Abstract
Escherichia coli strain MG1655 was chosen for sequencing because the few mutations it carries (ilvG rfb-50 rph-1) were considered innocuous. However, it has a number of growth defects. Internal pyrimidine starvation due to polarity of the rph-1 allele on pyrE was problematic in continuous culture. Moreover, the isolate of MG1655 obtained from the E. coli Genetic Stock Center also carries a large deletion around the fnr (fumarate-nitrate respiration) regulatory gene. Although studies on DNA microarrays revealed apparent cross-regulation of gene expression between galactose and lactose metabolism in the Stock Center isolate of MG1655, this was due to the occurrence of mutations that increased lacY expression and suppressed slow growth on galactose. The explanation for apparent cross-regulation between galactose and N-acetylglucosamine metabolism was similar. By contrast, cross-regulation between lactose and maltose metabolism appeared to be due to generation of internal maltosaccharides in lactose-grown cells and may be physiologically significant. Lactose is of restricted distribution: it is normally found together with maltosaccharides, which are starch degradation products, in the mammalian intestine. Strains designated MG1655 and obtained from other sources differed from the Stock Center isolate and each other in several respects. We confirmed that use of other E. coli strains with MG1655-based DNA microarrays works well, and hence these arrays can be used to study any strain of interest. The responses to nitrogen limitation of two urinary tract isolates and an intestinal commensal strain isolated recently from humans were remarkably similar to those of MG1655.
A number of laboratories have initiated studies of genome-wide expression in Escherichia coli strain MG1655 (3, 19), for which a complete genome sequence is available (4). As we performed simple experiments with the isolate of MG1655 that we received from the E. coli Genetic Stock Center (CGSC 6300), whose genotype was listed as ilvG rfb-50 rph-1, we became aware of several problems with this strain that limited its usefulness for physiological studies. Its rph-1 allele, which is polar on pyrE and causes slight uracil limitation in batch culture (27), causes problematic limitation in glycerol-limited continuous culture. Moreover, we report here that CGSC 6300 carries a large deletion around the fnr regulatory gene (allele fnr-267) and thus is unable to grow by anaerobic respiration. This phenotype was first observed as far back as 1986 (V. Stewart, unpublished data). The deletion is not present in the complete genome sequence reported for MG1655 (4), and hence the strain held by the Blattner laboratory was different. Indeed, the strain we received from them differed from CGSC 6300 not only in this regard but several others, including the facts that it gave rise to a mixture of large and small colonies on enriched and glucose-minimal media and that neither colony type grew on glycerol as the sole carbon source. As noted in the sequence (4), MG1655 carries an insertion in the eut operon, whose products are required for growth on ethanolamine as a carbon or nitrogen source.
Using DNA microarrays, we observed apparent cross regulation of gene expression between galactose catabolism and that of lactose and N-acetylglucosamine, effects that would not have been readily detected by other means. However, when we explored the bases for these effects, we found that MG1655 (CGSC 6300 unless otherwise noted) grew slowly on galactose, and apparent cross-regulatory effects were due to the appearance of suppressor mutations that allow fast growth.
We here present a compendium of the growth defects and selected other properties of MG1655 (CGSC 6300), which we analyzed in different degrees of detail. We also document differences between the Stock Center isolate and other strains designated MG1655 which we obtained from other sources. In agreement with previous studies (2), we show that cDNAs from a variety of E. coli strains hybridize well to MG1655-based DNA microarrays and hence that these arrays can be used to perform genomic and physiological studies of any strain of interest. Given the problems with “MG1655,” it may be advisable for laboratories to continue using the parental E. coli strains that they have characterized.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All strains used in this work are listed in Table 1. Different strains called MG1655 were obtained from four sources: (i) the E. coli Genetic Stock Center (CGSC 6300); (ii) the American Type Culture Collection (ATCC 47076); (iii) Mitchell Singer, originally from the laboratory of Carol Gross (NCM3430); and (iv) Frederick Blattner (NCM3629). Prototrophic E. coli K-12 strain NCM3722 was derived from strain NCM196 (ntrA7) by transduction to glutamine prototrophy with P1vir phage grown on donor strain CGSC 6300. NCM196 (ntrA7) was generated by mutagenesis with a frameshift mutagen, and NCM3722 comes as close as possible to reconstructing its parental strain, an E. coli K-12 wild type. That wild-type strain was unfortunately lost, as were the details of its history.
TABLE 1.
E. coli strains and plasmids
| Strain or plasmid | Relevant genotype or descriptiona | Constructionb or reference |
|---|---|---|
| Strains | ||
| MG1655 | a.k.a. NCM3105; see text and Table 7 | E. coli Genetic Stock Center (CGSC 6300) |
| MG1655 | a.k.a. NCM3628; see text | American Type Culture Collection (ATCC 47076) |
| MG1655 | a.k.a. NCM3430; see text | Mitchell Singer and Carol Gross |
| MG1655 | a.k.a. NCM3629; see text | Frederick Blattner |
| CAG12080 | F− λ−rph-1 mhpC281::Tn10 | E. coli Genetic Stock Center |
| CAG12163 | F− λ−rph-1 zib-207::Tn10 | E. coli Genetic Stock Center |
| W2244c | ΔlacZ39 rpsL110 (strR) | E. coli Genetic Stock Center (8) |
| M7044 | lacY328(Am) relA1 rpsL150 (Strr) spoT1 | E. coli Genetic Stock Center (38) |
| AD110 | Δ(lac-pro) supE thi malF::Tn10 F′ (traD36 proAB lacIqlacZΔM15) | H. Nikaido |
| AD111 | Δ(lac-pro) supE thi malG::Tn10 F′ (traD36 proAB lacIqlacZΔM15) | H. Nikaido |
| pop4062 | F−araD139 Δ(argF-lac)U169 rpsL150 relAI flbB5301 deoC1 ptsF25 rbsR malT::Tn10 | O. Danot |
| GM15 | malS::Tn10 malE::Kanr | W. Boos |
| TE2680 | F− λ− IN(rrnD-rrnE)1 ΔlacX74 rpsL galK2 recD1903::Tn10d-Tet trpDC700::putPA1303::[Kans-Camr-lac] | 11 |
| IBPC534 | nagA::Tetr | 45 |
| IBPC536 | nagC::Tetr | 45 |
| IBPC540 | nagD::Tetr | 45 |
| IBPC542 | nagE::Kanr | 45 |
| IBPC546 | nagB::Kanr | 45 |
| IBPC571 | nagB2 zbf-507::Tn10 asnB::Tn5 | 44 |
| IBPC590 | Δnag(EBACD)::Tetr | 44 |
| VH1000 | ΔlacIZ | V. J. Hernandez via C. L. Turnbough |
| RK4353 | fnr+ | 55 |
| RK4942 | ycj-630::Tn10 | 55 |
| RK5279 | fnr-250 | 55 |
| NCM3350 | malF::Tn10 | P1(AD110) → NCM3105d |
| NCM3351 | malG::Tn10 | P1(AD111) → NCM3105d |
| NCM3364 | malT::Tn10 | P1(pop4062) → NCM3105d |
| NCM3384 | mhpC281::Tn10 | P1(CAG12080) → NCM3105d |
| NCM3385c | ΔlacZ39 mhpC281::Tn10 | P1(NCM3384) → W2244d,e,f |
| NCM3386 | lacY328(Am) mhpC281::Tn10 | P1(NCM3384) → M7044d,g,h |
| NCM3387c | mhpC281::Tn10 ΔlacZ39 | P1(NCM3385) → NCM3105d,e,f |
| NCM3388 | mhpC281::Tn10 lacY328(Am) | P1(NCM3386) → NCM3105d,g,h |
| NCM3410 | malE::Kanr | P1(GM15) → NCM3105i |
| NCM3414 | Δlac1Z zib-207::Tn10 | P1(CAG12163) → VH1000d |
| NCM3415j | rph-1 zib-207::Tn10 | P1(NCM3414) → NCM3105d |
| NCM3416j | zib-207::Tn10 | P1(NCM3414) → NCM3105d |
| NCM3426 | recD1903::Tn10d-Tet | P1(TE2680) → NCM3105d |
| NCM3467 | recD1903::Tn10d-Tet ΔycjT::Kanr | See Materials and Methods |
| NCM3468 | ΔycjT::Kanr | P1(NCM3467) → NCM3105i |
| NCM3556 | Galactose fast-grower, LacZ− Lac(Con) | See Materials and Methods and Results |
| NCM3558 | Galactose fast-grower | See Materials and Methods |
| NCM3559 | Galactose fast-grower, Lac(Con) | See Materials and Methods |
| NCM3560 | Galactose fast-grower, Lac(Con) | See Materials and Methods |
| NCM3601 | seq404 (UTI isolate) | See Materials and Methods |
| NCM3610 | seq915 (UTI isolate) | See Materials and Methods |
| NCM3611 | 10002-006 (intestinal commensal) | See Materials and Methods |
| NCM3722 | E. coli K-12 prototroph | See Materials and Methods |
| NCM3748 | lacA::Kanr | See Materials and Methods |
| NCM3749 | mhpC281::Tn10 lacA::Kanr | P1(NCM3748) → NCM3384g,i |
| NCM3750c | mhpC281::Tn10 ΔlacZ39 lacA::Kanr | P1(NCM3748) → NCM3387e,f,i |
| NCM3786 | nagA::Tetr | P1(IBPC534) → NCM3105d,k |
| NCM3787 | nagC::Tetr | P1(IBPC536) → NCM3105d |
| NCM3789 | nagE::Kanr | P1(IBPC542) → NCM3105i |
| NCM3817 | nagB::Kanr | P1(IBPC546) → NCM3105i,l |
| NCM3818 | zbf-507::Tn10 asnB::Tn5 nagB2 | P1(IBPC571) → NCM3105d,l |
| NCM3819 | zbf-507::Tn10 asnB::Tn5 | P1(IBPC571) → NCM3105d,m |
| NCM3820 | Δnag(EBACD)::Tetr | P1(IBPC590) → NCM3105d,l |
| NCM3821 | nagD::Tetr | P1(IBPC540) → NCM3105d |
| NCM3845 | Galactose fast-grower, NagA− | See Materials and Methodsk |
| NCM3851 | nagE::KanrnagA::Tetr | P1(IBPC534) → NCM3789d,i,k |
| NCM3852 | nagE::KanrnagC::Tetr | P1(IBPC536) → NCM3789d,i |
| NCM3954 | eut+ | See Materials and Methods |
| NCM3955 | eut+ | See Materials and Methods/PICK> |
| Plasmids | ||
| pNoTA/T7 | PCR cloning vector | 5′ → 3′ Inc. |
| pUC4K | pUC4 carrying a kanamycin resistance cassette | Amersham-Pharmacia Biotech |
| pK03 | Gene replacement vector; repAts, cat, sacB | 33 |
| pJES1333 | PCR fragment containing ycjUTS in pBluescript-KS | See Materials and Methods |
| pJES1336 | pBluescript-KS containing ycjU′-ΔycjT::Kanr-′ycjS | See Materials and Methods |
| pJES1411 | PCR fragment containing lacA in pNoTA/T7 | See Materials and Methods |
| pJES1413 | EcoRI-SacI deleted from polylinker of pJES1411 | See Materials and Methods |
| pJES1415 | pJES1413 containing lacA::Kanr | See Materials and Methods |
| pJES1506 | pK03 containing lacA::Kanr | See Materials and Methods |
| pBR(NagA) | pBR322 containing nagA | 45 |
| pBR(NagC) | pBR322 containing nagC | 45 |
a.k.a., also known as.
For strains constructed by phage P1vir-mediated transduction, the donor is listed after P1vir and the recipient is listed after the arrow.
The length of the in-frame deletion in lacZ was estimated by PCR to be approximately 1,100 to 1,200 bp. The unique restriction sites for EcoRV, DraIII, AvaII, and SacI, all of which are approximately in the middle of the lacZ gene, are absent.
Selected for tetracycline resistance.
Screened for white colonies (lacZ null mutants) on X-Gal and IPTG.
Screened for blue colonies (lacZ+) on X-Gal and IPTG.
Screened for failure to grow on melibiose at 42°C (lacY) (36).
Selected for kanamycin resistance.
rph allele checked by sequence analysis.
Tested for the ability to grow on glucosamine but not N-acetylglucosamine as the sole carbon source.
Tested for failure to grow on glucosamine and N-acetylglucosamine as the sole carbon sources.
Screened for growth on glucosamine and N-acetylglucosamine as the sole carbon sources (Nag+).
Cultures were grown aerobically at 37°C in appropriately supplemented minimal medium lacking N and C sources (N−C− medium) (7, 18), unless otherwise indicated. The nitrogen source was 10 mM NH4Cl and the carbon source (0.4%, wt/vol) was as indicated. Glycerol was used at 0.2% (vol/vol). For glycerol-limited chemostats, N−C− medium was supplemented with 5 mM glycerol and 10 mM NH4Cl (54). Isopropyl-β-d-thiogalactopyranoside (IPTG; Denville Scientific Inc., catalogue no. C18280-3]) was used at the concentrations indicated. Tetracycline and kanamycin were used on plates at 10 and 25 μg/ml, respectively. Transduction by phage P1vir was performed as described previously (52). Selections and screens are given in the footnotes to Table 1.
Repair of rph-1 mutation in MG1655 (CGSC 6300).
The rph-1 lesion in CGSC 6300 was repaired in several steps. The zib-207::Tn10 insertion (39) was introduced into VH1000 (rph+) (14) by P1vir-mediated transduction to yield NCM3414. The linkage between the insertion and rph is ≈10%. P1vir phage grown on NCM3414 were used to introduce the rph+ allele into CGSC 6300 to yield NCM3416. Selection was done for tetracycline resistance, and screening was done for improved growth on N−C− medium with glucose as the carbon source. The presence of the rph+ allele in NCM3416 and the rph-1 allele in congenic strain NCM3415 was confirmed by sequencing.
Isolation of lac mutants from MG1655 (CGSC 6300) as galactose fast-growers.
When MG1655 (CGSC 6300) was passed three times on minimal medium containing glucose or glycerol as the carbon source, its doubling time remained constant at 67 and 98 min, respectively. After being passed three times on galactose, its doubling time decreased to 86 min (see Results). At this point, 96 clones were purified on Luria-Bertani (LB) and screened on minimal medium with galactose or glucose as the carbon source and containing the indicator 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to monitor β-galactosidase activity. Like CGSC 6300, most clones were white to pale blue. However, 16 were blue on glucose, whereas CGSC 6300 was white, and were dark blue on galactose, whereas CGSC 6300 was pale blue. Two strains, NCM3559 and NCM3560, were studied further, as was a control strain, NCM3558, which was a galactose fast-grower but looked like CGSC 6300 on indicator plates. The population of galactose fast-growers was also screened on LB containing X-Gal and 0.5 mM IPTG to induce expression of the lactose operon. Three clones of the 300 screened were white, whereas CGSC 6300 was blue. One, NCM3556, was studied further. No white colonies were seen in a population of CGSC 6300 passed once on galactose, and none were seen in populations passed three times on glucose or glycerol. Lesions in strains NCM3556, NCM3559, and NCM3560 were tested for linkage to the lac operon by P1vir-mediated transduction. The strains were transduced to tetracycline resistance on LB plates containing X-Gal with phage grown on lac+ strain NCM3384 (mhpC281::Tn10), and transductants were screened for the appropriate change in color.
Isolation of a nagA null mutant of MG1655 (CGSC 6300) as a galactose fast-grower.
MG1655 (CGSC 6300) was passed four times on minimal medium containing galactose as the carbon source. Then a portion of the culture was plated on rich medium (LB), and colonies were tested for growth on N-acetylglucosamine (GlcNAc) as the carbon source. One strain that failed to grow on GlcNAc, NCM3845, was chosen for further characterization. PCR amplification (nagA1 primer, 5′-GTTACCGTTAACGATGGTCTTGG; nagA2 primer, 5′-CCGGATCTTTACCGGCCACG) of the coding region of the nagA gene in NCM3845 yielded a fragment similar in size (1.1 kb) to that obtained from MG1655 (CGSC 6300), indicating that the lesion was not a large deletion. PCRs were performed with 1 μl of a 10-fold-diluted overnight LB culture of the strains tested in a 25-μl reaction volume (0.3 U of AmpliTaq enzyme, 1× reaction buffer, 200 μM deoxynucleoside triphosphate mix, and 300 nM each primer) at 94°C for 2 min, followed by 30 cycles of 30 s at 94°C, 30 s at 45°C, and 3 min at 72°C.
Isolation of Eut+ strains.
Parental strains were grown on LB and then streaked on N−C− plates containing 0.4% glycerol, 10 mM ethanolamine hydrochloride, and 0.2 μM cyanocobalamin (CN-B12; Sigma). Plates were incubated at 37°C. No Eut+ colonies were observed on plates that lacked cyanocobalamin.
Construction of a lacA mutant.
Strain NCM3748 (lacA::Kanr) was obtained by insertion of a kanamycin resistance cassette 255 bp downstream of the translational start codon for the lacA gene of MG1655 (CGSC 6300), as described below. (i) The lacA gene with short flanking sequences of the lacY (24-bp) and cynX (52-bp) genes was amplified by PCR from genomic DNA isolated from MG1655 (CGSC 6300) with primers cynX1 (5′-CCCTGCGTTTTGCACCAG; position 155 bp downstream of the stop codon for lacA) and lacY3 (5′-CGTCAGGTGAATGAAGTCGC; position 90 bp upstream of lacA). PCRs were performed with 34 ng of genomic DNA in a 25-μl reaction volume (1 U of AmpliTaq enzyme, 1× reaction buffer, 200 μM deoxynucleoside triphosphate mix, and 320 nM each primer). They were initiated at 94°C for 5 min, followed by 30 cycles of 30 s at 94°C, 30 s at 50°C, and 2 min at 72°C. (ii) The 856-bp amplified DNA fragment was ligated according to the instructions of the manufacturer into the pNo Ta/T7 shuttle vector (Prime PCR Cloner; 5′→3′, Inc.) to yield plasmid pJES1411. The identity of the PCR fragment was confirmed by sequence analysis. (iii) To eliminate the EcoRI site present in the polylinker, pJES1411 was digested with EcoRI and SacI, made blunt-ended with mung bean nuclease, and self-ligated to yield pJES1413. (iv) The 1.3-kb EcoRI fragment from plasmid pUC4K (Amersham Pharmacia Biotech), which carries a kanamycin resistance cassette, was ligated into pJES1413 which had been linearized with ApoI to yield pJES1415. (v) The 2.2-kb PmeI fragment of pJES1415, containing the lacA::Kanr construct, was moved into the SmaI site of the gene replacement vector pK03 (33), resulting in pJES1506. pK03 contains a temperature-sensitive origin of replication and the selectable markers sacB and cat. (vi) The lacA gene of MG1655 (CGSC 6300) was replaced as described previously (20, 33).
From the kanamycin-resistant transformants, one clone with the correct phenotype (Kanr, Cams, and sucrose resistant [growth on LB medium without NaCl and containing 6% sucrose (5)]) was chosen for further analysis. The presence of the lacA::Kanr insertion in this strain, NCM3748, was confirmed by PCR and by measuring transacetylase activity (not shown). That the insertion had indeed replaced the wild-type copy of lacA was confirmed by Southern analysis with full-length lacA and Kanr probes. Genomic DNAs were digested with BglI, HindIII, or both (data not shown).
Construction of a ΔycjT::Kanr deletion-disruption.
Strain NCM3467 (ΔycjT::Kanr) which carries a deletion of 686 bp of the ycjT gene (,2267 bp total; deletion starts 544 bp downstream of the translational start codon) and an insertion of a kanamycin resistance cassette, used for selection, was obtained from the recD strain NCM3426 (see Table 1) by the following steps. (i) The ycjT gene with flanking sequences of the ycjS and ycjU genes was amplified from strain MG1655 (CGSC 6300) with primers ycjT1 (5′-ACTAAGCTTAGATCCTGCCCAGGCGTACC) and ycjT2 (5′-AGTTCTAGAGCCAGAAGGCCGATAACCGC) and the Expand high-fidelity PCR system (Boehringer Mannheim-Roche). PCRs were performed with 34 ng of genomic DNA from MG1655 (CGSC 6300) in a 100-μl reaction volume (2.6 U of Expand enzyme mix, 1× reaction buffer, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphate mix, and 300 nM each primer). They were initiated at 94°C for 2 min, followed by 30 cycles of 30 s at 94°C, 30 s at 50°C, and 5 min at 72°C. The primers introduced a HindIII restriction site at the 5′ end and an XbaI site at the 3′ end of the fragment. (ii) The 4-kb PCR product was digested with HindIII and XbaI and ligated into pBluescript-KS (Stratagene) which had also been digested with HindIII and XbaI to yield plasmid pJES1333. (iii) The identity of the insert was confirmed by sequence analysis. A difference from the published sequence, apparently due to a mutation introduced by PCR amplification, was detected downstream of the SacII restriction site in ycjU. (iv) The 1.3-kb EcoRI fragment from plasmid pUC4K was made blunt-ended with the Klenow fragment of DNA polymerase I and ligated into pJES1333 which had been digested with EcoRV and MscI (deleting 686 bp of ycjT) to yield pJES1336. (v) The 4.6-kb KpnI-SacII (upstream of the possible mutation noted above) fragment of pJES1366, carrying ycjS′-ΔycjT1366::Kanr-′ycjT was transformed into the recD strain NCM3426 to yield strain NCM3467 (kanamycin resistant). The presence of the ΔycjT1366::Kanr deletion-insertion was confirmed by PCR (data not shown) and moved by P1vir-mediated transduction into other strains (Table 1).
Determination of fnr phenotypes of various strains.
Strains were tested anaerobically for growth on a variety of media containing appropriate electron donors and acceptors and for evolution of gas from glucose (see Results). The well-characterized fnr+ and fnr strains RK4353 and RK5279, respectively, were used as controls (55). Lesions in strains with phenotypic defects were tested for linkage to ycj-630::Tn10 by P1vir-mediated transduction. The ycj-630::Tn10 allele is 90% linked to fnr (55).
Analysis of fnr region of various strains by PCR.
To determine the basis of the Fnr− phenotype of MG1655 (CGSC 6300) (see Results), we attempted to amplify the fnr gene (b1344) and the chromosomal region surrounding it by PCR. PCR products were analyzed on 0.8% agarose gels and stained with ethidium bromide. PCR amplification of the fnr gene with the b1334 ORFmer primer set (Fnr-A/C) of Sigma Genosys produced a single PCR product of the expected size (≈0.75 kbp) for the Singer-Gross isolate of MG1655 (NCM3430), but a fragment of ≈3.0 kbp for the isolate of MG1655 from the CGSC (CGSC 6300). Control PCRs performed in the absence of one primer established that the 3′ primer (Fnr-C) amplified a nonspecific fragment of the same size (≈3.0 kbp).
New primers were designed (fnr-5′, 5′-CGCCATGAAGGTTATCTT-3′; fnr-3′, 5′-CCTTCTGCCAGATCAATA-3′). PCRs were performed with 40 ng of genomic DNA in a 30-μl reaction volume (1 U of Taq polymerase [Perkin-Elmer], 1× reaction buffer, 125 μM deoxynucleoside triphosphate mix, and 350 nM each primer). They were initiated at 94°C for 4 min, followed by 25 cycles of 1 min at 94°C, 1 min at 50°C, and 80 s at 72°C. A single PCR product of the expected size (≈1.0 kbp) was amplified from Fnr+ strain NCM3430, and no product was obtained from Fnr− strain CGSC 6300. PCR amplification of fnr with our primer set and of every other gene from b1330 to b1346 with the ORFmer primer sets of Sigma-Genosys yielded a single product of the expected size for each gene from NCM3430. (The primer pair for b1336 was an exception: it failed). However, products were obtained only for b1330 and b1331 and for b1345 and b1346 from CGSC 6300. No products were detected for b1332 through b1344. The entire fnr region was amplified by long-range PCR according to the manufacturer's instructions (Expand PCR Long Range; Boehringer-Roche) with 40 ng of genomic DNA, 3.75 U of Expand polymerase, 1× reaction buffer 2, 500 μM deoxynucleoside triphosphate mix, and 300 nM each primer in a 50-μl reaction) with forward primer b1331 and reverse primer b1345 at 94°C for 120 s and then for 10 cycles at 94°C for 10 s, 60°C for 30 s, and 68°C for 15 min, and 15 additional cycles with an increase of 20 s per cycle at 68°C (see Fig. 2).
FIG. 2.
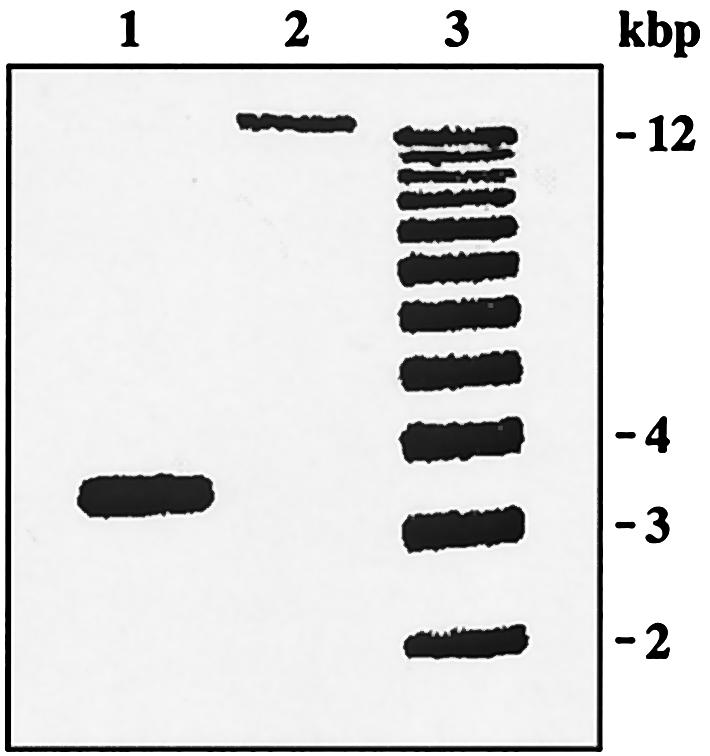
PCR amplification of fnr region from different MG1655 isolates. The fnr region was amplified from the CGSC isolate of MG1655 (CGSC 6300; lane 1) and the isolate obtained from M. Singer and C. Gross (NCM3430; lane 2) (see Materials and Methods). The sizes of the molecular standards in lane 3 are noted to the right. The genes deleted in the CGSC isolate (b1332 to b1344) are, respectively, ynaJ (open reading frame conserved in E. coli and Salmonella enterica), uspE (ydaA; UspA paralog) (17), fnr (Crp family activator of anaerobic respiratory gene transcription), ogt (O-6-alkylguanine-DNA/cysteine-protein methyltransferase), abgT (ydaH; p-aminobenzoyl-glutamate transport; oxidized folate recycling) (23), abgB (ydaI; p-aminobenzoyl-glutamate; oxidized folate recycling) (23), abgA (ydaJ; p-aminobenzoyl-glutamate; oxidized folate recycling) (23), abgR (ydaK; p-aminobenzoyl-glutamate regulator, LysR-type) (23), ydaL (open reading frame conserved in enterobacteria), ydaM (open reading frame conserved in E. coli), ydaN (open reading frame conserved in enterobacteria), dbpA (ATP-dependent RNA helicase), and ydaO (open reading frame conserved in enterobacteria). The deletion is flanked by tns5_4 (b1331), which codes for IS5 transposase, and ydaP (b1345), a rac prophage which codes for a putative prophage integrase.
Determination of β-galactosidase and thiogalactoside transacetylase activities.
The β-galactosidase assays were performed as previously described (21). Thiogalactoside transacetylase (LacA) activity was determined as described previously (36). Cells pelleted from various culture volumes were suspended in 0.2 ml of buffer (50 mM Tris [pH 7.9], 10 mM EDTA) and sonicated. Differential rates of enzyme synthesis (in units per milliliter per unit of optical density at 600 nm [OD600]) were obtained by determining the slopes of plots of enzyme activity versus the optical density of the culture at 600 nm (21, 26).
Determination of α-amylase activity.
The α-amylase activity assay was adapted from that of Freundlieb et al. (13) and modified. After centrifugation, cell pellets were washed and suspended in ice-cold sodium phosphate buffer (100 mM, pH 7.0). Cells were permeabilized with chloroform and sodium dodecyl sulfate (0.002% [wt/vol]) and warmed to 27°C for 5 min. Reactions were started by the addition of 4-nitrophenyl-α-d-hexa-(1→ 4)-glucopyranoside (PG6; Fluka Chemical Corp.), which was dissolved in the above buffer, to a final concentration of 1 mM. Reactions were stopped with Na2CO3, and cells were removed by centrifugation. The α-amylase activity was calculated as 1,000 × A405/(t × v) (36). Activity reflects the sum of the activities of the MalT-inducible periplasmic α-amylase MalS, the MalT-inducible cytoplasmic maltodextrin glucosidase MalZ (56), and the MalT-independent cytoplasmic α-amylase AmyA (47).
Clinical isolates of E. coli.
Twelve isolates obtained from the Tang Health Center at the University of California, Berkeley, were kindly provided by A. Manges and L. Riley in the School of Public Health. They had been identified as E. coli based on their ability to catabolize lactose (red colonies on MacConkey-lactose plates) and their ability to produce indole from tryptophan (35). Strains were frozen within three passages of the time of isolation. Strains NCM3601 (Tang identifier Seq404) and NCM3610 (Tang identifier Seq915) were isolated from patients with acute urinary tract infections (UTI), whereas NCM3611 (Tang identifier 10002-006) was an intestinal commensal strain isolated from the partner of a patient with an acute UTI. Each of these strains was isolated from a separate individual. Isolates from the Tang Center failed to grow on minimal medium, and many required nicotinic acid. Tang Center isolates readily yielded prototrophic derivatives, and derivatives of the strains listed above were chosen for microarray experiments because they grew well on minimal medium. Isolates from the Tang Center were resistant to phage P1vir.
Glass slide DNA microarray analysis.
Cells were grown to mid-exponential phase, and growth was then quenched with 1/10 volume of 5% phenol in ethanol (57, 60). RNA extraction, synthesis of cDNAs containing fluorescent nucleotide analogues, hybridization to microarrays, scanning of fluorescence, and normalization were done essentially as described previously (60). Data are displayed in genome images (57, 60).
Operon organization is taken from the RegulonDB database (http://www.cifn.unam.mx/Computational_Genomics/regulondb/) with the modifications of Zimmer et al. (60). Genes are listed in the order transcribed, with the exception of those in the flagellar or chemotaxis regulon. For the latter, the direction of transcription of individual operons is indicated with arrows. For comparisons of CGSC 6300 and clinical isolates of E. coli grown on glucose with either glutamine or NH4Cl as the nitrogen source, data from three experiments with CGSC 6300 were averaged. Data from three experiments performed once each with prototrophic derivatives of clinical isolates NCM3601, NCM3610, and NCM3611 were also averaged.
Affymetrix DNA microarray analysis.
E. coli K-12 strains NCM3722 and MG1655 (CGSC 6300) were grown in N−C− medium with either 0.4% glucose or 0.4% glycerol as the carbon source and either 10 mM NH4Cl or 2.5 mM arginine as the nitrogen source. Strains were adapted to growth in the appropriate medium by preculturing to the early stationary phase. Experimental cultures were incubated in a water bath at 37°C in 500- or 1,000-ml Erlenmeyer flasks containing 1/10 volume of medium. The starting OD600 was 0.025, and cells were harvested in mid-exponential phase (OD600 = 0.4 to 0.5) by adding 1/10 culture volume of 95% ethanol-5% phenol. Cell suspensions (25-ml portions) were then subjected to centrifugation at 6,000 rpm for 5 min, and pellets were frozen on dry ice. They were stored at −80°C prior to use.
Gene expression was assessed on Affymetrix E. coli Antisense GeneChip arrays (Affymetrix, Inc., Santa Clara, Calif.). The detailed protocol, which was slightly modified from that supplied to us by Affymetrix, Inc., is available at http://nature.berkeley.edu/≈opaliy/papers/MG1655.html. Briefly, total RNA was prepared from E. coli cells by phenol-chloroform extraction, and 15 μg of total RNA was used to synthesize cDNA. After purification with a Qiagen cDNA purification kit (Qiagen, Inc.), the cDNA was fragmented with DNase I. The fragmented cDNA was end biotinylated by terminal transferase with biotinylated ddATP. Hybridization of the fragmented, biotinylated cDNA to a chip, washing, staining, and scanning of the chip were done on an Affymetrix workstation as described previously (Expression Analysis Technical Manual; Affymetrix, Inc., Santa Clara, Calif.). Raw data files were analyzed by the statistical algorithm in MAS 5.0 (Affymetrix, Inc.) with the default parameters. All experiments were scaled globally to the same target intensity of 1,500 with only the probe sets for E. coli genes and not intergenic regions. Data obtained from different strains grown on the same medium were compared to determine differences in mRNA levels. Results were exported into Microsoft Excel for further analysis.
RESULTS
Growth defects caused by rph-1 allele of MG1655.
The rph-1 allele decreases the expression of pyrE by 100-fold and leads to pyrimidine limitation in minimal medium (27). We confirmed that growth of MG1655 (CGSC 6300) in glucose-minimal medium in batch culture was stimulated by addition of uracil (27) or by P1vir-mediated transductional repair of the rph-1 allele (14, 39) (see Materials and Methods).
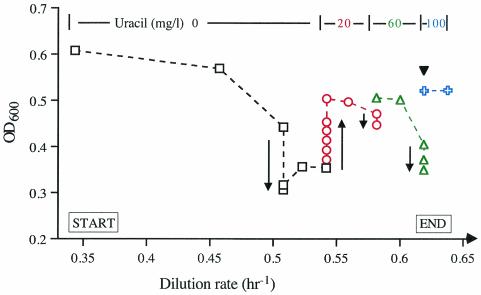
When we attempted to grow MG1655 (CGSC 6300) in glycerol-limited continuous culture, growth defects due to internal uracil (pyrimidine) starvation became more problematic (Fig. 1). At a very low dilution rate of 0.34 h−1, the culture reached steady state at an OD600 of 0.6, approximately that expected, and exhausted glycerol from the medium (<0.02 mM remaining) (Fig. 1 and legend). However, at a faster dilution rate of 0.51 h−1, the OD600 decreased, that is, the culture started to wash out. Surprisingly, however, at dilution rates in this range (0.52 to 0.54 h−1), it established a new steady state (OD 0.36 to 0.4) in which large amounts of glycerol remained in the medium (1.3 to 1.6 mM of the 5 mM total that was provided in the reservoir).
FIG. 1.
MG1655 becomes internally limited for uracil in a glycerol-limited chemostat. Cell yield (OD600; dotted lines) was plotted as a function of the dilution rate, which was varied from slowest (left) to fastest (right). At the dilution rates indicated on the line at the top, the medium in the reservoir was supplemented with uracil at the concentrations indicated. Symbols: black squares, unsupplemented; red circles, 20 mg/liter; green triangles, 60 mg/liter; blue crosses, 100 mg/liter. The directions of changes in OD600 at fixed dilution rates are indicated by arrows. When uracil was removed from the medium in the reservoir at the end of the experiment, the OD600 remained high (solid inverted triangle).
Knowing that the growth rate in batch culture was stimulated slightly by feeding uracil, we hypothesized that the internal pyrimidine pool concentration might have become limiting at these dilution rates. Adding uracil to the medium (to 20 mg/liter) increased the cell yield (OD to 0.5 units) and resulted in consumption of the glycerol (to <0.02 mM). After several further cycles of increasing the dilution rate and the concentration of uracil added to the medium, subsequent decreases in the uracil concentration (from 100 to 4 mg/liter) at a fixed fast dilution rate (0.62 h−1) no longer resulted in decreased optical density of the culture. (It remained at 0.55.) Hence, the strain had apparently acquired a mutation(s) that suppressed the pyrimidine requirement. When four clones taken from the chemostat at this point were grown in batch culture, their doubling times indicated that they had also acquired a mutation(s) that enabled them to grow faster than MG1655 or its rph+ derivative on glycerol as the carbon source (not shown) (24).
MG1655 (CGSC 6300) carries an fnr deletion.
Strains carrying fnr null alleles exhibit specific defects in anaerobic respiration: they fail to use fumarate or nitrate as an electron acceptor for respiration. They also fail to respire with nitrite or dimethyl sulfoxide as the electron acceptor, and they synthesize little formate-hydrogen lyase (due to Fnr-dependent expression of nickel uptake). They cannot use glycerol as an electron donor for anaerobic growth but grow reasonably well on glucose (fermentation) and also show essentially normal aerobic respiratory metabolism.
We determined the Fnr phenotype of various strains by testing anaerobically for growth on glycerol-fumarate (growth on defined medium), for formate-nitrate respiration (phenotype on MacConkey-nitrate [MN] indicator medium), and for gas production from glucose (55). We confirmed genetic linkage to fnr by testing for transductional linkage to a Tn10 element near the fnr locus (55). In each case, experimental strains were compared to well-characterized controls (55) (see Materials and Methods). MG1655 (CGSC 6300) failed to grow on glycerol-fumarate plates, formed small dark red colonies on MacConkey-nitrate plates, and evolved little gas from glucose. Introduction of an fnr+ allele from strain RK4353 (55) by P1vir-mediated transduction restored growth on glycerol-fumarate and allowed the formation of large salmon-colored colonies on MN medium.
Both phenotypic and genetic analyses indicated that a number of additional strains held at the CGSC, some related to MG1655 and some not, were also Fnr−. These included BD792 (CGSC 6159), which, like MG1655, is W1485F− W1485F+ (CGSC 5024); the original wild-type E. coli K-12 strain EMG2 of Clowes and Hayes (CGSC 4401); and WG1 of Lederberg (CGSC 5073). However, an isolate of MG1655 obtained through Mitchell Singer from the laboratory of Carol Gross (NCM3430) was Fnr+, as were isolates obtained from the American Type Culture Collection (ATCC 47076) and the Blattner laboratory (both colony types; see below). An E. coli K-12 wild-type strain used in our laboratory (NCM3722) was also Fnr+. It appears that storage at or shipping from the CGSC may predispose strains to become Fnr− (37).
PCR-based amplification of the fnr region from MG1655 (CGSC 6300) and the Singer isolate (NCM3430) showed that CGSC 6300 carries a deletion of this region (Fig. 2). PCR amplification of individual genes in the region from CGSC 6300 indicated that the deletion extends from gene b1332 (included) to b1344 (included) and results in the loss of 13 genes (legend to Fig. 2) and about 14 kbp. Interestingly, the genes flanking the deletion code for a transposase (b1331) and a prophage integrase (b1345) (4). Amplification of the fnr region from the Blattner isolate of MG1655 (both colony types; see below) and ATCC 47076 indicated that both were fnr+ (not shown). This was also true of K-12 wild-type strain NCM3722.
MG1655 grows slowly on galactose.
When E. coli MG1655 (CGSC 6300) was transferred from enriched medium to minimal medium with galactose as the carbon source or from minimal-glucose medium to minimal-galactose medium, it grew slowly (doubling time of 200 to 300 min; Table 2). As it was adapted to galactose by serial transfers, the culture grew more rapidly (doubling time of 70 to 80 min). The fast-growing population had unchanged doubling times on glucose or glycerol as the carbon source (data not shown). Clones isolated from the fast-growing population grew rapidly on galactose immediately upon transfer from other carbon sources or media (data not shown), indicating that they were genetically altered. Several were characterized further (see below).
TABLE 2.
Doubling times on galactose and activities of β-galactosidase (LacZ) and thiogalactoside transacetylase (LacA) of MG1655 (CGSC 6300) and strains derived from it
| Strain | Phenotype | Galactosea
|
Glycerola
|
Glucosea
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DT (min) | LacZ (U/ml/OD600) | LacA (U/ml/OD600) | DT (min) | LacZ (U/ml/OD600) | LacA (U/ml/OD600) | DT (min) | LacZ (U/ml/OD600) | LacA (U/ml/OD600) | ||
| MG1655b | 260 | 20 | ≤5 | 90 | 10 | ≤5 | 70 | ≤10 | ≤5 | |
| MG1655 adapted to galactosec | Fast growth on galactose | 75 | 440 | 10 | 90 | 420 | 10 | 70 | 145 | ≤5 |
| NCM3559 | Lac(Con) | 85 | 3,200 | 60 | 90 | 7,600 | 145 | 70 | 2,800 | 80 |
| NCM3560 | Lac(Con) | 80 | 2,900 | 50 | 95 | 670 | 6 | 70 | 180 | ≤5 |
| NCM3556 | Lac− | 80 | ≤10 | 55 | ||||||
| NCM3558d | Lac+ | 80 | ≤10 | ≤5 | ||||||
Minimal medium containing 0.4% C source. DT, doubling time. LacZ (β-galactosidase) and LacA (thiogalactoside transacetylase) activities are given in differential rates in units per milliliter per OD600 unit (see Materials and Methods) (21, 26). For data obtained on galactose, strains were grown on glucose and transferred to galactose.
When MG1655 was grown on glucose and transferred to galactose plus 0.1 mM IPTG, its doubling time and LacZ and LacA activities were 80 min and 2,900 and 60 U/ml/OD600 unit, respectively. When MG1655 was passed three times on either glucose or glycerol and then transferred to galactose, its doubling times and LacZ activities were the same as those shown in the table for each carbon source.
Passed three times on galactose as the C source.
Probably carries a suppressor mutation outside the lac region.
Phenotypes of various MG1655 isolates.
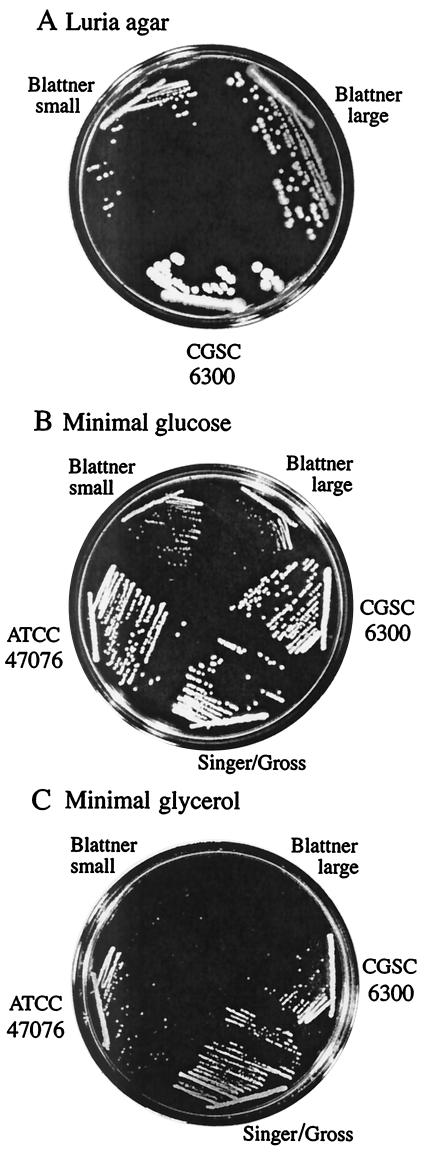
The MG1655 strain that we received from the Blattner laboratory (NCM3629) (4) gave a mixture of large and small colonies on enriched medium (Fig. 3A). When restreaked, the small colonies remained small, whereas the large colonies gave rise to large and small colonies. Hence, the small colonies appear to arise from the large ones. Neither colony type grew as rapidly as CGSC 6300, ATCC 47076, or the Singer-Gross isolate (NCM3430) on N−C− minimal medium with glucose as the carbon source (Fig. 3B), and unlike the other isolates, neither colony type of the Blattner isolate grew on N−C− medium with glycerol as the carbon source (Fig. 3C).
FIG. 3.
Appearance of different isolates of MG1655 on several media. MG1655 was obtained from the CGSC (CGSC 6300), the American Type Culture Collection (ATCC 47076), Mitchell Singer (from the laboratory of Carol Gross) (NCM3430), and the Blattner laboratory (NCM3629). Upon receipt, the isolate from the Blattner laboratory gave a mixture of large and small colonies on both LB and minimal glucose agar (see text). The properties of both are shown. Media were LB (A); N−C− medium with glucose (0.4%) and NH4Cl (10 mM) (B); and N−C− medium with glycerol (0.2%) and NH4Cl (10 mM) (C). Cells were streaked on LB before being transferred to the medium indicated and were incubated aerobically at 37°C for 1 to 2 days.
Gene expression of MG1655 (CGSC 6300) on lactose and galactose versus glucose.
In establishing DNA microarray studies of MG1655 (CGSC 6300), we observed induced expression of the lactose operon in glycerol-grown cells with the gratuitous inducer IPTG (57). To extend these studies, we compared expression profiles for cultures growing exponentially on lactose or galactose as the sole carbon source to those for cultures growing on glucose (Table 3). Growth on either lactose or galactose resulted in higher levels of expression of: galP (b2943), which codes for a low-affinity galactose transporter; the mglBAC operon (b2150 to b2148), which codes for a high-affinity transporter for β-methylgalactosides; galS (b2151), the regulator of mglBAC (16); and the galactose catabolic operon galETKM (b0759 to b0756) (Table 3, Fig. 4). Growth on galactose also resulted in higher levels of expression of the melBAR operon (b4120 to b4118), which codes for a transporter for α-galactosides, an α-galactosidase, and a regulator.
TABLE 3.
Genes induced ≥2-fold when MG1655 (CGSC 6300) was grown on lactose or galactose as the C source instead of glucose
| b no. | Genea | RNA ratiob
|
|
|---|---|---|---|
| Lactose vs. glucose | Galactose vs. glucosec | ||
| b0344 | lacZ | 57 | 4.1 |
| b0343 | lacY | 21 | 3.5 |
| b0342 | lacA | 21 | 4.6 |
| b0759 | galE | 12 | 6.4 |
| b0758 | galT | 10 | 3.6 |
| b0757 | galK | 2.1 | 1.5 |
| b0756 | galM | 2.9 | 1.4 |
| b2151 | galS | 1.3 | 3.3 |
| b2943 | galP | 2.8 | 3.9 |
| b2150 | mglB | 3.6 | 13 |
| b2149 | mglA | 2.6 | 7.3 |
| b2148 | mglC | 0.8 | 4.9 |
| b4120 | melB | 1.0 | 2.3 |
| b4119 | melA | 1.3 | 4.5 |
| b4118 | melR | 1.1 | 1.1 |
| b4035 | malK | 4.1 | 0.8 |
| b4036 | lamB | 4.6 | 0.6 |
| b4037 | malM | 1.4 | 1.0 |
| b4034 | malE | 5.2 | 0.7 |
| b4033 | malF | 1.6 | 0.7 |
| b4032 | malG | 1.5 | 1.0 |
| b3417 | malP | 3.9 | 0.3 |
| b3416 | malQ | 2.9 | 0.6 |
| b0679 | nagE | 0.9 | 3.3 |
| b0678 | nagB | 1.1 | 4.4 |
| b0677 | nagA | 1.1 | 3.7 |
| b0676 | nagC | 1.1 | 1.8 |
| b0675 | nagD | 1.6 | 1.0 |
| b3730 | glmU | 1.3 | 0.6 |
| b3729 | glmS | 1.1 | 0.4 |
The gene listed first in each group is the first gene in the operon.
If any gene in the operon showed a ratio of ≥2 in one of the comparisons, data are presented for all genes in the operon under both conditions. The glmUS operon is included because it is NagC regulated (see text). Electrophoresis of the PCR product for MalS that was printed on the microarrays showed that there was only a small amount of product of the correct size and a large amount of another product.
Population of galactose fast-growers.
FIG. 4.
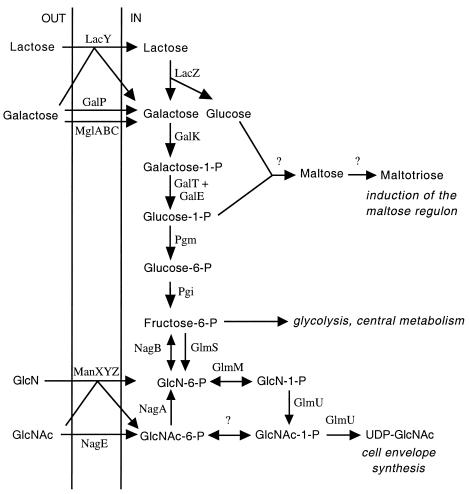
Diagram of the utilization of lactose, galactose, and N-acetylglucosamine (GlcNAc) as sole carbon sources and related pathways discussed in this work. The gene products indicated are LacY, lactose transporter; LacZ, β-galactosidase; GalP, low-affinity galactose transporter; MglABC, high-affinity galactose transporter; GalK, galactokinase; GalT, galactose 1-phosphate uridylyltransferase; GalE, UDP-galactose-4-epimerase; Pgm, phosphoglucomutase; Pgi, phosphoglucoisomerase; NagE, GlcNAc transporter; NagA, GlcNAc-6-phosphate deacetylase; NagB, GlcN-6-phosphate deaminase; ManXYZ, glucosamine transporter; GlmS, GlcN-6-phosphate synthase; GlmM, phosphoglucosamine mutase; GlmU, GlcN 1-phosphate acetylase; and UDP-GlcNAc synthase. As indicated by a bent arrow, LacY is known to transport galactose (15, 50), and elevated LacY expression apparently allows fast growth of MG1655 (CGSC 6300) on galactose as the carbon source (see text). Cells grown on lactose as the carbon source apparently synthesize inducers of the maltose regulon endogenously. Such synthesis from glucose and glucose 1-phosphate has been proposed previously (1, 9, 51), although the enzymes involved have not been defined. The product of gene ycjT does not appear to be solely responsible for endogenous synthesis of inducer (see text). NagC (not shown) regulates both the synthesis and degradation of GlcNAc (see Discussion). Inactivation of NagC allows fast growth of MG1655 on galactose, apparently by decreasing endogenous synthesis of GlcN 6-phosphate and thus increasing the flux of fructose 6-phosphate through central metabolism (see Discussion).
Growth on lactose resulted in a high level of expression of the lactose operon and, unexpectedly, in a two- to fivefold-elevated expression for three operons in the maltose regulon (6), those that code for a transporter for maltose and maltosaccharides (the malK-lamB-malM [b4035 to b4037] and malEFG [b403 to b4032] operons) and an operon that codes for catabolic enzymes (the malPQ operon [b3417 to b3416]) (Table 3). As reported (59), growth on galactose resulted in a somewhat increased level of expression of the lac operon (b0344 to b0342) (but see below). In addition, it resulted in elevated levels of expression of the nagBACD (b0678 to b0675) and nagE (b0679) operons (but see below). The products of these operons are responsible for catabolism of N-acetylglucosamine (Fig. 4) (45, 58).
Cross-induction of mal regulon by growth on lactose.
The fact that induction of expression of the mal regulon did not occur with IPTG indicated that it was not mediated directly by the lactose repressor, LacI. The fact that it did not occur on glycerol or galactose as the carbon source (Table 3 and not shown) provided evidence that it was not due simply to relief of catabolite repression. To test its dependence on MalT, the activator of the maltose regulon, and maltose transport components, we first confirmed that induction could be detected at the protein level both for MalE, the periplasmic maltose binding protein (not shown), and for MalS plus MalZ, periplasmic and cytoplasmic α-amylases, respectively (Table 4) (6, 12, 13). The α-amylase activity, which was ninefold higher on glycerol plus maltose than on glycerol, was also 2.5 times higher on lactose than on glycerol. In a strain carrying a null allele in malT, the increases in α-amylase activity on glycerol plus maltose and on lactose were eliminated, indicating that expression of malS plus malZ during growth on lactose is MalT dependent. Similarly, increased expression of the maltose binding protein on lactose was MalT dependent (not shown).
TABLE 4.
Activity of α-amylase in MG1655 (CGSC 6300) and derivatives grown on lactose and other carbon sources
| Strain | Genotype | α-Amylase activitya (U/ml/OD600)
|
||
|---|---|---|---|---|
| Lactose (0.4%) | Glycerol (0.2%) | Glycerol + maltose (0.2% + 0.2%) | ||
| MG1655 | Parent | 28 ± 1 | 11 ± 1 | 97 ± 3 |
| NCM3364 | malT::Tn10 | 10 | 14 | 17 |
| NCM3350 | malF::Tn10 | 38 | 21 | 15 |
| NCM3351 | malG::Tn10 | 36 | 15 | 12 |
Induction by MalT depends on the coinducer maltotriose (or the absence of the cytoplasmic maltose transport component MalK) (6, 28). To determine whether the coinducer was a contaminant of lactose, we introduced lesions in the cytoplasmic membrane components required for transport of maltose and maltosaccharides (malF::Tn10 or malG::Tn10 allele) into MG1655. Whereas these mutations eliminated the increase in α-amylase activity on glycerol plus maltose, confirming that they prevented maltose transport, they caused no decrease in activity on lactose (Table 4), indicating that induction of malS on lactose was apparently not due to the presence of contaminating coinducer in the lactose. Rather, it appeared to be due to synthesis of the coinducer endogenously. These results were confirmed qualitatively for the maltose binding protein (not shown).
Decker et al. (9) proposed that maltose and maltotriose may be synthesized endogenously from glucose and glucose 1-phosphate (Fig. 4) by a maltose phosphorylase. Hence, we inactivated ycjT (see Materials and Methods), which codes for a protein with 45% similarity over its entire length to MapA, a maltose phosphorylase from Lactobacillus sanfranciscensis (Blast website, http://www.ncbi.nlm.nih.gov/blast/) (10). Based on levels of the maltose binding protein (assessed by Western blotting; data not shown), the ycjT null allele did not affect induction of the maltose regulon on lactose, nor did it affect growth on maltose.
Apparent induction of lac operon by growth on galactose.
Our microarray experiments were performed before we knew that MG1655 (CGSC 6300) was acquiring mutations to fast growth in galactose. In a population of cells growing rapidly on galactose, we observed four- to fivefold-higher levels of expression of the lac operon than in cells grown on glucose (Table 3). Higher expression of the lac operon was not observed in a culture grown on glycerol or in a population of cells growing slowly on galactose (not shown). Similar results were obtained by enzyme assays (Table 2) except that the degree of increase in β-galactosidase activity in the population of galactose fast-growers (400 versus 10 U/ml/OD600) was at least 40-fold. We have noted such discrepancies between microarray data and enzyme assays previously (57).
To study the basis for higher expression of the lac operon in a culture of MG1655 “adapted to” galactose and growing on it rapidly, we screened 96 purified clones for a constitutive Lac phenotype (see Materials and Methods). Sixteen had this phenotype. Lesions in the two strains tested (NCM3559 and NCM3560) were linked by P1vir-mediated transduction to the lac operon (see Materials and Methods). These strains grew rapidly on galactose and had high β-galactosidase (LacZ) and galactoside transacetylase (LacA) activities when grown on galactose, glycerol, or glucose as the carbon source (Table 2, but note the differences between the strains). The proportion of such strains (≈15%) accounted nicely for the β-galactosidase activity observed in a population of galactose fast-growers derived from MG1655 (CGSC 6300) (3000 × 0.15 = 450).
In addition, we tested 300 clones from a population of galactose fast-growers for a LacZ− phenotype on LB agar plus IPTG, the inducer of the lac operon, and X-Gal. Three were white, indicating that they lacked β-galactosidase activity. The fact that none of the clones from a culture grown on glucose or glycerol as the carbon source was white indicated that this phenotype was associated with fast growth on galactose. The lesion in one such strain (NCM3556) was shown to be linked to the lac operon. The strain grew rapidly on galactose, and although it had no detectable β-galactosidase activity, it had high levels of transacetylase (Table 2). Such a phenotype can be accounted for by a small nonpolar deletion encompassing lacO and the beginning of lacZ (49). Together, the results indicated that high levels of expression of lacY and/or lacA resulted in fast growth on galactose in the MG1655 background. As expected, a galactose fast-grower that resembled its parental strain on both types of plates containing X-Gal (NCM3558) had very low levels of β-galactosidase and galactoside transacetylase activity when grown on galactose. Thus, not all galactose fast-growers had increased lac expression.
To determine the roles of individual lac genes in fast growth of MG1655 (CGSC 6300) on galactose, we employed strains with well-characterized mutations in lacZ, lacY, or lacA (insertion constructed for this work; see Materials and Methods). These strains and one carrying both the lacZ and lacA mutations were grown on galactose plus IPTG or glycerol plus IPTG to induce expression of the lac operon (Table 5). The MG1655 parental strain (not adapted to galactose) grew rapidly on galactose plus IPTG, in agreement with the view that expression of one or more lac genes was sufficient to confer this ability. Only the strain with the (polar) lacY mutation grew slowly, indicating that high activity of the lactose permease probably accounted for improved growth on galactose. Enzyme assays (not shown) verified the genotypes of the strains, and a control experiment indicated that all mutant strains had the same doubling time as their parent on glycerol plus IPTG (Table 5).
TABLE 5.
Doubling times on galactose plus IPTG of MG1655 (CGSC 6300) and derivatives carrying defined lac mutationsa
| Strain | Relevant genotype | Doubling time (min)
|
|
|---|---|---|---|
| Galactose + IPTG | Glycerol + IPTG | ||
| NCM3384 | Wild type | 110 | 100 |
| NCM3387 | ΔlacZ | 110 | 100 |
| NCM3388 | lacY(Am) | 225 | 95 |
| NCM3749 | lacA::Kanr | 110 | 100 |
| NCM3750 | ΔlacZ lacA::Kanr | 115 | 100 |
Minimal medium contained 0.4% galactose or 0.2% glycerol as a C source plus 0.1 mM IPTG. Cells were transferred to these media from minimal medium containing 0.4% glucose or 0.2% glycerol, respectively.
Apparent induction of nag operons by growth on galactose.
In a population of MG1655 (CGSC 6300) cells growing rapidly on galactose, there was a three- to fourfold increase in RNA levels for the nagA, nagB, and nagE products with respect to values on glucose (Table 3). These genes code for the catabolic enzymes N-acetylglucosamine-6-phosphate deacetylase and glucosamine-6-phosphate deaminase and for an N-acetylglucosamine-specific transport component, an enzyme II of the phosphotransferase system, respectively (Fig. 4). The nagBACD and nagE operons are divergently transcribed, and levels of induction for nagB and nagA are known to be higher than those for nagC and nagD due to higher basal levels of expression of the latter from internal promoters (43). The fact that induction did not occur on glycerol or lactose (not shown and Table 3) indicated that it was specific to galactose and not likely to be accounted for simply by relief of catabolite repression. Induction also occurred on galactose when the medium was supplemented with uracil (not shown).
To study induction further, we introduced defined nag mutations into MG1655 and determined their effect on its doubling time on galactose (Table 6). Every lesion known to result in derepression of the nag operons (45) (nagC, nagA, and nagB insertion [due to polarity on nagA], but not a point mutation in nagB or an insertion in nagD or nagE) resulted in fast growth on galactose. nagC codes for the Nag repressor, and the absence of NagA activity results in accumulation of the coinducer N-acetylglucosamine 6-phosphate (45). To determine whether high levels of nagE or nagD were required for rapid growth, we tested the effect of deletion of both nag operons (ΔnagEBACD). To our surprise, this also resulted in fast growth on galactose (NCM3820), indicating that no product of the nag operons, including the transporter, was required.
TABLE 6.
Doubling times on galactose of MG1655 (CGSC 6300) derivatives carrying defined nag lesionsa
| Strain | Relevant genotype | Doubling time (min) |
|---|---|---|
| MG1655 | Wild type | 280 |
| NCM3789 | nagE::Kanr | 300 |
| NCM3817 | nagB::Kanr | 85 |
| NCM3818 | nagB2 | 240 |
| NCM3786 | nagA::Tetr | 80 |
| NCM3787 | nagC::Tetr | 85 |
| NCM3821 | nagD::Tetr | 260 |
| NCM3820 | Δnag(EBACD)::Tetr | 80 |
| NCM3851 | nagA::TetrnagE::Kanr | 85 |
| NCM3852 | nagC::TetrnagE::Kanr | 80 |
| NCM3105/pBR322 | Wild type | 270 |
| NCM3105/pBRnagC+ | nagC+/nagC+ | 250 |
| NCM3787/pBR322 | nagC::Tetr | 85 |
| NCM3787/pBRnagC+ | nagC::Tetr/nagC+ | 155 |
| NCM3820/pBR322 | Δnag(EBACD)::Tetr | 80 |
| NCM3820/pBRnagC+ | Δnag(EBACD)::Tetr/nagC+ | 80 |
Precultures were grown on glucose. NCM3845 (nagA166), which was isolated from MG1655 as a galactose fast-grower, had the same doubling time as NCM3786 (nagA::Tetr) (75 min), whereas MG1655 had a doubling time of 240 min in this experiment.
To be certain that the transporter was not required, we constructed strains carrying double insertions, nagC nagE or nagA nagE. Both grew rapidly on galactose. We confirmed these results in two ways. First, we isolated a strain with a nagA lesion in a population of galactose fast-growers derived from MG1655 (see Materials and Methods). This strain failed to grow on N-acetylglucosamine as the carbon source but grew on glucosamine, and its growth defect was complemented by a nagA+ allele (not shown). Second, we confirmed that loss of nagC (or its function) was required for fast growth on galactose by showing that a nagC+ allele partially complemented the nagC derivative of MG1655 (NCM3787) to restore slower growth on galactose but that it did not complement the ΔnagEBACD derivative (NCM3820) because this strain accumulates coinducer internally (Table 6) (45).
Comparison of strain MG1655 (CGSC 6300) to E. coli K-12 strain NCM3722 on glass slide DNA microarrays.
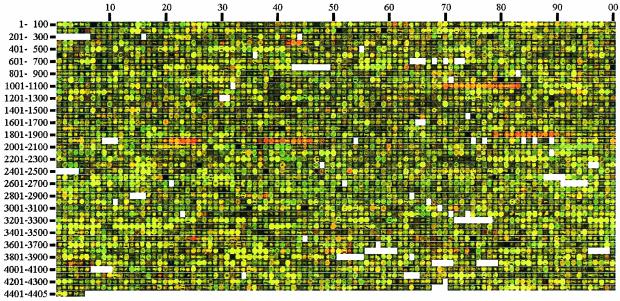
Given the difficulties with MG1655 isolates, we tested whether MG1655-based DNA microarrays could be used to probe gene expression in other E. coli strains. We first compared mRNA levels in an E. coli K-12 strain with no known mutational lesions (NCM3722; see Materials and Methods) to those in strain MG1655 (CGSC 6300) for cells grown to the mid-exponential phase in LB (Fig. 5). To be sure there would be at least one difference between the strains, we induced expression of the lactose operon in NCM3722 by adding IPTG (to 100 μM) for 25 min before harvest.
FIG. 5.
Genome image of a DNA microarray comparison of RNA levels in prototrophic E. coli K-12 strain NCM3722 and MG1655 (CGSC 6300). Strains were grown in LB medium, and expression of the lactose operon was induced in NCM3722 by addition of 100 μM IPTG 25 min before harvest. cDNAs from NCM3722 and MG1655 were labeled with Cy5 (red) or Cy3 (green), respectively, and spots in fluorescence scanning images obtained after hybridization were rearranged in genome order (57, 60). The b number centuries are indicated to the left, and decades are indicated on top. Blanks represent b numbers that do not correspond to open reading frames or no longer exist.
As is apparent from a genome image of the data, the K-12 strain NCM3722 not only showed higher levels of expression of the lac operon (b0341 to b0344) but had strikingly higher mRNA levels for the flagellar and chemotaxis regulon (b1070 to b1083 [flgNMA/BCDEFGHIJ/flgKL], b1878 to b1892 [flhEAB/cheZYBR-tap-tar/cheWA-motBA/flhCD], b1920 to b1926 [fliYZA/fliC/fliDST], and b1937 to b1950 [fliE/fliFGHIJK/fliLMNOPQR]). NCM3722 had ≥2-fold-higher levels of mRNA for most of the genes in this regulon (mean and median differences between the strains of 4.2 and 2.7, respectively, for 50 genes). By contrast, mRNA levels for the galactitol operon gatYZABCDR2 (b2096 to b2090) were lower in NCM3722 than in MG1655. Information and data on these and other differences between the two strains are available at http://nature.berkeley.edu/≈opaliy/papers/MG1655.html.
In most instances, differences in mRNA levels between the K-12 strain and MG1655 were probably due to differences in gene expression (see Discussion for, e.g., the flagellar and chemotaxis regulon and the galactitol operon). However, in some cases the presence of an insertion element may prevent expression in one of the strains or a gene(s) may be deleted (see Discussion). Like MG1655 (CGSC 6300), the K-12 strain NCM3722 showed higher levels of expression of the maltose regulon when grown on lactose than glucose (not shown).
Comparison of strain MG1655 (CGSC 6300) to E. coli K-12 strain NCM3722 on Affymetrix chips.
Comparison was made for cells grown on N−C− medium with glucose and NH4Cl as the C and N sources, respectively, for cells grown with glycerol and NH4Cl as the C and N sources, respectively, and for cells grown with glycerol and arginine as the C and N sources, respectively. In all cases, mRNA levels for genes of the flagellar and chemotaxis regulons were much higher for the K-12 strain than for MG1655. After averaging over the media, the mean and median differences were 54- and 30-fold, respectively, for the 50 genes indicated above. Affymetrix arrays confirmed lower levels of mRNA in the K-12 strain for the galactitol operon. Information and data on these and other differences between the strains are available at http://nature.berkeley.edu/≈opaliy/papers/MG1655.html.
Comparison of strain MG1655 (CGSC 6300) to clinical isolates of E. coli on glass slide DNA microarrays.
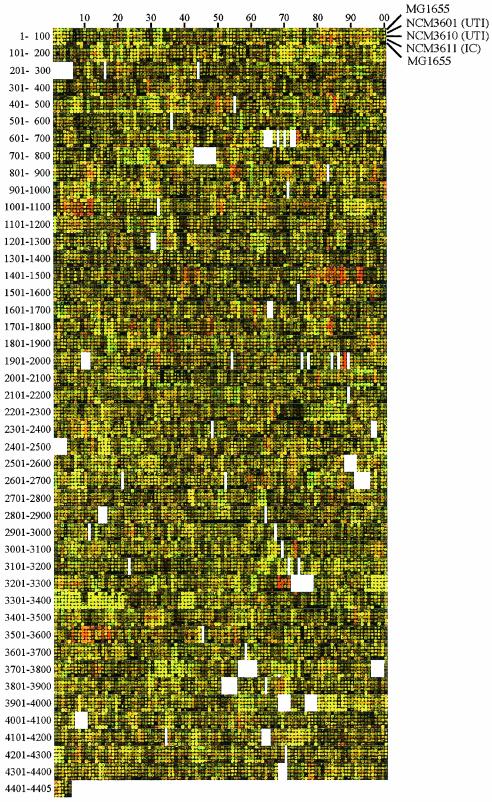
To determine whether use of the MG1655 arrays could be extended to E. coli strains isolated recently from humans and whether such strains would have physiological responses similar to those of laboratory strains, we worked with two urinary tract isolates and an intestinal commensal strain (see Materials and Methods). These had been identified as E. coli based solely on the fact that they were Lac+ on indicator plates and produced indole from tryptophan. We compared mRNA levels for cells grown on glutamine as the sole nitrogen source, a nitrogen-limiting condition, to those for cells grown on ammonium. Differences in gene expression under these two conditions were compared to those for strain MG1655 (CGSC 6300) (Fig. 6).
FIG. 6.
Aligned genome images of DNA microarrays of MG1655 (CGSC 6300) and prototrophic derivatives of clinical isolates of E. coli. The strains were MG1655 (CGSC 6300), rows 1 and 5; derivative of NCM3601 (a urinary tract isolate), row 2; derivative of NCM3610 (a urinary tract isolate), row 3; derivative of NCM3611 (an intestinal commensal isolate), row 4. They were grown in N−C− medium with 0.4% glucose as the carbon source and 5 mM glutamine or 10 mM NH4Cl as the nitrogen source. The cDNAs from cells grown with glutamine or NH4Cl were labeled with Cy5 (red) or Cy3 (green), respectively. Operons thought to be in the extended NtrC/Nac regulon (60) that are more highly expressed on glutamine in all strains include glnK-amtB (b0450 to b0451), glnHPQ (b0809 to b0811), potFGHI (b0854 to b0857), ycdGHIJKLM (b1006 to b1012), ydcSTUVW (b1440 to b1444), ddpXABCDE (b1483 to b1488), yeaGH (b1783 to b1784), yedL (b1932), cbl (b1987), nac (b1988), argT (b2310), ygjG (b3073), and glnALG (b3868 to b3870). See text for discussion of other differences in expression. A different print was used for the experiment in row 5 than for the others.
Examination of aligned genome images and comparison of red-to-green (R/G) ratios indicated that differences in mRNA levels in all four strains under these two conditions were strikingly similar. Specifically, expression of 17 genes thought to be activated by nitrogen regulatory protein C (NtrC) or the nitrogen assimilation control protein (Nac) was higher on glutamine than on ammonium (R/G ratio, ≥2). These genes extended to 13 operons containing 37 genes (see legend to Fig. 6) (60). Sixteen additional genes not under NtrC/Nac control were also more highly expressed on glutamine in all strains (R/G ratio, ≥2). Two that are readily rationalized were the gene for a putative glutaminase (b0485) and that for the glutamine-dependent asparagine synthetase (b0674). Differences in expression of the gadBC operon (b1493 to b1492) were very striking, as were those for operons in the hdeAB region (b3504 to b3517), about which we have commented previously (60). Expression of 14 genes in 11 operons was decreased on glutamine in all four strains (R/G ratio, ≤0.5). These operons code for enzymes of the tricarboxylic acid cycle, carbohydrate transporters, the F1-F0 ATPase, and ribosomal proteins. Decreases in their expression may reflect global sensing of nitrogen-limited slow growth. Further analysis of expression differences between cells grown on glutamine and ammonium and between the urinary tract isolates and MG1655 is available at http://nature.berkeley.edu/≈opaliy/papers/MG1655.html.
Ethanolamine utilization (eut) operon.
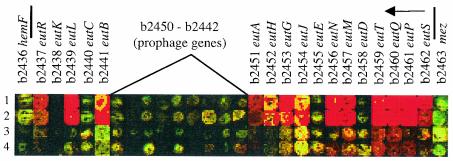
As expected from the sequence of MG1655 (4, 29), which indicated that a cryptic prophage was inserted between two genes whose products are both required to initiate ethanolamine degradation, eutA (b2451) and eutB (b2441), CGSC 6300 failed to grow on ethanolamine as the nitrogen source with glycerol as the carbon source. This was also true of E. coli K-12 strain NCM3722, but both strains readily gave rise to Eut+ derivatives (see Materials and Methods). Comparison of strain NCM3722 to itself when grown on ethanolamine plus ammonium versus ammonium alone showed about twofold increases in expression of several eut genes upstream of the site of the cryptic prophage insertion in MG1655 (eutS-eutN [b2462 to b2456]) but not of genes downstream (Fig. 7, rows 3 and 4) (http://nature.berkeley.edu/≈opaliy/papers/MG1655.html). (Cyanocobalamin was present in both cultures, because utilization of ethanolamine depends on vitamin B12, which is not synthesized aerobically [29].)
FIG. 7.
Portions of genome images showing expression of the eut operon under two conditions. The borders of the eut operon are indicated (lines), along with the direction of transcription (arrow) and the site of insertion of the cryptic prophage in MG1655 (4). For the experiment in rows 1 and 2, NCM3954, a Eut+ derivative of E. coli K-12 strain NCM3722, was grown aerobically on N−C− medium with 0.4% glycerol as the carbon source and 10 mM ethanolamine (Cy5 label) or NH4Cl (Cy3 label) as the nitrogen source. Cultures were supplemented with cyanocobalamin (0.2 μM) in both cases. The images are from two prints present on the same glass slide. Rows 3 and 4, E. coli K-12 strain NCM3722 was grown aerobically on N−C− medium with 5 mM ethanolamine plus 5 mM NH4Cl (indocarbocyanine) or with 10 mM NH4Cl alone (indodicarbocyanine). Cyanocobalamin (0.2 μM) was present in both cases. See text for discussion of individual genes.
The failure of strain NCM3722 to express all genes upstream of the site of the cryptic prophage in MG1655, namely eutM to eutA (b2457 to b2451), indicated that the lesion in NCM3722 might be upstream of that in CGSC 6300. This has not been tested further. Comparison of NCM3954, a Eut+ derivative of NCM3722, to itself when grown on ethanolamine or ammonium as the sole nitrogen source showed 4- to 15-fold increases in expression of eut genes both upstream and downstream of the cryptic prophage in CGSC 6300 (Fig. 7, rows 1 and 2). These findings are in agreement with the view that the lesion (probably an insertion) in strain NCM3722 had excised precisely in NCM3954 and that b2442 to b2450 (black spots in rows 1 and 2 of Fig. 7) code for openreading frames of the cryptic prophage inserted in CGSC 6300. Based on the absence of a spot for eutK (b2438) in rows 1 and 2 of Fig. 7, it is likely that there was a problem with synthesis of the PCR product for this gene. However, it is not clear whether this was also the case for eutD (b2458), eutE (b2455), and eutC (b2440), or whether we failed to see increased expression of these genes for another reasons. Like EutA and EutB, the EutC protein is required to initiate ethanolamine degradation (29). When grown on ethanolamine as the nitrogen source, NCM3954 also showed increased expression of several genes in the NtrC regulon (glnK, ddpXABCDW, yedL, argT, nac, and glnA) and of asnB and ompF (data not shown; http://nature.berkeley.edu/≈opaliy/papers/MG1655.html). Expression of eut genes in NCM3955, a Eut+ derivative of CGSC 6300, was similar to that in NCM3954 (data not shown).
DISCUSSION
Awareness of the growth defects of MG1655 (CGSC 6300) (Fig. 1 to 3 and 7, Tables 2 and 6), some of which can be corrected genetically, is essential to using this strain appropriately for studies of E. coli physiology. Unfortunately, MG1655 (CGSC 6300) has more growth defects than would be anticipated even from its amended genotype (ilvG rfb-50 rph-1 fnr-267 eut) (e.g., a pyrimidine requirement for growth in continuous culture and slow growth on galactose as the sole carbon source) (Table 7). Compounding the problem, isolates called MG1655 in different laboratories and stock centers differ from one another (Fig. 3). To avoid confusion in the E. coli community, it will be essential to specify as clearly as possible which of these isolates is being studied. The isolate used by the Blattner laboratory for sequencing was deposited with the American Type Culture Collection as ATCC 700926 (University of Wisconsin genome project strain RF 75357) and with the CGSC as CGSC 7740. Complementary DNAs from other E. coli strains hybridize well to the MG1655 DNA microarrays (2) (Fig. 5 and 6), and hence using other strains is a good alternative.
TABLE 7.
Genotype and phenotype of MG1655 (CGSC 6300)
| Lesion | Phenotype (reference) |
|---|---|
| ilvG | Growth inhibited by valine (31) |
| rfb-50a | Sensitive to phage P1 |
| rph-1 | Partial pyrimidine requirement |
| fnr-267b | Defects in anaerobic respirations |
| eut | Unable to grow on ethanolamine as the carbon or nitrogen source |
| ? | Slow growth on galactose |
| ? | Slow growth on glycerol (24) |
| ? | Poor motility (32) |
wbbL50::IS5, also called yefJ50 (48).
Δ(ynaJ-yda)267.
Comparison of expression profiles of a prototrophic E. coli K-12 strain used in our laboratory (NCM3722) and MG1655 (CGSC 6300) revealed that NCM3722 had markedly higher levels of mRNA for flagellar and chemotaxis genes. These differences were readily seen in genome images of the data from glass slides (Fig. 5) and were confirmed on Affymetrix arrays (effects were of greater magnitude; see Results). In fact, NCM3722 is motile, whereas MG1655 is only poorly motile (32; G. Prasad and S. Kustu, unpublished data). The role of the LysR-type regulator LrhA in repressing transcription of the flagellar/chemotaxis regulon of MG1655 and restricting its motility has recently been explored (32). We do not know whether NCM3722 carries a lesion in lrhA. The known gatR insertion (gatR::IS3) in MG1655 and another strain in its lineage, W3110 (3), presumably inactivates the Gat repressor and results in high levels of expression of the galactitol operon (40). This may account for higher levels of galactitol mRNA in MG1655 than in NCM3722.
By contrast to the above regulatory differences, differences in mRNA levels for the rfb genes and genes in the adjacent glf-wbbHIJK-yefJ operon (higher in MG1655) may be due to the presence of different rfb insertions and deletions in the two strains (34). We have not tested this possibility further. Finally, detection of the fnr deletion on microarrays was problematic for two reasons. First, detection depends on gene expression. Most of the genes in the fnr region other than fnr itself have not been well studied, and hence it is not known whether they are expressed under the growth conditions we have used. Lack of expression would affect the results on both glass slides and Affymetrix arrays (which did, however, detect differences between NCM3722 and CGSC 6300). Second, since 1999 our PCR products for glass slides have been made by a West Coast consortium (laboratories of N. Cozzarelli, C. Gross, and S. Kustu) from the CGSC isolate of MG1655 (with Sigma-Genosys primers), and we have noted that genes in the fnr region (b1332 to b1344) gave “failed PCR products.” Preliminary inspection indicated that other failed PCR products also tended to occur in clusters and affect genes that coded for open reading frames. We do not know whether any of the other clusters correspond to deletions in CGSC 6300. Oligonucleotides for Affymetrix arrays were designed from the published sequence of MG1655 (Affymetrix, Inc., Santa Clara, Calif.) and hence would include the fnr region.
The cDNAs from two urinary tract isolates and an intestinal commensal strain of E. coli recently isolated from humans hybridized well to MG1655 microarrays. Moreover, the responses of these recent isolates to nitrogen limitation were very similar to those of MG1655 (Fig. 6). The results indicated that studies of the central metabolism and physiology of laboratory strains are likely to be pertinent to pathogenic and commensal strains, too, and that the responses of such strains can also be studied directly on MG1655 microarrays.
One of the growth defects of MG1655 (CGSC 6300) not apparent from its genotype is slow growth on galactose (Table 2). Apparent cross regulation of gene expression between galactose and lactose metabolism—i.e., induction of the lac operon during growth on galactose (59) (Table 3)—and apparent cross-regulation between galactose and N-acetylglucosamine were actually due to the appearance of fast-growing mutants. Some (≈15%) of these galactose-fast-growing mutants had high levels of lac expression (Table 2). Studies with MG1655 derivatives carrying defined lac mutations showed that lacY, which codes for the lactose permease, is the only lac gene required (Table 5). LacY is known to transport galactose (15, 22, 50), and presumably increased galactose transport accounts for the fast growth.
MG1655 derivatives that lacked NagC, the repressor of the N-acetylglucosamine catabolic operons, or had high levels of N-acetylglucosamine 6-phosphate, the coinducer, also grew fast on galactose (Table 6). One such strain was isolated as a fast-growing suppressor. Unlike the case for lactose, none of the products of the nag operons was required for fast growth on galactose, and hence it appears that the absence of NagC or the absence of NagC function per se is what is necessary. NagC is also an activator of the glmUS operon, whose products are needed for biosynthesis of N-acetylglucosamine as a component of the cell envelope (Fig. 4) (41, 46). Decreased biosynthesis may result in increased availability of the precursor of N-acetylglucosamine, fructose 6-phosphate, and hence in increased flux through central carbon metabolism (Fig. 4). This may account for fast growth on galactose. The only other known effect of the absence of NagC is slightly increased expression (≈30%) of the manXYZ operon (42), which codes for components of the phosphotransferase system that have a wide substrate specificity for transport of hexoses. We have not investigated a possible role for ManXYZ in transport of galactose (Fig. 4).
By contrast to the cases above, cross regulation of gene expression between lactose and maltose (Table 3) does not seem to have a trivial explanation (Table 4) and occurred not only in MG1655 (CGSC 6300) but also in the unrelated E. coli K-12 strain NCM3722. Indeed, it has been observed previously (1, 9, 51). It is probably accounted for by endogenous generation of inducing maltosaccharides from glucose and glucose 1-phosphate, both products of lactose catabolism (Fig. 4A). Low-level induction of the maltose regulon in lactose-grown cells may be programmed to occur in E. coli, or at least not selected against, because lactose and maltosaccharides are found together in the mammalian intestine.
Finally, studies of MG1655 in glycerol-limited continuous culture (Fig. 1) provided a dramatic example of the principle that enteric bacteria do not perceive two unrelated nutrient limitations simultaneously (25, 30). As the availability of glycerol was increased by increasing the dilution rate, the cells became internally limited for uracil (pyrimidines) due to their defect in pyrE function. This was reflected in a marked decrease in cell yield and the appearance of suppressor mutations. For work with MG1655 in continuous culture, it is advisable to repair the rph-1 lesion or to supplement the medium with uracil.
Acknowledgments
Wally van Heeswijk and Eric Soupene contributed equally to this work.
We thank W. Boos, G. M. Church, O. Danot, A. Manges, H. Nikaido, L. Riley, and C. Turnbough for strains or plasmids and V. Wendisch and D. Zimmer for help with the initial phases of this project.
This work was supported by Public Health Service grant GM36877 from the National Institute of General Medical Sciences to V.S. and by National Institutes of Health grant GM38361 and a grant from the Torrey Mesa Research Institute, Syngenta Research and Technology, San Diego, Calif., to S.K.
REFERENCES
- 1.Adhya, S., and M. Schwartz. 1971. Phosphoglucomutase mutants of Escherichia coli K-12. J. Bacteriol. 108:621-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachman, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 4.Blattner, F. R., G. Plunkett, 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli with the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1458. [DOI] [PubMed] [Google Scholar]
- 6.Boos, W., and H. Shuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broach, J., C. Neumann, and S. Kustu. 1976. Mutant strains (nit) of Salmonella typhimurium with a pleiotropic defect in nitrogen metabolism. J. Bacteriol. 128:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook, A. L. J. 1962. Recombination studies of lactose nonfermenting mutants of Escherichia coli K-12. Genetics 47:1335-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decker, K., R. Peist, J. Reidl, M. Kossmann, B. Brand, and W. Boos. 1993. Maltose and maltotriose can be formed endogenously in Escherichia coli from glucose and glucose-1-phosphate independently of enzymes of the maltose system. J. Bacteriol. 175:5655-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrmann, M. A., and R. F. Vogel. 1998. Maltose metabolism of Lactobacillus sanfranciscensis: cloning and heterologous expression of the key enzymes, maltose phosphorylase and phosphoglucomutase. FEMS Microbiol. Lett. 169:81-86. [DOI] [PubMed] [Google Scholar]
- 11.Elliott, T. 1992. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J. Bacteriol. 174:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freundlieb, S., and W. Boos. 1986. α-amylase of Escherichia coli, mapping and cloning of the structural gene, malS, and identification of its product as a periplasmic protein. J. Biol. Chem. 261:2946-2953. [PubMed] [Google Scholar]
- 13.Freundlieb, S., U. Ehmann, and W. Boos. 1988. Facilitated diffusion of p-nitrophenyl-α-D-maltohexaoside through the outer membrane of Escherichia coli. Characterization of LamB as a specific and saturable channel for maltooligosaccharides. J. Biol. Chem. 263:314-320. [PubMed] [Google Scholar]
- 14.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278:2092-2097. [DOI] [PubMed] [Google Scholar]
- 15.Ganesan, A. K., and B. Rotman. 1966. Transport systems for galactose and galactosides in Escherichia coli. I. Genetic determination and regulation of the methylgalactoside permease. J. Mol. Biol. 16:42-50. [DOI] [PubMed] [Google Scholar]
- 16.Geanacopoulos, M., and S. Adhya. 1997. Functional characterization of roles of GalR and GalS as regulators of the gal regulon. J. Bacteriol. 179:228-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gustavsson, N., A. A. Diez, and T. Nystrom. 2002. The universal stress protein paralogues of Escherichia coli are coordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 43:107-117. [DOI] [PubMed] [Google Scholar]
- 18.Gutnick, D., J. M. Calvo, T. Klopotowski, and B. N. Ames. 1969. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J. Bacteriol. 100:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guyer, M. S., R. R. Reed, J. A. Steitz, and K. B. Low. 1981. Identification of a sex-factor-affinity site in E. coli as γδ. Cold Spring Harb. Symp. Quant. Biol. 45:135-140. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, L., E. Soupene, A. Ninfa, and S. Kustu. 1998. Physiological role for the GlnK protein of enteric bacteria: relief of NifL inhibition under nitrogen-limiting conditions. J. Bacteriol. 180:6661-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogema, B. M., J. C. Arents, R. Bader, K. Eijkemans, H. Yoshida, H. Takahashi, H. Aiba, and P. W. Postma. 1998. Inducer exclusion in Escherichia coli by non-PTS substrates: the role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Mol. Microbiol. 30:487-498. [DOI] [PubMed] [Google Scholar]
- 23.Hussein, M. J., J. M. Green, and B. P. Nichols. 1998. Characterization of mutations that allow p-aminobenzoyl-glutamate utilization. J. Bacteriol. 180:6260-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibarra, R. U., J. S. Edwards, and B. O. Palsson. 2002. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420:186-189. [DOI] [PubMed] [Google Scholar]
- 25.Ingraham, J. L., O. Maaløe, and F. C. Neidhardt. 1983. Growth of the bacterial cell. Sinauer Associates, Sunderland, Mass.
- 26.Jacob, F., and J. Monod. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3:318-356. [DOI] [PubMed] [Google Scholar]
- 27.Jensen, K. F. 1993. The Escherichia coli K-12 “wild-types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiley, P. J., and H. Beinert. 1998. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341-352. [DOI] [PubMed] [Google Scholar]
- 29.Kofoid, E., C. Rappleye, I. Stojiljkovic, and J. Roth. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubitschek, H. E. 1970. Introduction to research with continuous cultures. Prentice-Hall, Englewood Cliffs, N.J.
- 31.Lawther, R. P., D. H. Calhoun, C. W. Adams, C. A. Hauser, J. Gray, and G. W. Hatfield. 1981. Molecular basis of valine resistance in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 78:922-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehnen, D., C. Blumer, T. Polen, B. Wackwitz, V. F. Wendisch, and G. Unden. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521-532. [DOI] [PubMed] [Google Scholar]
- 33.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, D., and P. R. Reeves. 1994. Escherichia coli K-12 regains its O antigen. Microbiology 140:49-57. [DOI] [PubMed] [Google Scholar]
- 35.Manges, A. R., J. R. Johnson, B. Foxman, T. T. O'Bryan, K. E. Fullerton, and L. W. Riley. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345:1007-1013. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Naas, T., M. Blot, W. M. Fitch, and W. Arber. 1995. Dynamics of IS-related genetic rearrangements in resting Escherichia coli K-12. Mol. Biol. Evol. 12:198-207. [DOI] [PubMed] [Google Scholar]
- 38.Newton, W. A., J. R. Beckwith, D. Zipser, and S. Brenner. 1965. Nonsense mutants and polarity in the lac operon of Escherichia coli. J. Mol. Biol. 14:290-296. [DOI] [PubMed] [Google Scholar]
- 39.Nichols, B. P., O. Shafiq, and V. Meiners. 1998. Sequence of analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J. Bacteriol. 180:6408-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nobelmann, B., and J. W. Lengeler. 1996. Molecular analysis of the gat genes from Escherichia coli and of their roles in galactitol transport and metabolism. J. Bacteriol. 178:6790-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plumbridge, J. 1995. Co-ordinated regulation of amino sugar biosynthesis and degradation: the NagC repressor acts as both an activator and a repressor for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J. 14:3958-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plumbridge, J. 1998. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol. Microbiol. 27:369-380. [DOI] [PubMed] [Google Scholar]
- 43.Plumbridge, J. 1996. How to achieve constitutive expression of a gene within an inducible operon: the example of the nagC gene of Escherichia coli. J. Bacteriol. 178:2629-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plumbridge, J. A. 1992. A dominant mutation in the gene for the Nag repressor of Escherichia coli that renders the nag regulon uninducible. J. Gen. Microbiol. 138:1011-1017. [DOI] [PubMed] [Google Scholar]
- 45.Plumbridge, J. A. 1991. Repression and induction of the nag regulon of Escherichia coli K-12: the roles of nagC and nagA in maintenance of the uninduced state. Mol. Microbiol. 5:2053-2062. [DOI] [PubMed] [Google Scholar]
- 46.Plumbridge, J. A., O. Cochet, J. M. Souza, M. M. Altamirano, M. L. Calcagno, and B. Badet. 1993. Coordinated regulation of amino sugar-synthesizing and -degrading enzymes in Escherichia coli K-12. J. Bacteriol. 175:4951-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raha, M., I. Kawagishi, V. Muller, M. Kihara, and R. M. Macnab. 1992. Escherichia coli produces a cytoplasmic α-amylase, AmyA. J. Bacteriol. 174:6644-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. H. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 49.Reznikoff, W. S., and J. N. Abelson. 1978. The lac promoter, p. 221-243. In J. H. Miller and W. S. Reznikoff (ed.), The operon. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 50.Rosenberg, D., and A. S. Keston. 1969. Loss of “galactose permease” and galactose sensitivity of E. coli associated with mutation in lac operon. Biochem. Biophys. Res. Commun. 37:165-170. [DOI] [PubMed] [Google Scholar]
- 51.Schwartz, M. 1967. Sur l'existence chez Escherichia coli K-12 d'une régulation commune à la biosynthèse des recepteurs du bacteriophage et au metabolisme du maltose. Ann. Inst. Pasteur (Paris) 113:685-704. [PubMed]
- 52.Silhavy, T., M. Berman, and L. Enquist. 1984. Experiments with gene fusions, p. 107-112. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 53.Silhavy, T., M. Berman, and L. Enquist. 1984. Experiments with gene fusions, p. 271. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 54.Soupene, E., H. Lee, and S. Kustu. 2002. Ammonium/methylammonium transport (Amt) proteins facilitate diffusion of NH3 bidirectionally. Proc. Natl. Acad. Sci. USA 99:3926-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart, V. 1982. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J. Bacteriol. 151:1320-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tapio, S., F. Yeh, H. A. Shuman, and W. Boos. 1991. The malZ gene of Escherichia coli, a member of the maltose regulon, encodes a maltodextrin glucosidase. J. Biol. Chem. 266:19450-19458. [PubMed] [Google Scholar]
- 57.Wendisch, V. F., D. P. Zimmer, A. Khodursky, B. Peter, N. Cozzarelli, and S. Kustu. 2001. Isolation of Escherichia coli mRNA and comparison of expression with mRNA and total RNA on DNA microarrays. Anal. Biochem. 290:205-213. [DOI] [PubMed] [Google Scholar]
- 58.White, R. J. 1968. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem. J. 106:847-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, H. C. P., and H. M. Kalckar. 1966. Endogenous induction of the galactose operon in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 55:622-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]