Abstract
Thirteen different serotypes of Listeria monocytogenes can be distinguished on the basis of variation in somatic and flagellar antigens. Although the known virulence genes are present in all serotypes, greater than 90% of human cases of listeriosis are caused by serotypes 1/2a, 1/2b, and 4b and nearly all outbreaks of food-borne listeriosis have been caused by serotype 4b strains. Phylogenetic analysis of these three common clinical serotypes places them into two different lineages, with serotypes 1/2b and 4b belonging to lineage I and 1/2a belonging to lineage II. To begin examining evolution of the genome in these serotypes, DNA microarray analysis was used to identify lineage-specific and serotype-specific differences in genome content. A set of 44 strains representing serotypes 1/2a, 1/2b, and 4b was probed with a shotgun DNA microarray constructed from the serotype 1/2a strain 10403s. Clones spanning 47 different genes in 16 different contiguous segments relative to the lineage II 1/2a genome were found to be absent in all lineage I strains tested (serotype 4b and 1/2b) and an additional nine were altered exclusively in 4b strains. Southern hybridization confirmed that conserved alterations were, in all but two loci, due to absence of the segments from the genome. Genes within these contiguous segments comprise five functional categories, including genes involved in synthesis of cell surface molecules and regulation of virulence gene expression. Phylogenetic reconstruction and examination of compositional bias in the regions of difference are consistent with a model in which the ancestor of the two lineages had the 1/2 somatic serotype and the regions absent in the lineage I genome arose by loss of ancestral sequences.
Listeria monocytogenes is a ubiquitous gram-positive bacterium that can cause life-threatening infections including meningitis, septicemia, abortion, and fetal death. Its primary route of transmission to humans is through contaminated food and despite the relatively low incidence of listeriosis in humans, outbreaks of listeriosis often have high associated morbidity, particularly among pregnant women, unborn fetuses, and immunocompromised individuals (24). These characteristics, coupled with its physiological durability and its ubiquitous distribution in nature have propelled L. monocytogenes to the forefront of food safety research and the regulatory arena.
Pathogenesis of listeriosis is a consequence of the organism's ability to invade and replicate within several different cell types in mammalian tissues, including intestinal, liver, and neural tissues. During the course of food-borne infections, the bacteria penetrate the intestinal lining through cell invasion and translocation and use the lymphatic system as a conduit to reach the main target tissues within the liver and spleen (41). Prolonged replication in the liver, facilitated by depressed cell mediated immunity, is thought to be an antecedent to spread of the bacteria to brain tissue and breach of the placental barrier in pregnant women (56).
Genetic analyses of listeriosis in the mouse model and in tissue culture models have identified two clusters of genes necessary for invasion and intracellular replication. The main cluster of virulence genes is located between the prs and ldh genes on the chromosome. Portions of the cluster can be found at the same genomic position in the species L. ivanovii and L. seeligeri, suggesting that it is ancestral to the genus (9, 13, 36). The cluster encodes a hemolysin (HlyA) and two phospholipases (PlcA and PlcB) that participate in lysis of the phagosome of host cells, an actin polymerizing protein (ActA) that promotes motility within the host cell cytoplasm, a metalloprotease (Mpl) that is required for activation of PlcB, and PrfA, which encodes a transcriptional regulator of the virulence genes (reviewed in references 46, 52, and 56). Internalin (inlA) and the internalin-like inlB gene comprise a second cluster of virulence genes and their products orchestrate the initial stages of attachment and host cell invasion in nonphagocytic cells (16, 21, 23).
L. monocytogenes strains display both genetic and serotypic diversity. Serotypic diversity arises from combinations of somatic and flagellar antigens (51), resulting in thirteen recognized serotypes within the species. Phylogenetic analyses have shown that these serotypes are distributed between two different lineages (1, 3, 7, 26, 27, 44, 47, 57) with serotype 4b, 1/2b, and 3b strains found predominantly in lineage I and serotype 1/2a, 1/2c, and 3c strains found predominantly in lineage II (32, 58). A third lineage, lineage III, was recently proposed due to the genetic distance of serotypes 4a and 4c and the fact that they are rarely isolated from humans (48, 58).
Despite the fact that both virulence gene clusters can be found in the genome of the different L. monocytogenes serotypes, infectivity or transmission rates of the different serotypes appears to be nonrandomly distributed since most cases of human listeriosis (>90%) are attributed to serotypes 1/2a, 1/2b, and 4b (18, 50). Of these, serotype 4b may be the most virulent; it accounts for nearly all of the outbreaks of human food-borne and perinatal listeriosis even though it is not found as commonly in foods or the environment as other serotypes (18, 32, 43, 57). Moreover, serotype 4b strains isolated from outbreaks comprise two genetically uniform subtypes compared to the diversity of serotype 4b strains isolated from the environment, suggesting that subpopulations within the 4b serotype may comprise one or more epidemic clones (54, 61).
Previous population genetic analyses and comparative genotyping studies have suggested that significant genome diversity exists among the different serotypes (1, 3, 7, 26, 27, 44, 47, 54, 57, 58, 61). Comparative genome analyses using subtractive libraries suggest that as much as 5% of the serotype 4b genome is not present in serotype 1/2a strains and that the patterns of diversity are congruent with serotype distribution (30). Recent studies of different serotypes by mixed genome microarray analyses (10) further suggests that lineage and serotype-specific patterns of genome diversity exist and can be used as a tool for rapid diagnostics.
To systematically develop a model for evolution of the genome in the different L. monocytogenes phylogenetic lineages, we have implemented a combination of experimental and computational approaches. Experimentally, shotgun DNA microarrays from representative strains of the different phylogenetic lineages are first used to identify lineage-specific and serotype-specific differences among the genome of lineage I, II and III populations. Subsequent computational analyses are then used to determine which differences arose by gene acquisition versus gene loss. In this report, we used this approach to compare the genomes of the three most common clinical serotypes relative to the serotype 1/2a genome. Our studies identified 16 lineage-specific Regions of Difference (RD) comprising 47 different genes that are conserved among all lineage II serotype 1/2a strains but absent in all lineage I populations tested. The RD encompass five different functional categories, the most represented being cell-surface associated functions. Phylogenetic reconstruction suggests a simple model in which contemporary lineage I and II populations descended from a common ancestor bearing teichoic acids that comprise the 1/2 somatic antigens. Based on this model and compositional bias of genes within the RD, a pathway for genome divergence is presented.
MATERIALS AND METHODS
Strain collection.
A total of 44 L. monocytogenes strains were chosen from a large collection of human clinical, food, and environmental isolates. The set included serotype 1/2a, 1/2b, 4b, and 3b strains along with one strain each of L. innocua, and L. welshimeri. Each of the strains was initially subjected to automated EcoRI ribotyping (32) and serotyping to place them within a phylogenetic lineage. The characteristics of these strains are shown in Table 1.
TABLE 1.
Strains used in this study
| Strain | Origin | Serotype | Comments | Lineage | Ribotype | Source |
|---|---|---|---|---|---|---|
| LM2875 | Food | 1/2b | Sporadic | I | 1042C | M. Samadpour |
| LM2325 | Hot dog | 1/2b | I | 1043 | M. Samadpour | |
| LM775 | Food | 1/2b | I | 1043 | M. Samadpour | |
| LM1325 | Seafood | 1/2b | I | 1052 | M. Samadpour | |
| LM2425 | Park | 1/2b | I | 1052 | M. Samadpour | |
| LM2550 | Chicken | 1/2b | I | 1052 | M. Samadpour | |
| LM700 | 1/2b | I | 1052 | M. Samadpour | ||
| LM1225 | Radishes | 1/2b | I | 1042B | M. Samadpour | |
| LM1916 | Food | 1/2b | I | 1042B | M. Samadpour | |
| LM3200 | Salad | 1/2b | I | 1042C | M. Samadpour | |
| LM225 | Food | 3b | I | 1052 | M. Samadpour | |
| LM2500 | Food | 3b | I | 1052 | M. Samadpour | |
| LM475 | Human | 4b | Outbreak | I | 1044A | M. Samadpour |
| LM1766 | Human | 4b | Sporadic | I | 1044A | M. Samadpour |
| LM1772 | Human | 4b | Sporadic | I | 1044A | M. Samadpour |
| LM468 | Human | 4b | Sporadic | I | 1044B | M. Samadpour |
| LM575 | Food | 4b | I | 1038B | M. Samadpour | |
| LM2075 | Food | 4b | I | 1042B | M. Samadpour | |
| 10403s | Human | 1/2a | Reference strain | II | 1030A | N. Freitag |
| LM2925 | Human | 1/2a | II | 16619 | M. Samadpour | |
| LM3125 | 1/2a | II | 16619 | M. Samadpour | ||
| LM2450 | Beef Frank | 1/2a | II | 1039A | M. Samadpour | |
| LM2850 | Food | 1/2a | II | 1039A | M. Samadpour | |
| LM1475 | Pork sausage | 1/2a | II | 1039C | M. Samadpour | |
| LM2700 | Beef | 1/2a | II | 1039C | M. Samadpour | |
| LM3150 | 1/2a | II | 1039C | M. Samadpour | ||
| LM3175 | 1/2a | II | 1039C | M. Samadpour | ||
| LM975 | Food | 1/2a | II | 1039C | M. Samadpour | |
| LM125 | 1/2a | II | 1053A | M. Samadpour | ||
| LM150 | Frank | 1/2a | II | 1053A | M. Samadpour | |
| LM3025 | Environmental | 1/2a | II | 1053A | M. Samadpour | |
| LM75 | Beef | 1/2a | II | 1053A | M. Samadpour | |
| LM117 | Frank | 1/2a | II | 1053A | M. Samadpour | |
| LM875 | Food | NTa | II | 1045B | M. Samadpour | |
| LM2125b | Clinical | NT | II | 1056A | M. Samadpour | |
| LM115 | Frank | NT | II | 1039C | M. Samadpour | |
| LM2950 | Environmental | NT | II | 1047 | M. Samadpour | |
| LM3225 | Goat | NT | II | 1047 | M. Samadpour | |
| LM950 | Food | NT | II | 1039C | M. Samadpour | |
| LM175 | Human | NT | II | 1039E | M. Samadpour | |
| LM1375 | Seafood | NT | II | 1045A | M. Samadpour | |
| LM50 | Beef | NT | II | 1062A | M. Samadpour | |
| LM130 | Environmental | NT | II | 1062A | M. Samadpour | |
| LM2650 | Squash | 3 | II | 116-741-S-3 | M. Samadpour | |
| LM470 | Human | 4b | Sporadic | III | 1061 | M. Samadpour |
| LM1700 | Food | L. innocua | 1009 | M. Samadpour | ||
| LM275 | Food | L. welshimeri | 1074 | M. Samadpour |
Non-Typeable.
DNA microarray analysis.
Genome diversity was examined with a DNA microarray derived from a shotgun library of the lineage II serotype 1/2a strain 10403s (4). Briefly, 10 μg of chromosomal DNA was nebulized (Invitrogen, Carlsbad, Calif.) and the resulting fragments of average size 1.5 kb were gel-purified and cloned into the pCR4-Blunt-TOPO vector (Invitrogen). A total of 4,000 clones were subsequently picked to independent wells of 96-well plates, and each cloned segment was amplified using T7 and T3 primers. Amplification reaction products were evaluated by agarose gel electrophoresis, resulting in successful amplification from 3,857 clones. The products were ethanol precipitated and the purified products were arrayed in duplicate into four subarrays (44 by 50) using an OmniGrid arrayer (GeneMachines, San Carlos, Calif.). To compare genome diversity, independent 2-μg aliquots of strain 10403s reference DNA and DNA from a given test strain were random primed using Cy-5 and Cy-3 dye-labeled nucleotides, respectively, with BioPrime DNA labeling kits (Life Technologies, Rockville, Md.). Concentrated labeled products from each reference test pair were hybridized in formamide-containing buffer (Array Hyb Low Temp; Sigma, St. Louis, Mo.) for 4 h at 47°C. Slides were washed once each in 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.03% sodium dodecyl sulfate, 0.2× SSC, and finally 0.05× SSC. Fluorescence intensities of the array addresses were determined using a ScanArray4000 multicolor microarray scanner and ScanArray software (Perkin-Elmer, Boston, Mass.).
Data analysis.
Images were analyzed with Imagene software (BioDiscovery, Inc., Marina Del Ray, Calif.). Background subtracted ratios were derived by subtracting median intensities of background pixels from mean pixel intensities of fluorescence signal on each channel. Data from all arrays was composited and transformed into binary elements using a Perl-based program, FormatALL (J. Wise and A. K. Benson, unpublished data). The binary transformation algorithm calculates ratios for each spot, eliminates the higher of the two readings from duplicate spots, log transforms remaining ratios, calculates mean and standard deviation for all remaining ratios from each array, and converts to binary based on user-defined standard deviation cutoffs. For the studies reported here, we converted to binary 1 if <2 standard deviations (SD) from the mean and binary 0 if >2 SD from the mean. Numerical analyses were confirmed by visual inspection of the images.
Cluster analysis and sorting of polymorphisms were performed with the MARKFIND program (J. Wise and A. K. Benson, unpublished data). MARKFIND implements the unweighted pair-group method with arithmetic means (UPGMA) (39) or neighbor joining (49) algorithms for cluster analysis and bootstrap analysis (19) for estimating statistical significance of the branches. The program also uses a novel algorithm for sorting polymorphic characters in the binary strings relative to user-specified groups of taxa (35). The FormatALL and MARKFIND programs as well as the microarray data sets are available upon request.
Mapping RD.
To identify RD distinguishing lineage I and lineage II strains, clones corresponding to relevant array addresses were subjected to DNA sequence analysis. Cycle sequencing was performed with labeled T3 and T7 primers and the reaction products were run on Li-Cor 4200 automated DNA sequencers. Sequences were aligned into contiguous segments using Sequencher software (Gene Codes, Inc., Ann Arbor, Mich.) and the contiguous segments were mapped onto the lineage II serotype 1/2a strain EGD genome (25) by BLAST search. The genetic basis for altered intensity ratios was determined by Southern blots using the sequenced segments from the appropriate clones to probe the entire strain set. DNA from each strain was digested with EcoRI, resolved by agarose gel electrophoresis, transferred to nylon membranes, and probed with purified fragments from the relevant clones that had been random primed with [32]P-labeled dATP. Blots were hybridized at 65°C in 0.5 M sodium phosphate (pH 7.4)-1% bovine serum albumin-7% sodium dodecyl sulfate and washed at 65°C in 40 mM sodium phosphate before autoradiography.
Compositional bias calculations.
To estimate whether RD were acquired or derived states, compositional bias was compared by two different methods. First, genome signature (δ*) comparisons, calculated from dinucleotide frequencies, were determined as described by Karlin (34). Briefly, segments of the EGD genome in 40-kb intervals were compared to the whole EGD genome using the δ* difference as a vector of the relative dinucleotide abundance frequency: δ* = 1/16 ∑ |ρXY(f) − ρXY(g)|, where ρXY(f) is the relative abundance of dinucleotide XY for genome segment f and ρXY(g) is the relative abundance of dinucleotide XY for the entire genome. δ* values greater than two standard deviations from the mean were interpreted as potentially alien sequences along the genome.
To measure compositional bias of individual genes, codon bias was compared using the method of Ma et al. (40), which compares the relative codon bias (B) of gene (f) to the entire genome (g) or subsets of specific genes: B(f|g) = ∑ Pa(f) [∑ |f(x,y,z) − g(x,y,z)|]. For codon x,y,z, the average frequency of that codon in gene f and gene set g is calculated and normalized such that for a given amino acid, ∑(x,y,z) = 1. Thus, the term Pa(f) is the average frequency of codon x,y,z for gene f − the average frequency of codon x,y,z in gene set g. For the bias (B) of f to be considered significantly different from g, median bias (M) was calculated for different sizes of genes in sets f and g, with a separate M calculated for each of five bins based on the number of codons in a gene: 50 to 150, 100 to 200, 200 to 300, 300 to 500, or 500+. These thresholds for significance are defined for the lengths 100, 150, 250, 400, and 600 codons as the M value of B(f|g) plus 0.10, and linearly interpolated between these points when applying the appropriate threshold value (S. Karlin and J. Mrazek, personal communication). For lengths >600 codons, the threshold remains constant. Linear extrapolation is used in the range 80 to 100 codons. Genes <80 codons long were excluded from the analysis. For a gene to be considered putatively alien (PA), gene f must fulfill the criteria B(f|C) ≥ M + 0.10, B(f|RP) ≥ M + 0.10, B(f|TF) ≥ M + 0.10, and B(f|CH) ≥ M + 0.10, where C is the entire genome, RP is the set of ribosome proteins, TF is the set of translation factors, and CH is the set of chaperones (40). To lower the threshold and potentially capture genes that are questionably alien, we used the minimal criteria that B(f|C) ≥ M + 0.10 and B(f|RP) ≥ M + 0.10.
RESULTS
DNA microarray analysis of L. monocytogenes serotypes 1/2a, 1/2b, and 4b populations.
To examine evolution of the genome in the three most common clinical L. monocytogenes serotypes, we used shotgun DNA microarray analysis to test diversity among multiple strains of 1/2a, 1/2b, 4b, and 3b serotypes. The use of multiple strains from each lineage allowed us to discriminate phylogenetically relevant patterns of genome variation (e.g., lineage and serotype specific) from strain-level and other patterns of genome variation. Genotypic analysis of the strain set by ribotyping (Table 1) showed that the strains comprise 19 different ribotypes, with the ribotypes comprising five of the nine known ribotype groups described by Jeffers et al. (32).
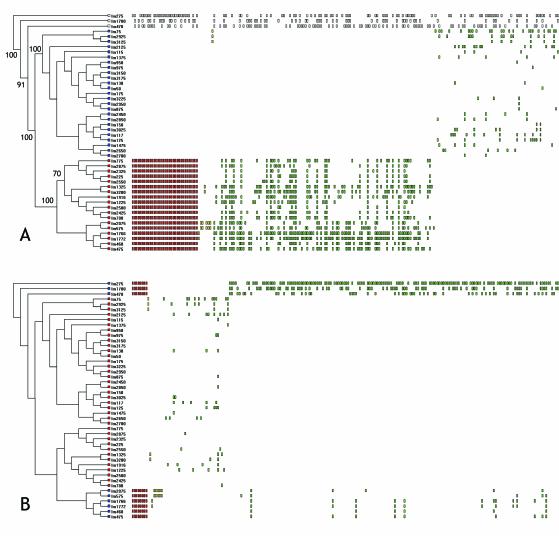
The array was fabricated from a shotgun clone library of the well studied serotype 1/2a strain 10403s (4). Hybridization of total genomic DNA from each of the L. monocytogenes strains and the L. welshimeri and L. innocua strains yielded a total of 730/3857 addresses that showed altered hybridization ratios from at least one strain. Cluster analysis of the binary-transformed fluorescence intensity ratios by UPGMA yielded a dendrogram shown in Fig. 1A. Bootstrap analysis demonstrated that the L. welshimeri, L. innocua, and lineage III L. monocytogenes strains were statistically distinct from the other L. monocytogenes strains. Lineage I serotype 4b, 1/2b, and 3b strains formed a statistically significant cluster clearly distinguished from lineage II serotype 1/2a strains. The lineage I strains were split into two subgroups that were statistically significant and congruent with serotype distribution; one cluster comprised serotype 1/2b and 3b strains while the other cluster comprised serotype 4b strains. Thus, cluster analysis of microarray data is in agreement with the clustering of data from other genotyping methods such as ribotyping and pulsed-field gel electrophoresis (PFGE) analysis, indicating that the phylogenies inferred from both low and high-resolution methods and from whole-genome and multilocus methods are concordant.
FIG. 1.
Analysis of genome content by reference microarray. (A)Dendrogram derived from UPGMA analysis of binary data from the L. monocytogenes serotype 1/2a array. UPGMA was performed on binary data generated from 44 strains of Listeria monocytogenes and one strain each of L. welshimeri and L. innocua. Tree length, using 730 polymorphic characters, was 1,941 steps with a consistency index of 0.3509 and homoplasy index of 0.6491. Bootstrap scores, using 1,000 repetitions of a UPGMA search, are shown on nodes scoring > 70%. Leaves of the tree from lineage I strains are colored red, those from lineage II are colored blue, and those from lineage III or other Listeria species are uncolored. Rectangles to the right of the strain names depict polymorphisms present in a given strain sorted by the MARKFIND program. Those polymorphisms conserved within all members of a lineage are colored red, those that are lineage-specific but nonconserved are colored green (polyphyletic) or yellow (monophyletic). The 489 polymorphisms distributed among strains in both lineages are not shown. (B) The same data as those in panel A were analyzed to identify polymorphisms exclusive to and conserved within serotype 4b L. monocytogenes strains and L. welshimeri and L. innocua. Red rectangles depict altered loci in serotype 4b strains and L. welshimeri and L. innocua, green rectangles indicate those polymorphisms shared among polyphyletic groups of strains within the serotype 4b cluster and among the serotype 4b and 1/2a clusters.
To identify lineage and serotype-specific genome differences, the polymorphisms were sorted into four classes based on their distribution relative to lineage I and lineage II strains. The class I array addresses comprised 34 of the 730 polymorphic array addresses and were consistently altered in all lineage I strains (Fig. 1A, red rectangles). The sorting algorithm also identified 207 polymorphic addresses that are lineage-specific but not consistently altered in all strains within a lineage (yellow and green rectangles in Fig. 1A). The remaining 489 polymorphic addresses were distributed among polyphyletic groups of strains in both lineages (not shown). When the data was sorted by lineage then serotype, we identified eight addresses that were present in all serotype 1/2a, 1/2b, and 3b strains but were absent in all serotype 4b and L. innocua and L. welshimeri strains (Fig. 1B, red rectangles).
Identification of lineage-specific and serotype-specific genome alterations.
To identify the lineage-specific and serotype-specific Regions of genome Difference (RD), 49 clones detecting lineage-specific hybridization patterns (34 that hybridized exclusively to all lineage II strains and 15 that hybridized exclusively to all but one lineage II strain) and 8 clones detecting serotype-specific patterns (hybridizing to all serotype 1/2a, 1/2b, and 3b strains) were subjected to DNA sequence analysis.
DNA sequences from the 57 clones formed 19 different contiguous segments, 10 of which comprised two or more overlapping clones. These contigs are referred to as regions of difference (RD). BLAST alignments of the RD against the serotype 1/2a genome sequence (25) showed that 16 of the RD aligned with the 1/2a genome sequence and comprise 47 different coding regions (Table 2) while the three remaining RD contained unique sequences not found in the strain EGD genome. BLAST analysis of the unique contiguous segments against the nonredundant database showed that RD17 had similarity to a segment of the A118 prophage of L. monocytogenes and L. innocua while the other two showed no significant similarity to GenBank entries.
TABLE 2.
Genes within RD conserved among serotype 1/2a strains
| RD | Gene (function) | Southern blot results froma:
|
BLAST result with unfinished 4bb | ||
|---|---|---|---|---|---|
| 1/2a strains | 1/2b strains | 4b strains | |||
| RD1 | Lmo0160 (putative LPXTG) | Present in most | Absent | Absent | None |
| RD1 | Lmo0161 (unknown) | Present in most | Absent | Absent | None |
| RD2 | Lmo0170 (unknown) | Present | Absent | Absent | None |
| RD2 | Lmo0171 (putative internalin LPXTG) | Present | Absent | Absent | None |
| RD3 | Lmo0409 (putative internalin LPXTG) | Present | Absent | Absent | None |
| RD4 | Lmo0421 (RodA paralog) | Present | Absent | Absent | None |
| RD5 | Lmo0525 (unknown) | Present in most | Absent | Absent | None |
| RD6 | Lmo0732 (putative LPXTG) | Present | Absent | Absent | None |
| RD6 | Lmo0736 (ribose 5-phosphate isomerase) | Present | Absent | Absent | None |
| RD7 | Lmo1060 (response regulator) | Present | Absent | Absent | None |
| RD7 | Lmo1061 (histidine kinase) | Present | Absent | Absent | None |
| RD7 | Lmo1062 (ABC permease) | Present | Absent | Absent | None |
| RD7 | Lmo1063 (similar to ABC [ATP binding]) | Present | Absent | Absent | None |
| RD7 | Lmo1064 (similar to membrane and transport) | Present | Absent | Absent | None |
| RD8 | Lmo1077 (similar to TagB) | RFLP I and II | RFLP I | RFLP III | None |
| RD8 | Lmo1078 (similar to sugar nucleotide transferase) | RFLP I and II | RFLP I and II | RFLP III | Present |
| RD8 | Lmo1079 (similar to YfhO) | RFLP I and II | RFLP I and II | Absent | None |
| RD8 | Lmo1080 (similar to teichoic acid biosynthetic protein GgaB) | RFLP I and II | RFLP I and II | Present | Present |
| RD8 | Lmo1081 (similar to sugar nucleotide transferase) | RFLP I and II | RFLP I and II | Absent | None |
| RD8 | Lmo1082 (similar to sugar nucleotide epimerase) | RFLP I and II | RFLP I and II | Absent | None |
| RD8 | Lmo1083 (similar to sugar nucleotide dehydratase) | RFLP I and II | RFLP I and II | Absent | None |
| RD8 | Lmo1084 (similar to sugar nucleotide reductase) | RFLP I and II | RFLP I and II | Absent | None |
| RD8 | Lmo1085 (similar to TagB) | RFLP I | RFLP I | RFLP II | None |
| RD9 | Lmo1258 (unknown) | Present in most | Absent | Absent | Present |
| RD9 | Lmo1259 (ProA) | Present in most | Absent | Absent | Present |
| RD9 | Lmo1260 (ProB) | Present in most | Absent | Absent | Present |
| RD10 | Lmo1913 (unknown) | RFLP I | RFLP I | RFLP II | Present |
| RD10 | Lmo1914 (unknown) | RFLP I | RFLP I | RFLP II | Present |
| RD10 | Lmo1915 (Malate dehydrogenase) | RFLP I | RFLP I | RFLP II | Present |
| RD11 | Lmo1967 (similar to tellurite resistance) | Present | Absent | Absent | None |
| RD11 | Lmo1968 (creatine aminohydrolase) | Present | Absent | Absent | None |
| RD11 | Lmo1969 (2-keto-3-deoxygluconate aldolase) | Present | Absent | Absent | None |
| RD11 | Lmo1970 (putative phosphotriesterase) | Present | Absent | Absent | None |
| RD11 | Lmo1971 (similar to PTS EnzIIC) | Present | Absent | Absent | None |
| RD11 | Lmo1972 (similar to PTS EnzIIB) | Present | Absent | Absent | None |
| RD11 | Lmo1973 (similar to PTS EnzIIA) | Present | Absent | Absent | None |
| RD11 | Lmo1974 (similar to GntR regulator) | Present | Absent | Absent | None |
| RD12 | Lmo2176 (similar to TetR) | Present | Absent | Absent | Present |
| RD12 | Lmo2177 (unknown) | Present | Absent | Absent | Insertion-deletion |
| RD12 | Lmo2178 (LPXTG) | Present | Absent | Absent | Insertion-deletion |
| RD13 | Lmo2550 (similar to glycosyl transferase) | Present | Present | Absent | None |
| RD13 | Lmo2551 (Rho) | Present | Present | Absent | Present |
| RD14 | Lmo2576 (LPXTG) | Present | Absent | Absent | None |
| RD15 | Lmo2603 (unknown) | Present | Absent | Absent | None |
| RD15 | Lmo2604 (unknown) | Present | Absent | Absent | None |
| RD16 | Lmo2786 (BvrC) | Present | Absent | Absent | None |
| RD16 | Lmo2787 (BvrB) | Present | Absent | Absent | None |
| RD16 | Lmo2788 (BvrA) | Present | Absent | Absent | None |
| RD17 | Similar to A118 prophage | Present | Absent | Absent | None |
| RD18 | Unknown | Present | Absent | Absent | None |
| RD19 | Unknown | Present | Absent | Absent | None |
Present, present in all; absent, absent in all; RFLP I or II, RFLP pattern I or pattern II, respectively.
Results from BLAST analysis (BLASTn) with the unfinished serotype 4b sequence (http://www.tigr.org). None, no significant alignment; insertion-deletion, insertion and deletion in the same region relative to EGD.
Southern blot analysis using the corresponding amplified segments from the shotgun library as probes showed that most probes from the 19 RD hybridized to all 1/2a strains but no 1/2b or 4b strains (Table 2). The exceptions were RD 8 and 10, where 1/2a and 1/2b strains shared identical restriction fragment length polymorphism patterns, and RD 13, where only 4b and L. innocua and L. welshimeri strains failed to hybridize. Thus, the majority of RDs detected by the microarray were due to differences in genome content or significant divergence between the lineages and the serotypes. BLAST analysis of the RD against the serotype 4b strain (http://www.tigr.org) yielded significant alignments only in RD8-RD10, RD12, and RD13, indicating that most are missing in the serotype 4b genome sequence or remain in unfilled sequencing gaps.
The largest class of genes within the RD is associated with the cell surface.
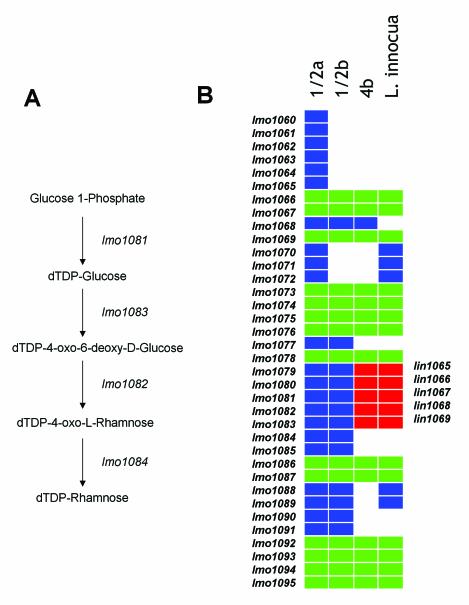
The 47 identifiable genes within the RD fell into six different functional categories, including cell wall-cell surface, small molecule transport, central metabolism, transcription regulation, and stress resistance. The largest functional category of genes included cell wall-surface associated genes. This category contains genes that are known or putatively associated with teichoic acid biosynthesis, genes encoding wall anchored proteins, and genes associated with wall biosynthesis. Teichoic acids are a major determinant of the somatic antigens in L. monocytogenes strains (20, 22, 38, 45, 55). One of the serotype 4b-specific RDs (RD13) encodes the gtcA gene that is necessary for galactose decoration of the teichoic acid of serotype 4 strains (45). The segment from the microarray spans the gctA and lmo2550 genes and comparison of the serotype 1/2a EGD genome sequence and the serotype 4b genome sequence in this region shows that the gtcA allele in EGD is quite divergent from the 4b allele and the adjacent gene, lmo2550, is not present. A second serotype-specific RD (RD8) includes a large segment spanning the region from lmo1077-lmo1088 relative to the EGD genome. Using the KEGG database (http://www.genome.ad.jp/kegg/), analysis of genes from the serotype 1/2a genome sequence within this region showed that four of the genes show significant similarity to a four gene biosynthetic pathway for dTDP-rhamnose (Fig. 2A). Since serotype 1/2a and 1/2b teichoic acids are decorated with rhamnose residues (55), we propose that this region encodes serotype-specific biosynthetic components for an activated sugar-nucleotide precursor. Consistent with the pattern of polymorphism determined by our microarray data, comparison of the 4b genome sequence in this region to the serotype 1/2a strain EGD genome sequence (Fig. 2B) demonstrates several differences between serotype 4b and 1/2a. In fact, the serotype 4b sequence in this region contains several genes that are closely related to L. innocua, notably the lin1065-lin1069 genes, suggesting that they are orthologous.
FIG. 2.
Polymorphism in putative teichoic acid biosynthetic genes between serotype 1/2 and 4b populations. (A) The putative four-gene biosynthetic pathway for dTDP-l-rhamnose encoded by lmo1081-lmo1084 is shown. (B) The map shows the alignment of genes in the region relative to the L. monocytogenes serotype 1/2a, 4b, and L. innocua genome sequences and the inferred presence and absence of genes in serotype 1/2b relative to the microarray data. The rectangles represent individual genes. White space indicates absence of a gene at that relative position in the genome and the colored squares indicate orthologous genes at the respective positions. Gene indices relative to the L. monocytogenes EGD genome are indicated to the left of the illustration (lmo1060-lmo1094) and genes from the L. innocua genome (lin1065-lin1069) are on the right. Genes shared by L. monocytogenes serotype 4b and L. innocua are in red.
In addition to serotypic determinants, at least five additional RD (RD1, RD2, RD3, RD6, and RD16) encode genes whose products are predicted to be displayed on the cell surface, based on the presence of the LPXTG wall-anchoring motif (lmo0160, lmo0171, lmo0409, lmo0732, and lmo2576). Three of these genes (lmo171, lmo0732, and lmo0409) encode proteins with leucine-rich repeats characteristic of the internalin family of proteins. The prototype members of this family, inlA and inlB, are known to mediate host cell invasion in nonphagocytic cells, however the role of additional internalin-like molecules, identified from molecular genetic and genome sequence analysis, are not known (16, 17, 23, 25). Collectively, the fact that these wall-anchored proteins are absent or significantly divergent between the two lineages and the diversity of putative functions associated with them implies that cell surface differences between the lineages may confer unique abilities of the bacteria to interact with host cells and the environment.
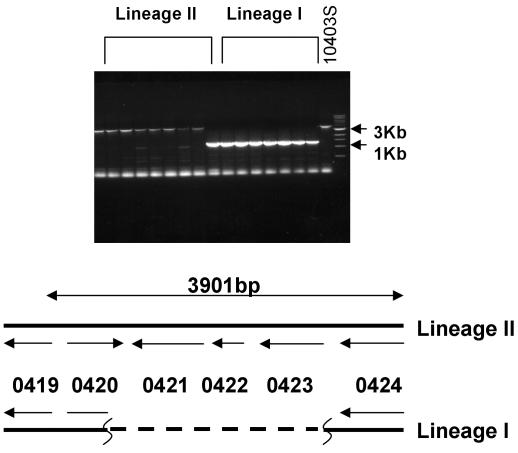
A third type of cell-surface related RD is represented by RD 4, which encodes a putative paralogue of the morphogenetic RodA-FtsW family of proteins. The RodA and FtsW proteins are essential cell division and cell shape proteins in E. coli and B. subtilis that direct elongation (RodA) and septation (FtsW) phases of cell wall growth and division (2, 5, 8, 11, 12, 15, 29, 42, 59, 60). Apparently the lmo0421 gene is not essential in lineage I strains; indeed, there are at least five other genes in the strain EGD genome that show similarity to the RodA-FtsW family. The apparent functional redundancy could reflect specialization of the rodA/ftsW paralogues under different environmental conditions. This hypothesis is supported by the positioning of lmo0421, which is located as the terminal gene of a putative three-gene operon containing lmo0422 and lmo0423. These additional genes encode an unknown protein and a putative sigma subunit of RNA polymerase, respectively. The Lmo0423 protein is similar to the extracytoplasmic function family of sigma factors that typically respond to stimuli that are external to the cell and they often direct expression of genes whose function is external to the cell (28). It is therefore tempting to speculate that organization and function of these genes may comprise a device to couple gene expression to some aspect of cell wall biosynthesis in lineage II strains. PCR analysis using primers within the flanking lmo0419 and lmo0424 genes yielded an expected 3.9-kb product from all serotype 1/2a strains but only a 1.3-kb product from serotype 1/2b and 4b strains (Fig. 3). DNA sequence analysis of the PCR product from serotype 1/2b and 4b strains shows that lmo0421, lmo0422, and lmo0423 are all absent from lineage I strains.
FIG. 3.
PCR analysis of the lmo0421-lmo0423 region. Agarose gel electrophoresis of PCR amplification products derived from PCR amplification of the lmo0420-lmo0424 region. Products from lineage I and lineage II strains are indicated. The control strain, lineage II strain 10403s, is indicated on the right end of the gel along with the 3- and 1-kb markers from the size standards. The map underneath depicts the region in lineage II and lineage I strains. The lineage I map is based on sequence analysis of PCR amplification products from two independent lineage I strains.
Small-molecule transport.
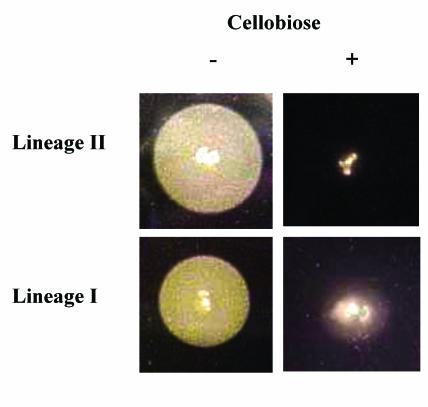
Three different RD (RD7, RD11, and RD16) encode proteins similar to transporter systems, including two that are similar to ABC-type transporters (RD7 and RD11) and a third encoding a novel PEP-dependent β-glucoside transport system (RD16). While function of the ABC-like transport systems is unknown, the PEP-dependent β-glucoside transport system encoded by RD 16 has previously been characterized in L. monocytogenes. This locus encodes the bvrABC system that couples β-glucoside transport to catabolite repression of the PrfA-dependent virulence genes (6). A recent study by Call et al. (10) also showed this region missing in lineage I strains and we therefore tested whether the characteristic of cellobiose-mediated repression of PrfA activity is lineage-specific. As shown in Fig. 4 and Table 3, the PrfA-dependent Phospholipase B (PlcB) activity was cellobiose repressed in strains from both lineages, suggesting that cellobiose-mediated repression is either polygenic in wild-type strains or it has evolved through independent systems in lineage I and II strains. In support of the former, Huillett et al. (31) have shown that additional loci contribute to cellobiose-mediated repression in the lineage II serotype 1/2c strain LO28. Whether polygenic or independently evolved, downregulation of virulence genes in the presence of β-glucosides, which are found principally in plant tissues, appears to be an important characteristic in L. monocytogenes.
FIG. 4.
Phospholipase activity of L. monocytogenes strains. Strains from lineage I and lineage II were spotted onto egg yolk agar to measure phospholipase activity. Colonies on the left hand panels were grown on agar with no cellobiose and those on the right hand panel were grown on agar with cellobiose. The opaque zones around the colonies grown on plates without cellobiose are derived from the activity of the PrfA-dependent phospholipase B.
TABLE 3.
Results for phospholipase plate assay
| Phospholipase activitya for strains grown:
|
||
|---|---|---|
| Without cellobiose | With cellobiose | |
| Lineage I | ||
| FSL W3-011 | ++ | − |
| FSL W3-018 | + | − |
| FSL W3-021 | + | − |
| FSL W3-026 | ++ | − |
| FSL W3-028 | ++ | − |
| FSL W3-040 | ++ | − |
| FSL W3-062 | + | − |
| Lineage II | ||
| FSL W3-007 | ++ | − |
| FSL W3-008 | ++ | − |
| FSL W3-031 | + | − |
| FSL W3-042 | + | − |
| FSL W3-045 | ++ | − |
| FSL W3-052 | + | − |
| FSL W3-053 | +/− | − |
Phospholipase activity was determined as the diameter of the zone of clearance on egg yolk plates with and without cellobiose added. Symbols: ++, average diameter, >1.0 cm; +, average diameter, 0.5 to 1.0 cm; +/−, average diameter, <0.5 cm; −, no zone of clearance. Each result represents at least two independent replicates.
Transcriptional regulation.
In addition to lmo0423 described above four other putative regulatory genes were absent in lineage I genomes. These are encoded within RD7, RD11, and RD12. The adjacent lmo1060 and lmo1061 genes within RD 7 are highly similar to two component system histidine protein kinases and response regulators, respectively. Their coding regions overlap by 25 bases and the pair of genes is flanked by putative factor-independent terminator sequences suggesting that they comprise a translationally coupled two-gene operon. RD11 and RD12 include members of the GntR (lmo1974) and TetR (lmo2176) families, respectively. Although function of any of these putative transcription factors is not known, their absence in lineage I strains would imply that significant differences exist in the patterns of gene expression between the two lineages.
Stress resistance.
Several genes absent from lineage I strains encode known or putative genes associated with stress resistance. Of note is a proline biosynthetic pathway encoded by lmo1259-lmo1260 (proAB), the lmo0421-lmo0423 region described above, and a putative tellurite resistance determinant encoded by lmo1967. The proline biosynthetic pathway encoded by the proAB genes catalyzes de novo synthesis of proline from intracellular pools of glutamate via gammaglutamyl kinase (proA) and gammaglutamyl phosphate reductase (proB). In the serotype 1/2c laboratory strain LO28, this pathway participates in osmotic protection in the absence of preferred osmoprotectants betaine and carnitine (53). Although both the microarray and southern blot data indicate that the proAB genes are absent in all lineage I strains that we tested, four different restriction fragment length polymorphism patterns were observed among the lineage II strains and three lineage II strains were missing the genes. Moreover, in contrast to genes in the other RD, the lineage-specific proAB genes are present in the unfinished genome sequence of the serotype 4b strain in the same relative position as the EGD genome sequence. These observations imply that this region is relatively labile and has been subject to deletion or alteration on multiple occasions during divergence of lineage I and lineage II strains. Analysis of the DNA sequence surrounding the region revealed no unusual compositional features that might facilitate loss of this segment.
Use of codon usage analysis and compositional bias to predict gene acquisition or loss.
There are two simple explanations for the origin of conserved lineage-specific and serotype-specific differences in genome content. First, genes within the RD may have been present in the most recent common ancestor of the two lineages, and subsequently lost during divergence of lineage I. Alternatively, the RD could have been acquired in lineage II. Of course, it is also possible that some may have been acquired and some lost. Assuming divergence of the two lineages was not recent, comparison of compositional bias between the RD and the rest of the genome can provide a means for inferring whether genes within the RD are recent additions to lineage II or ancestral sequences lost in lineage I. To this end, we measured compositional bias by two independent statistics, dinucleotide frequency and codon usage.
Dinucleotide genome signatures, measured as the δ* vector of the 16 dinucleotide frequencies, have been used to successfully identify and characterize islands, particularly pathogenicity islands (34). When the δ* values in 40 kb are calculated for the L. monocytogenes strain EGD genome sequence, three regions show atypical patterns (Table 4). These putative alien (PA) regions correspond to nucleotides 720,001 to 760,000, encoding a contingent of flagellar genes, nucleotides 1,120,001 to 1,160,000 encoding a remnant of a conjugal transposon, and nucleotides 2,360,001 to 2,400,000 encoding a prophage-like segment. None of the RD lie within the PA regions, suggesting that they have ancestral dinucleotide signatures. Although RD8—which includes the highly polymorphic segment lmo1077-lmo1085 encoding a putative pathway for TDP-rhamnose synthesis—is adjacent to the PA interval at nucleotides 1,120,000 to 1,160,000 (lmo1086-lmo1124), the fact that its dinucleotide signature is not significantly different from the genome and the fact that the region is conserved in the divergent 1/2a and 1/2b serotypes supports the hypothesis that the lmo1076-lmo1085 region is ancestral and the transposon is a recent acquisition in a portion of the 1/2a lineage.
TABLE 4.
Overlapping regions of deviant compositional bias in the L. monocytogenes genome
| Significant δ* intervala | PA genesb |
|---|---|
| 720,001-760,000 | lmo0711, lmo0713, lmo0721, lmo0722, lmo0723 |
| 1,120,001-1,160,000 | lmo1110, lmo1113, lmo1124 |
| 2,360,001-2,400,000 | lmo2279, lmo2302, lmo2319, lmo2327 |
Nucleotide coordinates relative to the strain EGD genome sequence (33) of δ* intervals having values more than 2 SD from the mean.
Putative alien genes (based on codon bias) within each of significant the δ* intervals.
As a second measure of foreignness, we also used codon usage statistics to determine if any genes within the RD met the criteria as PA. As demonstrated by Karlin and Mrazek (33), codon usage can appear atypical if genes are putatively highly expressed (PHX) or if they are PA. When the normalized codon usage frequencies for each gene in the L. monocytogenes genome were compared to the entire genome and the ribosome proteins, 120 different genes met the minimal criteria as PA. Of the 120, 12 fell within the three PA segments that had unique dinucleotide signatures (Table 4), indicating that the two independent methods identified a common subset of genes as PA. However, like the dinucleotide signatures, no genes within any of the RD met the minimal PA criteria based on codon bias. Since neither of these independent measures of compositional bias provide any evidence for recent acquisition of genes within the RD, we conclude that these genes are likely ancestral to the genus and that the RD arose by loss of these ancestral sequences during divergence of the lineage I populations.
DISCUSSION
Comparative genome analyses of L. monocytogenes serotypes using various types of fragment analyses, multilocus sequencing, and genome content analyses consistently divide the three most common serotypes among two main phylogenetic lineages (1, 3, 7, 26, 27, 44, 47, 57). A recent study by Call et al. (10) also revealed that low-density DNA microarrays derived from multiple serotypes could resolve lineage-specific and serotype-specific genome segments and provide a simple method for genotyping and serotyping. Our study, aimed at understanding evolution of the genome in the phylogenetic lineages, demonstrates substantial lineage-specific differences in genome content. Although phylogenetic analysis of data from microarray based studies and other methods of genome comparison yield concordant results, our data sorting algorithm demonstrates that monophyletic signal with regard to genome content is by far a minor proportion of the polymorphic loci that were detected (Fig. 1). In fact, over half of the polymorphic signals were distributed among polyphyletic groups. This observation supports the hypothesis that relative to the rate of genome alteration, periodic selection or clonal expansion only rarely fixes genome events in L. monocytogenes populations.
Despite evidence for only rare periodic selection, the potential for genome diversification may be significant in this organism. This characteristic was recently reported from studies of strain variation among isolates derived from independent patients of a single listeriosis outbreak (14). The authors reported variation in cell surface antigens and suggested that the variation could be the consequence of selection imposed by the immune system. In testing independent isolates from a single outbreak, we have also found that variation in genome content can be detected using our microarray. In these experiments, we probed 11 strains isolated from independent patients of a multistate outbreak that occurred in 1998. Each of the isolates was epidemiologically linked to the same contaminated food source and had been passed no more than 8 times during isolation from clinical samples, transfer to state public health laboratories, and transfer to a single laboratory for molecular typing. Each of the 11 strains shared an identical ribotype and shared a common PFGE pattern with only two strains differing by a single band. The pattern of polymorphism detected in the microarray demonstrated that despite the similarity by PFGE, each could be distinguished (Fig. 5). In fact, 158 different polymorphisms could be detected that distinguished the strains from one another, 76 of which were shared by more than half of the strains, indicating that some of the polymorphic events were commonly occurring. Southern blot data from five randomly chosen polymorphic loci detected among the outbreak strains showed that they arose from deletions (J. Nietfeldt and A. K. Benson, unpublished). Assuming that the outbreaks studied by us here and by Clark et al. (14) did not arise from food contaminated by multiple populations, these data support a view that the L. monocytogenes genome may have a propensity to undergo gene deletion under selective conditions associated with passage through individual human hosts or during laboratory cultivation from clinical samples. It will be interesting to determine if a common set of loci are affected and whether this is a general characteristic that occurs during isolation of L. monocytogenes or passage through human hosts.
FIG. 5.
Analysis of genome content among strains derived from a single outbreak. DNA microarray analysis was performed on 11 different L. monocytogenes strains that were isolated from independent patients of a single outbreak. Each strain shared identical PFGE profiles and ribotypes. The dendrogram was rendered by UPGMA analysis and the data sorted based on the two groups (blue and red leaves) using the MARKFIND program.
Though the data from the two lineages and from outbreak strains seem to support the potential for rapid diversification of the genome, there is little evidence for a common underlying mechanism. Among the 19 different RD that we identified, only two (RD2 and RD8) are adjacent to elements that could promote gene loss or rearrangement and only one (RD12) contained repeated elements and a complex pattern of insertion/deletion. In general the strain EGD genome is generally lacking in mobile genetic elements since only 11 copies of insertion sequences, transposases, or integrases can be identified on the basis of sequence similarity. Only one intact prophage (A118) and one cryptic prophage can be found in the EGD genome and we identified only one prophage-like segment among the lineage-specific RD, suggesting that prophage are not substantial sources of genome variability. Given the lack of evidence for the mechanisms of genome diversification and the lack of evidence for the RD being acquired states, it is yet unclear precisely how the RD arose. Analysis of the regions adjacent to the RD did not provide evidence for unusual composition or other distinguishing features such as repeated sequences.
Evolution of serotype 1/2a, 1/2b, and 4b genomes and lineage-specific functional differences.
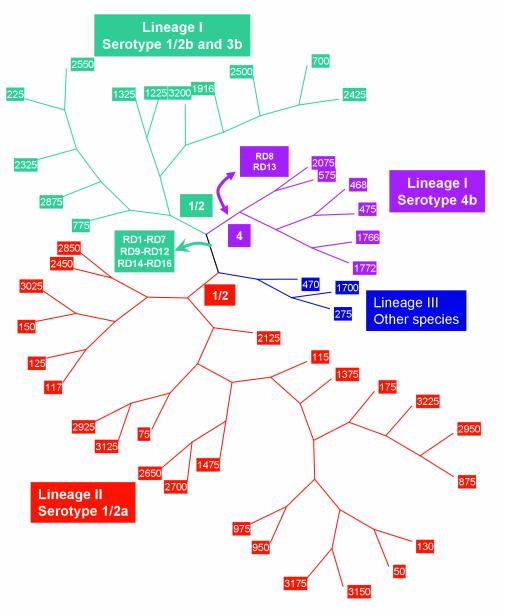
By combining our data with information from the genome sequences of serotype 1/2a and 4b L. monocytogenes strains and the Clip 11612 strain of L. innocua, we propose a model for evolution of the serotype 1/2a, 1/2b, and 4b genomes. The model is illustrated in Fig. 6, using an unrooted cladogram to depict the phylogenetic relationships. Since serotype 1/2b, 3b, and 4b strains in lineage I share 14 different RD distinguishing them from lineage II serotype 1/2a strains, it is clear that they share a recent common ancestor. However, to reconcile the finding that lineage I serotype 1/2b and lineage II serotype 1/2a populations share the same somatic antigens and that they share RD8 and RD13, the simplest model predicts that the ancestor of lineage I and lineage II strains had the serotype 1/2 somatic antigen. Genes in the RD8 and RD 13 regions are known or putatively associated with biosynthesis of serotype 1/2 and serotype 4 somatic antigens. Thus, the simplest model would hold that the serotype 4b population descended relatively recently from a serotype 1/2b ancestor and that the serotype switch occurred by gene acquisition/replacement in these regions in the ancestor of the serotype 4b populations in lineage I. Since the serotype 4b genome sequence and the L. innocua genome sequence are similar at RD8 and since the gtcA and gltAB regions are also shared among L. monocytogenes serotype 4b and some L. innocua strains (37, 38), it seems plausible that the 4b serotype arose by multiple gene transfer events from L. innocua into a serotype 1/2b-like ancestor.
FIG. 6.
Model for evolution of the L. monocytogenes genome. The unrooted dendrogram was rendered from UPGMA analysis of the microarray data. Branches corresponding to lineage I serotype 1/2b and 3b strains are colored green, lineage I serotype 4b colored purple, and lineage II serotype 1/2a colored red. The branch containing the lineage III strain and other species is colored blue. Branch points where genome deletions and acquisitions are postulated to have arisen are indicated with boxes colored according to the affected lineage.
Prior to divergence of the 4b population, the ancestor of lineage I underwent several genome alterations in RD1 to RD7, RD9 to RD12, and RD14 to RD19. Codon usage analysis and dinucleotide signature calculations identified an overlapping set of genes as PA in the serotype 1/2a genome, however neither method provided evidence for genes within the RDs being PA. This implies that the RD we detected arose through deletion of ancestral sequences in the 1/2b lineage. Based on our study design, we were only able to identify genes that were unique to lineage II strains. Using a subtractive library, Herd and Kocks (30) showed that nearly 5% of their clones were unique to the serotype 4b genome, indicating that lineage I strains also carry a substantial amount of lineage-specific genes. Indeed, the recent study from Call et al. (10) using small multiple-genome shotgun libraries also supports this view. Similarly, using a shotgun microarray derived from a serotype 4b strain, our laboratory has also found a significant number of genes that are specific to the lineage I and lineage III genomes (C. Zhang and A. K. Benson, unpublished), indicating that differences between the lineage I, II, and III genome occurred through multiple gene acquisition and deletions events.
With the addition of data from our study, there are now several independent comparative genome studies of L. monocytogenes that consistently show a collective bias of toward cell surface-related differences in genome content among the L. monocytogenes serotypes and Listeria species (10, 25, 30). This seems to point toward selection favoring different combinations of cell surface characteristics, perhaps distinguishing the way the lineages and species interact at the cell surface with host cells or the external environment. In addition to cell surface molecules, at least five different putative transcription regulators were specific to the 1/2a lineage, implying that there are likely lineage-specific patterns of gene expression. The potential for lineage-specific transcription patterns is particularly highlighted in the instance of the lineage II-specific lmo0421-lmo0423 region encoding a putative extracytoplasmic function sigma factor and a putative gene with similarity to the rodA family. It therefore remains possible that selection has favored differentiation of the phylogenetic lineages on the basis of cell surface and gene expression properties. Thus, understanding the functional differences imparted by these genotypic differences may provide insight into the ecologies of the lineages, the types of selective pressures that may have shaped them, and the basis for apparent differences in transmission rates or infectivity characteristics of the different lineages.
Acknowledgments
We thank Lewis Graves for serotyping and Etsuko Moriyama, Sam Karlin, and Jan Mrazek for advice on computing compositional bias. We also thank Robert Hutkins and James Alfano for critical comments on the manuscript.
This work is funded in part by grants from the USDA National Research Initiative Competitive Grants Program (award 2002-35201-12649 to A.K.B.) and the National Institutes of Health (award R01GM63259 to M.W.).
Footnotes
A contribution of the University of Nebraska Agricultural Research Division, Lincoln (journal series no. 14131).
REFERENCES
- 1.Aarts, H. J., L. E. Hakemulder, and A. M. Van Hoef. 1999. Genomic typing of Listeria monocytogenes strains by automated laser fluorescence analysis of amplified fragment length polymorphism fingerprint patterns. Int. J. Food Microbiol. 49:95-102. [DOI] [PubMed] [Google Scholar]
- 2.Begg, K. J., and W. D. Donachie. 1985. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J. Bacteriol. 163:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibb, W. F., B. Schwartz, B. G. Gellin, B. D. Plikaytis, and R. E. Weaver. 1989. Analysis of Listeria monocytogenes by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations. Int. J. Food Microbiol. 8:233-239. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes: the influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 5.Botta, G. A., and J. T. Park. 1981. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J. Bacteriol. 145:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehm, K., M.-T. Ripio, J. Kreft, and J.-A. Vasquez-Boland. 1999. The bvr locus of Listeria monocytogenes mediates virulence gene repression by β-glucosides. Infect. Immun. 181:5024-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosch, R., J. Chen, and J. B. Luchansky. 1994. Pulsed-field fingerprinting of Listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl. Environ. Microbiol. 60:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burman, L. G., J. Raichler, and J. T. Park. 1983. Evidence for diffuse growth of the cylindrical portion of the Escherichia coli murein sacculus. J. Bacteriol. 155:983-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, S., and M. Wiedmann. 2001. Characterization of the prfA virulence gene cluster insertion site in non-hemolytic Listeria spp.: probing the evolution of the Listeria virulence gene island. Curr. Microbiol. 43:271-277. [DOI] [PubMed] [Google Scholar]
- 10.Call, D. R., M. K. Borucki, and T. E. Besser. 2003. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J. Clin. Microbiol. 41:632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canepari, P., G. Botta, and G. Satta. 1984. Inhibition of lateral wall elongation by mecillinam stimulates cell division certain cell division conditional mutants of Escherichia coli. J. Bacteriol. 157:130-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canepari, P., C. Signoretto, M. Boaretti, and L. Del Mar. 1997. Cell elongation and septation are two mutually exclusive processes in Escherichia coli. Arch. Microbiol. 168:152-159. [DOI] [PubMed] [Google Scholar]
- 13.Chakraborty, T., T. Hain, and E. Domann. 2000. Genome organization and the evolution of the virulence gene locus in Listeria species. Int. J. Med. Microbiol. 290:167-174. [DOI] [PubMed] [Google Scholar]
- 14.Clark, E. E., I. Wesley, F. Fielder, N. Promadej, and S. Kathariou. 2000. Absence of serotype-specific surface antigen and altered teichoic acid glycosylation among epidemic-associated strains of Listeria monocytogenes. J. Clin. Microbiol. 38:3856-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Pedro, M. A., J. C. Quintela, J.-H. Joltje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dramisi, S., I. Biswas, E. Magiun, L. Braun, P. Mastroeni, and P. Cossart. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of InlB, a surface protein of the internalin multigene family. Mol. Microbiol. 16:251-261. [DOI] [PubMed] [Google Scholar]
- 17.Dramsi, S., P. Dehoux, M. Lebrun, P. L. Goossens, and P. Cossart. 1997. Identification of four new members of the internalin multigene family of Listeria monocytogenes EGD. Infect. Immun. 1997. 65:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feslenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 17:368-376. [DOI] [PubMed] [Google Scholar]
- 20.Fielder, F., J. Seger, A. Schrettenbrunner, and J. P. R. Seeliger. 1984. The biochemistry of murein and cell wall teichoic acids in the genus of Listeria. Syst. Appl. Microbiol. 5:360-376. [Google Scholar]
- 21.Finlay, B. B., and P. Cossart. 1997. Exploitation of mammalian host cell functions by bacterial pathogens. Science 276:718-725. [DOI] [PubMed] [Google Scholar]
- 22.Fuji, H. K. Kamisango, M. Nagaoka, K. Uchikawa, I. Sekikawa, K. Yamamoto, and I. Azuma. 1985. Structural study of teichoic acids of Listeria monocytogenes types 4a and 4d. J. Biochem. 97:833-891. [DOI] [PubMed] [Google Scholar]
- 23.Gaillard, J., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 24.Gellin, B. G., and C. V. Broome. 1989. Listeriosis. JAMA 261:1313-1320. [PubMed] [Google Scholar]
- 25.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couvelin, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vaszquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 26.Graves, L. M., B. Swaminathan, M. W. Reeves, S. B. Hunter, R. E. Weaver, B. D. Pikaytis, and A. Schuchat. 1994. Comparison of ribotyping and multilocus enzyme electrophoresis for subtyping of Listeria monocytogenes isolates. J. Clin. Microbiol. 32:2936-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutekunst, K. A., B. P. Holloway, and G. M. Carlone. 1992. DNA sequence heterogeneity in the gene encoding a 60-kilodalton extracellular protein of Listeria monocytogenes. Can. J. Microbiol. 38:865-870. [DOI] [PubMed] [Google Scholar]
- 28.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 29.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 30.Herd, M., and C. Kocks. 2001. Gene fragments distinguishing an epidemic-associated strain from a virulent prototype strain of Listeria monocytogenes belong to a functional subset of genes and partially cross-hybridize with other Listeria species. Infect. Immun. 69:3972-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huillet, E., S. Larpin, P. Pardon, and P. Berche. 1999. Identification of a new locus in Listeria monocytogenes involved in cellobiose-dependent repression of hly expression. FEMS Microbiol. Lett. 174:265-272. [DOI] [PubMed] [Google Scholar]
- 32.Jeffers, G. T., J. L. Bruce, P. McDonough, J. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 33.Karlin, S., and J. Mrazek. 2000. Predicted highly expressed genes of diverse prokaryotic genomes. J. Bacteriol. 182:5238-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlin, S. 2001. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 9:335-343. [DOI] [PubMed] [Google Scholar]
- 35.Kim, J., J. Nietfeldt, J. Ju, J. Wise, N. Fegan, P. Desmarchelier, and A. K. Benson. 2001. Ancestral divergence, genome diversification, and phylogeographic variation in subpopulations of sorbitol-negative, β-glucuronidase-negative enterohemorrhagic Escherichia coli O157. J. Bacteriol. 183:6885-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreft, J., J.-A. Vazquez-Boland, E. Ng, and W. Goebel. 1999. Virulence gene clusters and putative pathogenicity islands in Listeria, p. 219-232. In J. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile genetic elements. American Society for Microbiology, Washington, D.C.
- 37.Lan, Z., F. Fielder, and S. Kathariou. 2000. A sheep in wolf's clothing: Listeria innocua strains with teichoic acid-associated surface antigens and genes characteristic of Listeria monocytogenes serogroup 4. J. Bacteriol. 182:6161-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei, X.-H., F. Fielder, Z. Lan, and S. Kathariou. 2001. A novel serotype-specific gene cassette (gltA-gltB) is required for expression of teichoic acid-associated surface antigens in Listeria monocytogenes serotype 4b. J. Bacteriol. 183:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, W.-H. 1997. Molecular evolution, p. 106-108. Sinauer, Sunderland, Mass.
- 40.Ma, J., A. Campbell, and S. Karlin. 2002. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184:5733-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marco, A. J., J. Altimira, N. Prats, S. Lopez, L. Dominguez, M. Domingo, and V. Briones. 1997. Penetration of Listeria monocytogenes in mice infected by the oral route. Microb. Pathog. 23:255-263. [DOI] [PubMed] [Google Scholar]
- 42.Matsuhashi, M., M. Wachi, and F. Ishino. 1990. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res. Microbiol. 141:89-103. [DOI] [PubMed] [Google Scholar]
- 43.McLauchlin, J. 1990. Distribution of serovars of Listeria monocytogenes isolated from different categories of patients with listeriosis. Eur. J. Clin. Microbiol. Infect. Dis. 9:210-213. [DOI] [PubMed] [Google Scholar]
- 44.Piffaretti, J.-C., H. Kressebuch, M. Aeschenbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Promadej, N., F. Fielder, P. Cossart, S. Dramsi, and S. Kathariou. 1999. Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel serogroup-specific gene. J. Bacteriol. 181:418-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Portnoy, D. A., T. Chakraborty, W. Goebel, and P. Cossart. 1992. Molecular determinants of Listeria monocytogenes pathogenesis. Infect. Immun. 60:1263-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rasmussen, O. F., T. Beck, J. E. Olsen, L. Dons, and L. Rossen. 1991. Listeria monocytogenes isolates can be classified into two major types according to the sequence of the listeriolysin gene. Infect. Immun. 59:3945-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein, and listeriolysin O genes. Microbiology 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 49.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 50.Schuchat, A., B. Swaminathan, and C. V. Broome. 1991. Epidemiology of human listeriosis. Clin. Microbiol. Rev. 4:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seelinger, H. P. R., and K. Hoehne. 1979. Serotypes of Listeria monocytogenes and related species. Methods Microbiol. 13:31-49. [Google Scholar]
- 52.Sheehan, B., C. Kocks, S. Dramsi, E. Gouin, A. D. Klarsfield, J. Mengaud, and P. Cossart. 1994. Molecular and genetic determinants of the Listeria monocytogenes infectious process. Curr. Top. Microbiol. 192:187-216. [DOI] [PubMed] [Google Scholar]
- 53.Sleator, R. D., C. G. Gahan, and C. Hill. 2001. Identification and disruption of the proBA locus in Listeria monocytogenes: role of proline biosynthesis in salt tolerance and murine infection. Appl. Environ. Microbiol. 67:2571-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tran, H. L., and S. Kathariou. 2002. Restriction fragment length polymorphisms detected with novel DNA probes differentiate among diverse lineages of serogroup 4 Listeria monocytogenes and identify four lineages in serotype 4b. Appl. Environ. Microbiol. 68:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uchikawa, K., I. Sekikawa, and I. Azuma. 1986. Structural studies on teichoic acids in cell walls of several serotypes of Listeria monocytogenes. J. Biochem. 99:315-327. [DOI] [PubMed] [Google Scholar]
- 56.Vasquez-Boland, J., M. Kuhn, P. Berche, T. Cahkraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vines, A., M. W. Reeves, S. Hunter, and B. Swaminathan. 1992. Restriction fragment length polymorphism in four virulence-associated genes of Listeria monocytogenes. Res. Microbiol. 143:281-294. [DOI] [PubMed] [Google Scholar]
- 58.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wientjes, F. B., E. Pas, P. E. M. Taschner, and C. L. Woldringh. 1985. Kinetics of uptake and incorporation of meso-diaminopimelic acid in different Escherichia coli strains. J. Bacteriol. 164:331-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wientjes, F. B., and N. Nanninga. 1989. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J. Bacteriol. 171:3412-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng, W., and S. Kathariou. 1995. Differentiation of epidemic-associated strains of Listeria monocytogenes by restriction fragment length polymorphism in a gene region essential for growth at low temperature (4°C). Appl. Environ. Microbiol. 61:4310-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]