wild-type strain 38, but neither activity was present in cell extracts of glucose-grown strain XC8 containing the galE1 pseudogene and disrupted galE2 (Table 5). Clearly, GalE2 is the only functional galactose 4-epimerase in parent strain 38. Significantly, cell extracts of strain XC8R, which contained complete galE1 and disrupted galE2, possessed UDP-galactose 4-epimerase activity, which was elevated in galactose-grown cells, but lacked UDP-N-acetylgalactosamine 4-epimerase activity (Table 5). Thus, complete and catalytically functional GalE1 is specific for unacetylated substrates, whereas the bifunctional GalE2 catalyzes the epimerization of both acetylated and unacetylated substrates.
TABLE 5.
Gal and GalNAc 4-epimerase activities in cell extracts of wild-type S. gordonii 38 and mutant strains grown in glucose- or galactose-containing medium
| Strain | Mutant | Carbon sourcea | Gal 4-epimerase activityb | GalNAc 4-epimerase activityc |
|---|---|---|---|---|
| 38 | Wild type | Glucose | 96.6 ± 1.5 | 26.0 ± 2.0 |
| Galactose | 32.2 ± 1.6 | 11.2 ± 0.9 | ||
| XC8 | galE2 | Glucose | <2d | <0.04d |
| Galactose | No growth | No growth | ||
| XC8Re | galE1 (complete) | Glucose | 19.4 ± 3.1 | <0.04d |
| Galactose | 183 ± 14.2 | <0.04d |
Each strain was grown in FMC medium containing 1% glucose or 1% galactose.
Nanomoles of UDP-Gal converted to UDP-Glc min−1 mg protein−1.
Nanomoles of UDP-GalNAc converted to UDP-GlcNAc min−1 mg protein−1.
No detectable activity.
Strain XC8R contains disrupted galE2 and complete galE1.
DISCUSSION
The identification of different genetic loci for type 2Gn RPS biosynthesis in S. gordonii 38 extends the characterization of the coaggregation receptor on this strain from the structural (39) to the molecular level. Probes for different genes in the cps19f cluster of S. pneumoniae (33) were used to identify three genes in the biosynthetic pathway of dTDP-Rha in a plasmid library of S. gordonii 38 genomic DNA. These genes in S. gordonii (i.e., rmlA, rmlC, and rmlB) were, however, not associated with a polysaccharide gene cluster, as in S. pneumoniae (33), but instead were organized like the rml genes of S. mutans (49, 50). The fortuitous detection of type 2Gn RPS on the partially sequenced viridans group streptococcus (48), presently designated S. gordonii AT, facilitated the identification of additional genes for RPS biosynthesis in S. gordonii 38, including those in the 2Gn RPS gene cluster and those outside this region for four nucleotide-linked sugars, namely, UDP-Glc, UDP-Gal, UDP-GalNAc, and dTDP-Rha (Fig. 5). The genomic database of strain AT has been replaced on The Institute of Genomic Research website (http://www.tigr.org/tdb/mdb/mdbinprogress.html) by the more complete database obtained from ongoing whole-genomic sequencing of S. gordonii Challis CH1. While strain Challis does not produce RPS, it does possess a GalNAc-binding surface adhesin that recognizes Gn types of RPS on streptococci such as strains AT and 38 (12, 19, 45). Thus, both available S. gordonii databases represent valuable resources for studies of the different genotypes that represent this species (22).
FIG. 5.
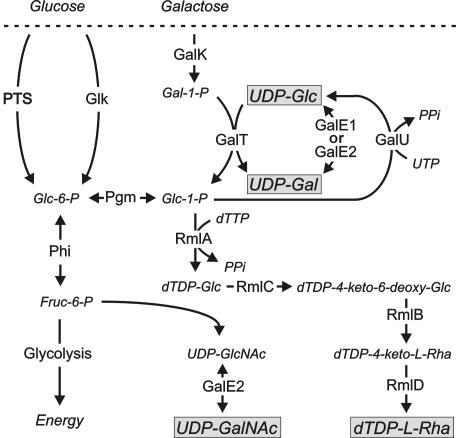
Pathways for the biosynthesis of UDP-Glc, UDP-Gal, UDP-GalNAc, and dTDP-Rha in S. gordonii 38 showing the proposed involvement of proteins encoded by different genes identified outside the RPS gene cluster. Complete GalE1 or GalE2 can interconvert UDP-Glc and UDP-Gal, but only GalE2 catalyzes the epimerization of UDP-GlcNAc and UDP-GalNAc. PTS, phosphoenol pyruvate-dependent sugar phosphotransferase system. Other enzymes include glucokinase (Glk), phosphoglucomutase (Pgm), and phosphohexose isomerase (Phi).
We anticipated that the supply of UDP-Glc and UDP-Gal for RPS production would depend on genes associated with the Leloir pathway of galactose metabolism (16). Indeed, growth of S. gordonii on galactose was abolished and RPS production was greatly reduced by insertional inactivation of galU, blocking the formation of UDP-Glc from UTP and glucose-1-phosphate (Fig. 5). However, the same effects were not observed from insertional inactivation of galE1, the last gene in the galactose operon, which was expected to encode the epimerase required for the conversion of UDP-Glc to UDP-Gal. Instead, growth of bacteria on galactose and RPS production were dependent on galE2, which was identified as the last gene in an operon that includes the first three genes of the dTDP-Rha biosynthetic pathway. The apparent inactivity of galE1 in strain 38 was explained by the presence of a natural mutation in this gene (i.e., the deletion of a single adenine) that effectively separated the coding region for the NAD+ binding domain of the encoded epimerase from the rest of the gene. Interestingly, galE1 of S. gordonii AT was also interrupted by a different mutation identified near the middle of this gene. Repair of the galE1 pseudogene in the galE2 mutant of strain 38 (i.e., strain XC8) restored growth on galactose but not RPS production. Significantly, cell extracts that contained functional GalE1, but not GalE2, catalyzed the interconversion of UDP-Glc and UDP-Gal but not the epimerization of UDP-GlcNAc and UDP-GalNAc. In contrast, extracts of wild-type strain 38 that contained functional GalE2 but not GalE1 catalyzed the epimerization of both acetylated and unacetylated substrates. Thus, the expression of complete galE1 or galE2 supports the growth of S. gordonii on galactose, but only the expression of galE2 supports RPS production, which requires UDP-GalNAc as well as UDP-Gal (Fig. 5). The presence of different galE1 pseudogenes in strains 38 and AT suggests that selective pressure for the maintenance of the complete gene in the galactose operon may have been removed by the expression of bifunctional galE2 during the evolution of GalNAc-containing surface receptors for oral biofilm formation.
A number of findings associate the polysaccharide gene cluster identified in the present investigation with RPS production. First, ermAM insertional mutagenesis of wefE, the 14th gene in the cluster and the 7th for a putative glycosyltransferase, abolished the presence of surface receptors for coaggregation with A. naeslundii and antigenically detectable RPS. These effects were not observed from disruption of the two genes further downstream. Second, RPS production was also abolished by insertional disruption of wchA, the fifth gene in the cluster and the first for a putative glycosyltransferase. These effects most likely reflect the essential role of wchA in RPS production but may also be explained by a polar effect of the ermAM insertion on the expression of essential downstream genes, such as wefE. Experiments were not performed to rule out the latter possibility. A polar effect was not, however, observed from insertion of the ermAM cassette into wzg, the first gene in the RPS cluster. RPS production by the resulting mutant, strain XC1, may have been reduced, but it was certainly not abolished (Table 3). The nonessential role of wzg in RPS production, although unexpected, is consistent with the putative regulatory role of this gene and with previous findings from studies with S. pneumoniae type 19F showing that deletion of cps19fA (i.e., wzg) reduced but did not abolish CPS production (35). Third, the involvement of the gene cluster in RPS production is evident from the predicted properties of the proteins encoded by the 10 nonregulatory genes, from wchA to wefE, which is the number that is necessary and sufficient for the synthesis, transport, and polymerization of the type 2Gn RPS heptasaccharide repeating unit and the production of UDP-Galf, a unique RPS precursor. Obviously, extension of the present findings will be required to define the specific role of each individual gene in the S. gordonii 38 cluster.
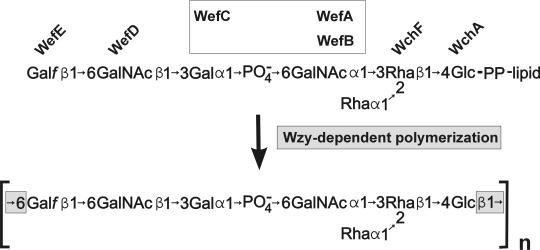
A working hypothesis of RPS biosynthesis can, however, be suggested (Fig. 6) from the predicted properties of the seven putative glycosyltransferases that are encoded by genes in this cluster (Table 2) and the structure of type 2Gn RPS (39). The first two putative glycosyltransferases have counterparts in S. pneumoniae (Table 2). In this species, WchA transfers glucose-1-phosphate from UDP-Glc to a lipid carrier, initiating the synthesis of different CPS repeating units (23, 52), and WchF has been implicated in the subsequent β1-4 transfer of l-Rha to Glc in CPS serotypes 2, 18C, and 23F (20). The same two steps are predicted for S. gordonii and serve to define the putative biosynthetic repeating unit of the polysaccharide (Fig. 6), which has one glucose unit per heptasaccharide repeat (39).
FIG. 6.
Model of RPS biosynthesis based on the predicted properties of the putative glycosyltransferases encoded by genes in the RPS cluster of S. gordonii 38 and the structure of strain 38 RPS. The identification of wchA as the first gene for a glycosyltransferase suggests that the transfer of glucose-1-phosphate to carrier lipid initiates RPS production. This and the presence of a single glucose in the strain 38 RPS heptasaccharide repeat (39) define the lipid-linked biosynthetic repeating unit. Specific glycosyltransferase activities are predicted for WchF, WefD, and WefE as indicated. The activities of the three remaining proteins (WefA, WefB, and WefC) are presumably associated with synthesis of the central region of the repeating unit (see the text). Wzy-dependent polymerization of heptasaccharide repeating units is predicted to account for the presence of Glcβ1-6Galf in the polysaccharide.
The specific steps associated with the third, fourth, and fifth putative glycosyltransferases (i.e., WefA, WefB, and WefC, respectively) are not yet clear (Fig. 6). The identification of WefA as a putative retaining glycosyltransferase is, however, expected from the homology noted between this protein and two members of glycosyltransferase family 4 (8 and http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html), namely, Eps6G of S. thermophilus and Cps2G of S. suis (Table 2). The predicted retaining activity of WefA could account for the transfer of α-d-GalNAc to Rha, but this activity is not anticipated for the transfer of α-l-Rha to Rha, which would require an inverting enzyme. A second potentially important insight comes from the weak homology noted between WefC and SacB of Neisseria meningitidis (Table 2). SacB is the putative ManNAcα-1-PO4 transferase that forms the homopolymeric meningococcal serogroup A capsule (44), suggesting that WefC may be the Galα-1-PO4 transferase that forms the phosphodiester linkage in the middle of the RPS repeating unit. If so, the remaining protein, WefB, may transfer α-l-Rha to Rha, forming the α-l-Rha branch (Fig. 6). This branch is the only structural feature that distinguishes the heptasaccharide repeat of type 2Gn RPS from the hexasaccharide repeat of S. oralis 34 type 1Gn RPS (4, 30, 39). Thus, the identity of the gene associated with this structural difference may emerge from ongoing studies to characterize the RPS gene cluster of strain 34.
The predicted activities of the two remaining putative glycosyltransferases (i.e., WefD and WefE, respectively) associate these proteins with the last two steps in synthesis of the proposed repeating unit (Table 2, Fig. 6). WefD is a putative inverting glycosyltransferase on the basis of its sequence homology with several members of glycosyltransferase family 2 (8 and http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html), including Cps14J, a Galβ-transferase of S. pneumoniae (24), and WefE is a putative galactofuranose transferase on the basis of its homology with E. coli WbbI (43). From these homologies, we predict that WefD is the GalNAcβ transferase that forms the host-like recognition motif (i.e., GalNAcβ1-3Gal) and that WefE is the galactofuranose transferase that completes the lipid-linked heptasaccharide. Finally, the present model of RPS biosynthesis (Fig. 6) associates the presence of Glcβ1-6Galf in the strain 38 polysaccharide (39) with Wzy-dependent polymerization of adjacent heptasaccharide repeating units.
Similarities between the S. gordonii RPS and S. pneumoniae CPS clusters, although striking in terms of gene composition and order (Table 2), are not sufficient at the nucleotide level to indicate the recent lateral transfer of genes for polysaccharide biosynthesis between these closely related species (21). Lateral transfer of CPS genes between strains of S. pneumoniae is, however, well established and thought to be of primary importance in the ongoing evolution of new CPS serotypes (13). Like pneumococci, the four RPS-producing species of oral viridans group streptococci are readily transformable. In addition, the latter bacteria are intimately associated with each other as well as with other members of the human oral biofilm community (19, 37, 38). The present identification of an RPS gene cluster from a human oral viridans group streptococcus provides a basis for further studies to assess the role of lateral gene transfer in the ongoing evolution of microbial surface receptors for mixed-species biofilm formation.
Acknowledgments
We thank Brian A. Dougherty, Steven R. Gill, and The Institute of Genomic Research for providing S. gordonii AT and access to the genomic database of this strain. Jacob Donkersloot, Tim Fritz, and Dennis J. Kopecko provided helpful comments during preparation and review of the manuscript.
REFERENCES
- 1.Abeygunawardana, C., C. A. Bush, and J. O. Cisar. 1991. Complete structure of the cell surface polysaccharide of Streptococcus oralis ATCC 10557: a receptor for lectin-mediated interbacterial adherence. Biochemistry 30:6528-6540. [DOI] [PubMed] [Google Scholar]
- 2.Abeygunawardana, C., C. A. Bush, and J. O. Cisar. 1991. Complete structure of the cell surface polysaccharide of Streptococcus oralis C104: a 600-MHz NMR study. Biochemistry 30:8568-8577. [DOI] [PubMed] [Google Scholar]
- 3.Abeygunawardana, C., C. A. Bush, and J. O. Cisar. 1990. Complete structure of the polysaccharide from Streptococcus sanguis J22. Biochemistry 29:234-248. [DOI] [PubMed] [Google Scholar]
- 4.Abeygunawardana, C., C. A. Bush, S. S. Tjoa, P. V. Fennessey, and M. R. McNeil. 1989. The complete structure of the capsular polysaccharide from Streptococcus sanguis 34. Carbohydr. Res. 191:279-293. [DOI] [PubMed] [Google Scholar]
- 5.Ajdic, D., I. C. Sutcliffe, R. R. Russell, and J. J. Ferretti. 1996. Organization and nucleotide sequence of the Streptococcus mutans galactose operon. Gene 180:137-144. [DOI] [PubMed] [Google Scholar]
- 6.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 7.Boels, I. C., A. Ramos, M. Kleerebezem, and W. M. de Vos. 2001. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 67:3033-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, J. A., G. J. Davies, V. Bulone, and B. Henrissat. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326:929-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chassy, B. M., and A. Giuffrida. 1980. Method for the lysis of gram-positive, asporogenous bacteria with lysozyme. Appl. Environ. Microbiol. 39:153-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cisar, J. O., P. E. Kolenbrander, and F. C. McIntire. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 24:742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cisar, J. O., A. L. Sandberg, C. Abeygunawardana, G. P. Reddy, and C. A. Bush. 1995. Lectin recognition of host-like saccharide motifs in streptococcal cell wall polysaccharides. Glycobiology 5:655-662. [DOI] [PubMed] [Google Scholar]
- 12.Cisar, J. O., A. L. Sandberg, G. P. Reddy, C. Abeygunawardana, and C. A. Bush. 1997. Structural and antigenic types of cell wall polysaccharides from viridans group streptococci with receptors for oral actinomyces and streptococcal lectins. Infect. Immun. 65:5035-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffey, T. J., M. C. Enright, M. Daniels, J. K. Morona, R. Morona, W. Hryniewicz, J. C. Paton, and B. G. Spratt. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol. Microbiol. 27:73-83. [DOI] [PubMed] [Google Scholar]
- 14.Creuzenet, C., M. Belanger, W. W. Wakarchuk, and J. S. Lam. 2000. Expression, purification, and biochemical characterization of WbpP, a new UDP-GlcNAc C4 epimerase from Pseudomonas aeruginosa serotype O6. J. Biol. Chem. 275:19060-19067. [DOI] [PubMed] [Google Scholar]
- 15.Estrela, A. I., H. M. Pooley, H. de Lencastre, and D. Karamata. 1991. Genetic and biochemical characterization of Bacillus subtilis 168 mutants specifically blocked in the synthesis of the teichoic acid poly(3-O-beta-D-glucopyranosyl-N-acetylgalactosamine 1-phosphate): gneA, a new locus, is associated with UDP-N-acetylglucosamine 4-epimerase activity. J. Gen. Microbiol. 137(Pt 4):943-950. [DOI] [PubMed] [Google Scholar]
- 16.Frey, P. A. 1996. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 10:461-470. [PubMed] [Google Scholar]
- 17.Garcia, E., D. Llull, R. Munoz, M. Mollerach, and R. Lopez. 2000. Current trends in capsular polysaccharide biosynthesis of Streptococcus pneumoniae. Res. Microbiol. 151:429-435. [DOI] [PubMed] [Google Scholar]
- 18.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, S. D., J. O. Cisar, A. L. Sandberg, and M. Kilian. 1994. Adhesive properties of viridans group streptococcal species. Microb. Ecol. Health Dis. 7:125-137. [Google Scholar]
- 20.Jiang, S. M., L. Wang, and P. R. Reeves. 2001. Molecular characterization of Streptococcus pneumoniae type 4, 6B, 8, and 18C capsular polysaccharide gene clusters. Infect. Immun. 69:1244-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 22.Kilian, M., L. Mikkelsen, and J. Henrichsen. 1989. Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven, 1946), Streptococcus oralis (Bridge and Sneath, 1982), Streptococcus mitis (Andrews and Horder, 1906). Int. J. Syst. Bacteriol. 39:471-484. [Google Scholar]
- 23.Kolkman, M. A., B. A. van der Zeijst, and P. J. Nuijten. 1997. Functional analysis of glycosyltransferases encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J. Biol. Chem. 272:19502-19508. [DOI] [PubMed] [Google Scholar]
- 24.Kolkman, M. A., W. Wakarchuk, P. J. Nuijten, and B. A. van der Zeijst. 1997. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol. Microbiol. 26:197-208. [DOI] [PubMed] [Google Scholar]
- 25.LeBlanc, D. J., L. N. Lee, and A. Abu-Al-Jaibat. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130-145. [DOI] [PubMed] [Google Scholar]
- 26.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, L., J. M. Tanzer, and F. A. Scannapieco. 2002. Identification and analysis of the amylase-binding protein B (AbpB) and gene (abpB) from Streptococcus gordonii. FEMS Microbiol. Lett. 212:151-157. [DOI] [PubMed] [Google Scholar]
- 28.Lunsford, R. D. 1995. Recovery of RNA from oral streptococci. BioTechniques 18:412-414. [PubMed] [Google Scholar]
- 29.Lunsford, R. D., and J. London. 1996. Natural genetic transformation in Streptococcus gordonii: comX imparts spontaneous competence on strain wicky. J. Bacteriol. 178:5831-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McIntire, F. C., C. A. Bush, S. S. Wu, S. C. Li, Y. T. Li, M. McNeil, S. S. Tjoa, and P. V. Fennessey. 1987. Structure of a new hexasaccharide from the coaggregation polysaccharide of Streptococcus sanguis 34. Carbohydr. Res. 166:133-143. [DOI] [PubMed] [Google Scholar]
- 31.McIntire, F. C., L. K. Crosby, A. E. Vatter, J. O. Cisar, M. R. McNeil, C. A. Bush, S. S. Tjoa, and P. V. Fennessey. 1988. A polysaccharide from Streptococcus sanguis 34 that inhibits coaggregation of S. sanguis 34 with Actinomyces viscosus T14V. J. Bacteriol. 170:2229-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mollerach, M., R. Lopez, and E. Garcia. 1998. Characterization of the galU gene of Streptococcus pneumoniae encoding a uridine diphosphoglucose pyrophosphorylase: a gene essential for capsular polysaccharide biosynthesis. J. Exp. Med. 188:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morona, J. K., R. Morona, and J. C. Paton. 1997. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol. Microbiol. 23:751-763. [DOI] [PubMed] [Google Scholar]
- 34.Morona, J. K., R. Morona, and J. C. Paton. 1999. Comparative genetics of capsular polysaccharide biosynthesis in Streptococcus pneumoniae types belonging to serogroup 19. J. Bacteriol. 181:5355-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morona, J. K., J. C. Paton, D. C. Miller, and R. Morona. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 35:1431-1442. [DOI] [PubMed] [Google Scholar]
- 36.Nyvad, B., and O. Fejerskov. 1987. Scanning electron microscopy of early microbial colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:287-296. [DOI] [PubMed] [Google Scholar]
- 37.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 38.Palmer, R. J., Jr., S. M. Gordon, J. O. Cisar, and P. E. Kolenbrander. 2003. Coaggregation-mediated interactions of streptococci and actinomyces in initial human dental plaque. J. Bacteriol. 185:3400-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reddy, G. P., C. Abeygunawardana, C. A. Bush, and J. O. Cisar. 1994. The cell wall polysaccharide of Streptococcus gordonii 38: structure and immunochemical comparison with the receptor polysaccharides of Streptococcus oralis 34 and Streptococcus mitis J22. Glycobiology 4:183-192. [DOI] [PubMed] [Google Scholar]
- 40.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Smith, H. E., M. Damman, J. van der Velde, F. Wagenaar, H. J. Wisselink, N. Stockhofe-Zurwieden, and M. A. Smits. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67:1750-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swartley, J. S., L. J. Liu, Y. K. Miller, L. E. Martin, S. Edupuganti, and D. S. Stephens. 1998. Characterization of the gene cassette required for biosynthesis of the (α1->6)-linked N-acetyl-D-mannosamine-1-phosphate capsule of serogroup A Neisseria meningitidis. J. Bacteriol. 180:1533-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi, Y., K. Konishi, J. O. Cisar, and M. Yoshikawa. 2002. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect. Immun. 70:1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi, Y., S. Ruhl, J. W. Yoon, A. L. Sandberg, and J. O. Cisar. 2002. Adhesion of viridans group streptococci to sialic acid-, galactose- and N-acetylgalactosamine-containing receptors. Oral Microbiol. Immunol. 17:257-262. [DOI] [PubMed] [Google Scholar]
- 47.Terleckyj, B., N. P. Willett, and G. D. Shockman. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 49.Tsukioka, Y., Y. Yamashita, Y. Nakano, T. Oho, and T. Koga. 1997. Identification of a fourth gene involved in dTDP-rhamnose synthesis in Streptococcus mutans. J. Bacteriol. 179:4411-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsukioka, Y., Y. Yamashita, T. Oho, Y. Nakano, and T. Koga. 1997. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J. Bacteriol. 179:1126-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaillancourt, K., S. Moineau, M. Frenette, C. Lessard, and C. Vadeboncoeur. 2002. Galactose and lactose genes from the galactose-positive bacterium Streptococcus salivarius and the phylogenetically related galactose-negative bacterium Streptococcus thermophilus: organization, sequence, transcription, and activity of the gal gene products. J. Bacteriol. 184:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Selm, S., M. A. Kolkman, B. A. van der Zeijst, K. A. Zwaagstra, W. Gaastra, and J. P. van Putten. 2002. Organization and characterization of the capsule biosynthesis locus of Streptococcus pneumoniae serotype 9V. Microbiology 148:1747-1755. [DOI] [PubMed] [Google Scholar]
- 53.Wang, L., S. Huskic, A. Cisterne, D. Rothemund, and P. R. Reeves. 2002. The O-antigen gene cluster of Escherichia coli O55:H7 and identification of a new UDP-GlcNAc C4 epimerase gene. J. Bacteriol. 184:2620-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wierenga, R. K., M. C. H. De Maeyer, and W. G. J. Hol. 1985. Interaction of pyrophosphate moieties with α-helixes in dinucleotide binding proteins. Biochemistry 24:1346-1357. [Google Scholar]
- 55.Yamashita, Y., Y. Tsukioka, Y. Nakano, K. Tomihisa, T. Oho, and T. Koga. 1998. Biological functions of UDP-glucose synthesis in Streptococcus mutans. Microbiology 144(Pt 5):1235-1245. [DOI] [PubMed] [Google Scholar]
- 56.Yamashita, Y., Y. Tsukioka, K. Tomihisa, Y. Nakano, and T. Koga. 1998. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J. Bacteriol. 180:5803-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]