Abstract
Hybrid proteins containing the β-autotransporter domain of the immunoglobulin A (IgA) protease of Neisseria gonorrhoea (IgAβ) and the partner leucine zippers of the eukaryotic transcriptional factors Fos and Jun were expressed in Escherichia coli. Such fusion proteins targeted the leucine zipper modules to the cell surface. Cells displaying the Junβ sequence flocculated shortly after induction of the hybrid protein. E. coli cells expressing separately Fosβ and Junβ chimeras formed stable bacterial consortia. These associations were physically held by tight intercell ties caused by the protein-protein interactions of matching dimerization domains. The role of autotransporters in the emergence of new adhesins is discussed.
Bacteria frequently adhere to specific molecular targets through the presentation of protein structures on their cell surface (typically fimbriae) with distinct surface-binding capacities (29, 44). Some potent adhesins found in a variety of gram-negative pathogens may, instead, be anchored directly to the outer membrane (OM), so the result of the attachment is an intimate target cell contact. A major class of such adhesins are displayed onto the cell surface by virtue of the protein secretion system known as autotransporters (ATs) (17). Examples of this type include proteins of Salmonella enterica (ShdA and MisL) (21, 22), Rickettsia rickettsii (recombinant OmpA) (30), Bordetella species (pertactin, Vag8) (10, 12), Haemophilus influenzae (Hap, Hia, Hsf; (11, 27, 41), and some virulent Escherichia coli strains (Ag43, AIDA-1, and TibA) (5, 24, 31), among others (16). Such proteins endow bacteria the ability to stick to firm surfaces (11, 21, 27) or to interact with other bacteria (9, 18, 46).
The AT secretion systems involve the export of two-module polypeptides. These include a C-terminal domain (called transporter or β module) which ends up being inserted as an oligomer in the OM, and the passenger domain, which is the protein moiety eventually presented on and anchored to the cell surface (17, 37, 49). Such a design seems to be naturally well suited for the emergence of novel adhesins with new specificities. This is because the protein export and anchoring mechanism appears to be tolerant to a large variety of protein structures (7). Taking advantage of this property several heterologous passenger polypeptides have been exposed on the cell surface fused to the β-domain of different members of the AT family, i.e., the mouse metallothionein (47), the B subunit of the cholera toxin (33), the β-lactamase (28), the bovine adrenodoxin (19), the carboxylesterase EstA from Burkholderia gladioli (43), recombinant scFv antibodies (48), or the fimbrial protein FimH (23), among others (39).
In this work, we have attempted to reprogram the adhesion properties of E. coli by displaying in its surface an entirely alien protein fold, with potential adhesin-like activity, that attaches distinctly to itself or to a second molecule presented by the target surface. To this end, we created a fusion protein containing the β-AT domain of the IgA protease of Neisseria gonorrhoeae, an archetypal AT protein (37) and the partner leucine zippers of eukaryotic transcription factors Fos and Jun (2). Fos and Jun have an intrinsic ability to heterodimerize through the strong interactions between their coiled coils of their protein folds (Jun can also produce homodimers, see below; (1, 2). When such hybrid proteins were expressed in E. coli, cells acquired novel adherence traits resulting in the self-association and clumping of otherwise planktonic bacteria in liquid media, or in formation of stable consortia between cells of strains expressing the dimerization domains, either being Jun/Jun or Jun/Fos. These cell to cell associations are caused by tight protein-protein interactions that connect matching dimerization domains. Our data shed some light on the evolutionary value of AT systems for the appearance of novel adhesion determinants.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli UT5600 (ΔompT proC leu-6 trpE38 entA (14) harboring pFosβ and pJunβ (see below) were grown at 37°C in LB agar plates (36) containing 2% glucose (wt/vol) and chloramphenicol (40 μg ml−1) or inoculated in the same liquid medium. For induction, cultures were adjusted to an optical density at 600 nm (OD600) of 0.5 in liquid LB medium without glucose, added with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated further for 3 h at 37°C. The sequences of the jun and fos leucine zipper were PCR amplified from plasmid pJUFO (8). The DNA segment corresponding to fos was amplified with primers Fos1 (5′-ATT ACT CGC GGC CCA GCC GGC CAT GGC GGA CCT GAC CGA CAC CCT G-3′) and Fos2 (5′-GCT CGA ATT CGG CCC CCG AGG CCT CGT GTG CCG CCA GGA TGA AC-3′). Primers Jun1 (5′-ATT ACT CGC GGC CCA GCC GGC CAT GGC GGA CCG GAT CGC CCG GCT C-3′) and Jun2 (5′-GCT CGA ATT CGG CCC CCG AGG CCT CGT GGT TCA TGA CTT TCTG TTT-3′) were similarly used for amplification of the jun leucine zipper sequence. The resulting PCR products were digested with SfiI and cloned into the same site of the plasmid pMTβ-1 (Cmr) (47). The plasmids thereby originated were named pFosβ and pJunβ. These expressed, respectively, fusions of the fos and jun leucine zippers, to the β domain of the IgA protease under the control of the lac promoter. The control plasmid pHEβ expresses a fusion protein between a polyhistidine tag and the IgA protease β domain (49).
Immunofluorescence microscopy.
For detecting the E tag on the surface of E. coli cells, bacteria were washed with phosphate-buffered saline (PBS) (3) and fixed in the same buffer containing 3% (wt/vol) paraformaldehyde. 80 μl of such cell suspensions were laid for 1 h on glass coverslips precoated with poly-l-lysine (1 mg ml−1). The glass slides were then blocked for 40 min in B buffer (PBS with 3% [wt/vol] bovine serum albumin). Samples were incubated for 1 h in the same buffer with an anti-E tag monoclonal antibody (MAb) (10 μg ml−1; Amersham Pharmacia Biotech). Each specimen was then washed five times with PBS and further incubated for 1 h with an anti-mouse IgG serum conjugated with Cy3 (Amersham Pharmacia Biotech) diluted at 1:100 in B buffer. Cells were then examined with epifluorescence microscopy as described elsewhere (48).
Western blotting and protease digestion.
For monitoring the presence of the hybrids in cell extracts, proteins samples were denatured and run in sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis as described before (48). Briefly, after electrophoresis the proteins were blotted onto polyvinylidene difluoride membranes (Immobilon-P; Millipore) and incubated with anti-E tag MAb conjugated with peroxidase (Amersham Pharmacia). The membranes were developed with a chemiluminescence reaction and exposed to an X-ray film. Protease accessibility analyses were carried out as explained in (49). In brief, induced E. coli cells were washed and resuspended in PBS, trypsin was added externally (1 μg ml−1). Samples were incubated for 25 min at 37°C and stopped by adding trypsin inhibitor (5 μg ml−1; Sigma). Total protein extracts from these cells were analyzed by Western blotting.
ELISA.
The level of surface display of the E-tagged leucine zippers was quantified as follows. For intact cells, E. coli cultures were harvested, washed in PBS and resuspended in the same buffer at a final OD600 of 2.0. Alternatively, bacteria were resuspended in PBS with 20 mM EDTA and lysed by sonication (48). Either intact or lysed cells were attached to the enzyme-linked immunosorbent assay (ELISA) plate (Maxisorb; Nunc) for 1 h at room temperature. Samples were then blocked with 3% (wt/vol) skim milk in PBS for 1 h, rinsed in PBS and added with 1 μg ml−1 of the anti-E tag MAb for 1 h. Plates were washed three times prior to incubation with anti-mouse IgG peroxidase conjugate (0.3 U ml−1; Roche). The ELISA were developed using o-phenylenediamine (Sigma). A similar protocol was used for assaying the accessibility of the peptidoglycan in E. coli expressing hybrid proteins. In this case, a rabbit serum anti-E. coli peptidoglycan (38) was used at 1:1,000, followed by treatment with a 1:500 dilution of protein A-peroxidase conjugate (Roche).
Cell aggregation assays.
Liquid cultures of E. coli strains bearing plasmids pFosβ, pJunβ, or pHEβ were separately grown to an OD600 of 0.5 and then induced with 0.5 mM IPTG for 3 h at 37°C under vigorous shaking (180 rpm). After that, 5-ml aliquots of each culture were mixed as indicated. The mixtures were then transferred to 10-ml tubes and cultivated without shaking. Triplicate 100-μl samples were withdrawn at 20-min intervals from the top of each tube for measurement of OD600 across time. Additionally induced samples were also taken for examination under the microscope. To this end, 15-μl aliquots of the cultures were deposited on the surface of a microscope slide covered with a thin layer of 1% (wt/vol) agarose in Tris 50 mM (pH 7.5) in order to immobilize the bacteria, and incubated at 37°C for 1 h in a water-saturated atmosphere. Coverslips were then placed on the samples, and subsequently the samples were visualized by phase-contrast microscopy.
RESULTS AND DISCUSSION
Expression of Fosβ and Junβ proteins in E. coli.
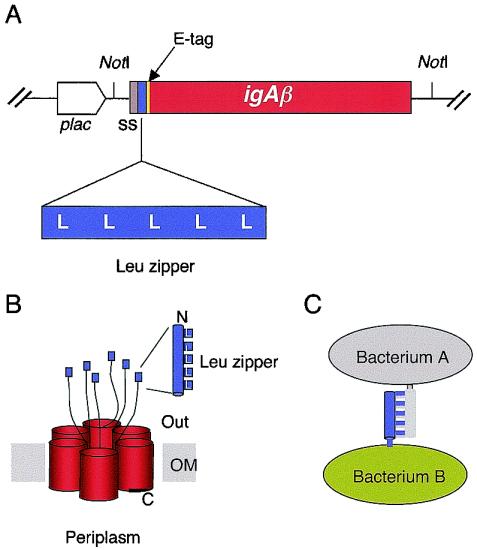
The rationale of the work described below is summarized in Fig. 1. We have constructed hybrid outer membrane proteins of E. coli containing the β-AT domain of the IgA protease of N. gonorrhoeae (25, 49) and the partner leucine zippers of the eukaryotic transcription factors Fos and Jun (1, 2). The relevant inserts of plasmids pFosβ and pJunβ are sketched in Fig. 1. These were designed to enable the production of fused two-module proteins affording the surface display of a passenger domain with hetero- or homodimerization capacity. The production of the hybrid proteins in vivo can be monitored by means of a short E-tag epitope engineered within the linker region between the AT domain (∼45 kDa) and the Fos/Jun modules (∼8 kDa). In addition, the vector adds the N-terminal signal sequence of pelB for an efficient secretion (20). Finally, these constructs place the expression of the hybrid proteins under the control of the lac promoter so that their production can be induced by IPTG and partially repressed by glucose.
FIG. 1.
Structure of Fosβ and Junβ hybrid proteins. (A) Organization of significant inserts in plasmids pFosβ and pJunβ, encoding, respectively, Fosβ and Junβ hybrids. The position of the pelB signal sequence (ss), the E tag, the leucine zippers, and the igAβ gene segments are indicated as well as the lac promoter (plac). Sizes are symbolic. (B) Predicted topology of the Fosβ and Junβ hybrids in the bacterial OM, with the leucine zipper domains exposed to the external medium and the oligomeric AT complex in the bacterial OM (49). (C) Putative interactions between two bacteria expressing Fosβ and Junβ proteins.
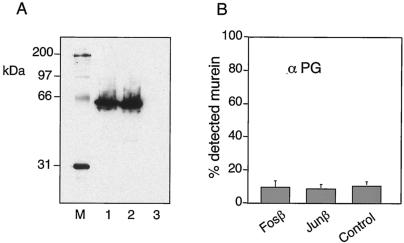
Once the constructs were made, we examined their production, cellular location and their performance in vivo. Fig. 2A shows the Western blots corresponding to whole cell extracts of IPTG-induced E. coli cells transformed with plasmids pFosβ and pJunβ. Bands of the expected size for the hybrid β-fusions were clearly produced when the blots were probed with an anti-E tag MAb. These bands were well defined, with no indication of instability or proteolytic degradation, thereby indicating that the Fosβ and the Junβ hybrids are produced as full-length polypeptides. Since overproduction of OM proteins may cause damage to the cell envelope resulting in an artifactual display of otherwise not exposed proteins, we ensured also membrane integrity in cells expressing Fosβ and the Junβ. To this end, we assayed the accessibility of the peptidoglycan in induced E. coli cells transformed with pFosβ and pJunβ. As shown in the ELISA of Fig. 2B, the same level of peptidoglycan detection was observed in E. coli control cells (nontransformed) than in those expressing the Fosβ and Junβ hybrids. This result indicated that production of the β-hybrid did not alter the integrity of the OM.
FIG. 2.
Expression of Fosβ and Junβ in the E. coli strain UT5600. (A) Immunoblot of whole-cell protein extracts from induced cultures expressing Fosβ (lane 1) or Junβ (lane 2) or nontransformed (lane 3). The blot was probed with anti-E tag MAb. Streptavidin-peroxidase conjugate was also added to detect the biotinylated protein standards (M). (B) Accessibility of the peptidoglycan of the same induced E. coli cells. The signals of intact cells, using an antipeptidoglycan serum (38), are shown as a percentage of lysed cells. Nontransformed E. coli strain UT5600 was used as control.
Presentation of Fos and Jun domains on the surface of E. coli cells.
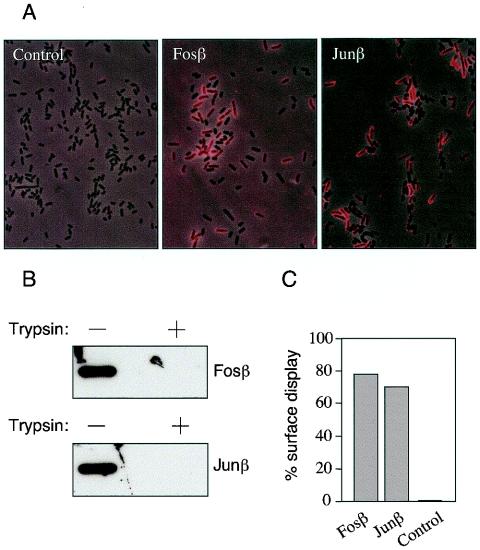
The targeting of the Fos and Jun sequences to the envelope of E. coli cells as fusions to the AT domain of the IgA protease was investigated. Once more, the E tag engineered in the corresponding fusion proteins was instrumental to tackle this concern. First, we attempted to visualize directly the location of the E tag on intact cells through epifluorescence microscopy (Fig. 3A). Cells from IPTG-induced cultures of E. coli cells harboring pFosβ, pJunβ, and a plasmidless control were fixed and treated first with the anti-E tag MAb described above and then with a Cy3-conjugate of anti-mouse IgG serum (Materials and Methods). Under the epifluorescence microscope, we observed a strong red signal over the cells transformed with the Fosβ- or Junβ-encoding plasmids, but not in the control. This was indicative of the display of the E tag on the cell surface. Since the E tag was engineered in pFosβ and pJunβ behind the leucine zippers (Fig. 1), it is necessarily secreted following the passenger Fos and Jun sequences, which must be exposed on the cell surface as well.
FIG. 3.
External localization of Fosβ and Junβ passenger domains. (A) Intact fixed bacteria expressing Fosβ or Junβ or nontransformed (control) bacteria were probed with anti-E tag MAb by indirect immunofluorescence. Binding of anti-E tag MAb to the surface-exposed epitopes was detected using a Cy3-conjugated secondary antibody (in red). These pictures are the result of superimposing of the phase contrast image, and the immunofluorescence image in which only those which expressed the antigen on their surface emit an optical signal. (B) Intact E. coli cells expressing Fosβ or Junβ were incubated (+) or not (−) with trypsin (1 μg/ml). The digestion of the Fosβ and Junβ exposed modules was monitored by Western blots using the anti-E tag MAb. (C) Quantification of the display of Fosβ and Junβ on the surface of intact E. coli cells by ELISA. The values shown are the average of three independent experiments and represent the percentage of the signal produced by intact (PBS-washed) cells with respect to the output obtained with an identical number of lysed cells.
Closer inspection of the images of Fig. 3A revealed that not every E. coli cell in the cultures bearing pFosβ or pJunβ produced epifluorescence. This could be explained on the basis of the stochastic induction that appears to operate on the lac promoter (4, 45). But also, it may well happen that not all the expressed Fosβ and Junβ proteins target the E tag (and the accompanying Fos and Jun modules) to the surface. To examine this possibility, we carried out a second assay to ascertain whether all or only part of the E tag of the fusion proteins was accessible from the outside. Intact cells expressing Fosβ and Junβ were treated with externally added trypsin. This protease is not able to penetrate into intact cells (Fig. 2B) and thus is only able to degrade exposed domains (26, 42, 49). After trypsin digestion cells were collected and their whole protein extracts analyzed by Western blotting with the anti-E tag MAb. The data shown in Fig. 3B indicates that 100% of the E tag signal disappears in samples treated with trypsin, thus demonstrating the full localization of the epitope on the cell outermost, accessible surface. In addition we performed ELISA in which we compared the levels of E tag which could be recognized by the anti-E Tag MAb in intact cells versus lysed cells. As shown in Fig. 3C, a minimum of 75% of the entire complement of E tags produced in vivo were detected in whole cells expressing Fosβ or Junβ. Since integrity of the outer membrane of cells expressing Fosβ or Junβ is preserved (see above; Fig. 2B) we conclude that the ELISA signals from whole-cell assays were due to the display of the E tags encoded by the hybrid proteins. The difference observed in the level of surface exposed passenger domains (100% measured by trypsin digestion versus 75% by ELISA) was maybe due to the interference in ELISA of the surface polysaccharides presented on intact E. coli cells.
Taken together, the results above show that all the produced Fosβ and Junβ proteins complete the AT secretion pathway and localize their N-terminal domains (encompassing the E-tag and the Fos/Jun moieties) on the cell surface. This is especially intriguing in the case of the Junβ protein, since (unlike Fos) the corresponding leucine zipper has a tendency to associate with itself, a property which could interfere with the secretory mechanism. However, this was clearly not the case, most likely because the mechanism of AT secretion also involves the formation of an oligomer (49) and tolerates the efficient translocation of folded protein domains towards the cell surface (E. Veiga, V. de Lorenzo, and L. A. Fernández, submitted for publication). The next step was thus to examine the biological activity of the secreted dimerization domains.
Cell adhesion properties mediated by surface display of leucine zippers.
If leucine zippers of the Jun and Fos sequences retained their ability to bring about strong protein-protein interactions when exposed on the surface of E. coli cells, then their expression should cause cell aggregation. On this basis, we set out several assays to monitor cell-to-cell adhesion mediated by the occurrence of dimerization of the corresponding leucine zippers. The background of these assays is that Fos domains can form heterodimers only with the partner Jun domains. On the contrary, Jun modules can form both homo- and heterodimers (1, 2).
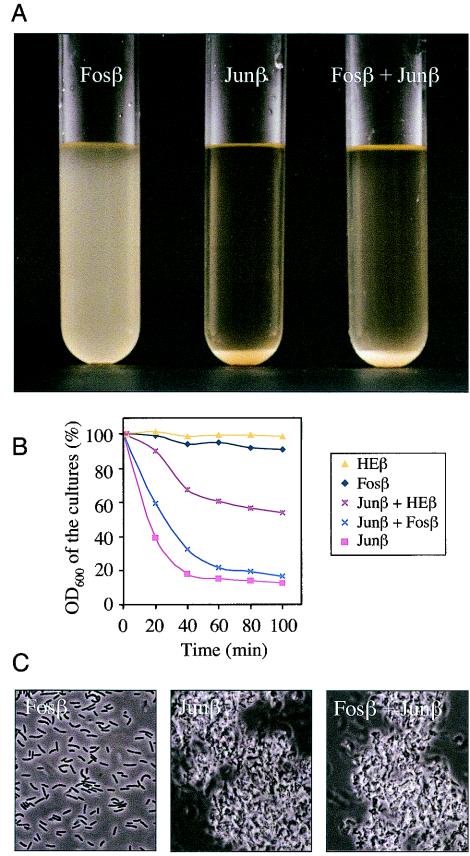
As shown in Fig. 4A, 90 min after induction (see Materials and Methods), E. coli cells expressing Fosβ remained in suspension, while the same cells bearing the pJunβ plasmid sedimented spontaneously (thereby quickly lowering the OD600 of the culture from 2.2 to 0.3; Fig. 4B). This was accompanied by formation of a thick bacterial precipitate. This bacterial precipitate could be resuspended by vortexing and the cultures recovered the same OD600 than that of the culture expressing Fosβ (data not shown). Interestingly, a mixed population of cells expressing Fosβ and Junβ cosedimented very rapidly as well (Fig. 4A and B) suggesting that the cells expressing the Junβ fusion pulled down entirely those presenting Fosβ.
FIG. 4.
Specific cell adhesion driven by the interaction of the surface exposed leucine zipper dimerization domains. (A) Samples of induced E. coli cells expressing Fosβ, Junβ or a mixture of the same number of cells from both populations (Fosβ + Junβ) were placed at the same OD600 in three different tubes without shaking. The photo was taken after 90 min. (B) Precipitation kinetics of E. coli cultures. The samples examined were E. coli cells expressing separately Fosβ, Junβ, and HEβ as well as mixtures of an equal number of cells expressing Junβ plus Fosβ or Junβ plus HEβ. The OD was measured from aliquots taken at ∼1 cm from the surface. The initial OD of the cultures, just after induction, was taken as 100%. (C) Phase-contrast microscopy images of induced E. coli cells placed onto microscopy slides without shaking after 90 min.
To determine whether the aggregation observed in the mixed population of Fosβ and Junβ expressing bacteria was due to specific interactions between the Fos and Jun leucine zippers we performed similar experiments using a control β fusion devoid of leucine zipper (HEβ). This control fusion consisted in a poly-His segment fused to the β domain of the IgA protease. Cells expressing HEβ remained in a planktonic state identical to those expressing Fosβ (Fig. 4B). The OD600 of a mixed culture of bacteria expressing Junβ and HEβ decreased only by half of the initial OD. This indicated that only part of the mix culture (probably this corresponding to the Junβ expressing bacteria) precipitated. On the contrary (Fig. 4A) a mixed population of cells expressing Fosβ and Junβ cosedimented to the same extent as the cells expressing Junβ alone. These data confirm that precipitation of the mixed culture of bacteria expressing Fosβ and Junβ was due to the heterodimeric interactions of Fos and Jun exposed leucine zipper domains.
Samples from the different cultures were also inspected by phase contrast microscopy as shown in Fig. 4C in order to visualize the bacterial associations that promoted the precipitation of the cultures. The resulting images were fully consistent with what was observed at a macroscopic scale, namely, that cells expressing Fosβ grew basically as a populations of individual cells, while those bearing pJunβ and the mixed population of bacteria expressing Fosβ and Junβ formed large aggregates. All these data showed unequivocally that the new cell surface properties were due to the adhesin-like activity created by the Fos/Jun leucine zippers.
Conclusion.
This report shows that protein dimerization domains (i.e., coiled coils of Jun/Fos leucine zippers) which are typical of some eukaryotic transcription factors become effective mediators of strong and distinct cell-cell attachment when fused to a carrier module of AT. Because of their unusual self-secretion mechanism, the β-domain of ATs such as the IgA protease of N. gonorrhoeae can mediate, with few restrictions, the presentation of a large variety of protein structures. The size of the pore formed by the oligomeric AT complex (49) allows the efficient secretion of folded Ig domains (submitted for publication). Interestingly, Ig domains are frequently found naturally in bacterial adhesins (6, 15, 32, 40).
Although the passenger domains of some AT proteins (typically the IgA protease) are released into the external medium (16), the AT β domains anchor in many cases protein modules which endow cells with novel adhesion properties. AT systems are highly similar in their C-terminal portion, which encompasses the β barrel plus the linker region necessary for secretion (25, 34). Yet, ATs are very divergent in their N-terminal part (i.e., the passenger domain). Thus, ATs are suited as scaffolds for the generation of novel adhesins. These could be selected in vivo through a simple attachment-mediated clonal selection similar to that observed in Igs (35) or the selection of clones of interest by phage display (13). Unlike fimbriae, the attachment properties of which are strongly limited by structural constrains (7, 39), ATs tolerate a wide range of protein modules that become displayed with the same structure that they had prior to become fused to the β domain (19, 23, 28, 33, 47, 48). Adventitious fusion of a protein fold typical of distant DNA binding proteins result in novel cell surface adhesion properties. In light of our results, it is tempting to speculate with the possibility that this recruitment of protein-protein interaction domains may occur also in nature for the generation of novel adhesins. Such aleatory fusions to AT domains could easily lead to functional switching of the recruited protein folds.
Acknowledgments
The technical work of Sofía Fraile is greatly appreciated.
This work was supported by EU contracts QLK3-CT-2002-01933, QLK3-CT-2002-01923, and INCO-CT-2002-1001; by grants BIO2001-2274 and BMC2002-03024 of the Spanish Ministerio de Ciencia y Tecnología (MCyT); by grant COLIRED-O157 (G03/025) of the Fondo de Investigaciones Sanitarias (FIS); and by the Strategic Research Groups Program of the Autonomous Community of Madrid.
REFERENCES
- 1.Abate, C., D. Luk, E. Gagne, R. G. Roeder, and T. Curran. 1990. Fos and jun cooperate in transcriptional regulation via heterologous activation domains. Mol. Cell. Biol. 10:5532-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abate, C., D. Luk, R. Gentz, F. J. Rauscher III, and T. Curran. 1990. Expression and purification of the leucine zipper and DNA-binding domains of Fos and Jun: both Fos and Jun contact DNA directly. Proc. Natl. Acad. Sci. USA 87:1032-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Becskei, A., and L. Serrano. 2000. Engineering stability in gene networks by autoregulation. Nature 405:590-593. [DOI] [PubMed] [Google Scholar]
- 5.Benz, I., and M. A. Schmidt. 1992. AIDA-I, the adhesin involved in diffuse adherence of diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol, Micriobiol. 6:1539-1546. [DOI] [PubMed] [Google Scholar]
- 6.Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S. J. Hultgren, and S. D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061-1066. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis, P. 2000. Expressing genes in different Escherichia coli compartments. Curr. Opin. Biotechnol. 11:450-454. [DOI] [PubMed] [Google Scholar]
- 8.Crameri, R., and M. Suter. 1993. Display of biologically active proteins on the surface of filamentous phages: a cDNA cloning system for selection of functional gene products linked to the genetic information responsible for their production. Gene 137:69-75. [DOI] [PubMed] [Google Scholar]
- 9.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 10.Emsley, P., I. G. Charles, N. F. Fairweather, and N. W. Isaacs. 1996. Structure of Bordetella pertussis virulence factor P. 69 pertactin. Nature 381:90-92. [DOI] [PubMed] [Google Scholar]
- 11.Fink, D. L., B. A. Green, and J. W. St. Geme III. 2002. The Haemophilus influenzae Hap autotransporter binds to fibronectin, laminin, and collagen IV. Infect. Immun. 70:4902-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finn, T. M., and D. F. Amsbaugh. 1998. Vag8, a Bordetella pertussis bvg-regulated protein. Infect. Immun. 66:3985-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths, A. D., and A. R. Ducan. 1998. Strategies for selection of antibodies by phage display. Curr. Opin. Bio/Technol. 9:102-108. [DOI] [PubMed] [Google Scholar]
- 14.Grodberg, J., and J. J. Dunn. 1988. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J. Bacteriol. 170:1245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamburger, Z. A., M. S. Brown, R. R. Isberg, and P. J. Bjorkman. 1999. Crystal structure of invasin: a bacterial integrin-binding protein. Science 286:291-295. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 18.Hendrixson, D. R., and J. W. St. Geme III. 1998. The Haemophilus influenzae Hap serine protease promotes adherence and microcolony formation, potentiated by a soluble host protein. Mol. Cell 2:841-850. [DOI] [PubMed] [Google Scholar]
- 19.Jose, J., R. Bernhardt, and F. Hannemann. 2002. Cellular surface display of dimeric Adx and whole cell P450-mediated steroid synthesis on E. coli. J. Biotechnol. 95:257-268. [DOI] [PubMed] [Google Scholar]
- 20.Keen, N. T., and S. Tamaki. 1986. Structure of two pectate lyase genes from Erwinia chrysanthemi EC16 and their high-level expression in Escherichia coli. J. Bacteriol. 168:595-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kingsley, R. A., R. L. Santos, A. M. Keestra, L. G. Adams, and A. J. Baumler. 2002. Salmonella enterica serotype Typhimurium ShdA is an outer membrane fibronectin-binding protein that is expressed in the intestine. Mol. Microbiol. 43:895-905. [DOI] [PubMed] [Google Scholar]
- 22.Kingsley, R. A., K. van Amsterdam, N. Kramer, and A. J. Baumler. 2000. The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding. Infect. Immun. 68:2720-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kjaergaard, K., H. Hasman, M. A. Schembri, and P. Klemm. 2002. Antigen 43-mediated autotransporter display, a versatile bacterial cell surface presentation system. J. Bacteriol. 184:4197-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kjaergaard, K., M. A. Schembri, H. Hasman, and P. Klemm. 2000. Antigen 43 from Escherichia coli induces inter- and intraspecies cell aggregation and changes in colony morphology of Pseudomonas fluorescens. J. Bacteriol. 182:4789-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klauser, T., J. Krämer, K. Otzelberger, J. Pohlner, and T. F. Meyer. 1993. Characterization of the Neisseria Igaβ-core. The essential unit for outer membrane targeting and extracellular protein secretion. J. Mol. Biol. 234:579-593. [DOI] [PubMed] [Google Scholar]
- 26.Klauser, T., J. Pohlner, and T. F. Meyer. 1990. Extracellular transport of cholera toxin B subunit using Neisseria IgA protease β-domain: conformation-dependent outer membrane translocation. EMBO J. 9:1991-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laarmann, S., D. Cutter, T. Juehne, S. J. Barenkamp, and J. W. St Geme. 2002. The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Mol. Microbiol. 46:731-743. [DOI] [PubMed] [Google Scholar]
- 28.Lattemann, C. T., J. Maurer, E. Gerland, and T. F. Meyer. 2000. Autodisplay: functional display of active β-lactamase on the surface of Escherichia coli by the AIDA-I autotransporter. J. Bacteriol. 182:3726-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, V. T., and O. Schneewind. 2001. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 15:1725-1752. [DOI] [PubMed] [Google Scholar]
- 30.Li, H., and D. H. Walker. 1998. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb. Pathog. 24:289-298. [DOI] [PubMed] [Google Scholar]
- 31.Lindenthal, C., and E. A. Elsinghorst. 2001. Enterotoxigenic Escherichia coli TibA glycoprotein adheres to human intestine epithelial cells. Infect. Immun. 69:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo, Y., E. A. Frey, R. A. Pfuetzner, A. L. Creagh, D. G. Knoechel, C. A. Haynes, B. B. Finlay, and N. C. Strynadka. 2000. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405:1073-1077. [DOI] [PubMed] [Google Scholar]
- 33.Maurer, J., J. Jose, and T. F. Meyer. 1997. Autodisplay: one-component system for efficient surface display and release of soluble recombinant proteins from Escherichia coli. J. Bacteriol. 179:794-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maurer, J., J. Jose, and T. F. Meyer. 1999. Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J. Bacteriol. 181:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meffre, E., R. Casellas, and M. C. Nussenzweig. 2000. Antibody regulation of B cell development. Nat. Immunol. 1:379-385. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Pohlner, J., R. Halter, K. Beyreuther, and T. F. Meyer. 1987. Gene structure and extracellular secretion of Neisseria gonorrhoea IgA protease. Nature 325:458-462. [DOI] [PubMed] [Google Scholar]
- 38.Quintela, J. C., M. A. de Pedro, P. Zöllner, G. Allmaier, and F. Garcia-del Portillo. 1997. Peptidoglycan structure of Salmonella typhimurium growing within cultured mammalian cells. Mol. Microbiol. 23:693-704. [DOI] [PubMed] [Google Scholar]
- 39.Samuelson, P., E. Gunneriusson, P. A. Nygren, and S. Stahl. 2002. Display of proteins on bacteria. J. Biotechnol. 96:129-154. [DOI] [PubMed] [Google Scholar]
- 40.Sauer, F. G., K. Futterer, J. S. Pinkner, K. W. Dodson, S. J. Hultgren, and G. Waksman. 1999. Structural basis of chaperone function and pilus biogenesis. Science 285:1058-1061. [DOI] [PubMed] [Google Scholar]
- 41.Scarselli, M., R. Rappuoli, and V. Scarlato. 2001. A common conserved amino acid motif module shared by bacterial and intercellular adhesins: bacterial adherence mimicking cell cell recognition? Microbiology 147:250-252. [DOI] [PubMed] [Google Scholar]
- 42.Schenkman, S., A. Tsugita, M. Schwartz, and J. P. Rosenbusch. 1984. Topology of phage lambda receptor protein. Mapping targets of proteolytic cleavage in relation to binding sites for phage or monoclonal antibodies. J. Biol. Chem. 259:7570-7576. [PubMed] [Google Scholar]
- 43.Schultheiss, E., C. Paar, H. Schwab, and J. Jose. 2002. Functional esterase surface display by the autotransporter pathway in Escherichia coli. J. Mol. Catalysis B Enzymatic 18:89-97. [Google Scholar]
- 44.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tolker-Nielsen, T., K. Holmstrom, L. Boe, and S. Molin. 1998. Non-genetic population heterogeneity studied by in situ polymerase chain reaction. Mol. Microbiol. 27:1099-1105. [DOI] [PubMed] [Google Scholar]
- 46.Torres, A. G., N. T. Perna, V. Burland, A. Ruknudin, F. R. Blattner, and J. B. Kaper. 2002. Characterization of Cah, a calcium-binding and heat-extractable autotransporter protein of enterohaemorrhagic Escherichia coli. Mol. Microbiol. 45:951-966. [DOI] [PubMed] [Google Scholar]
- 47.Valls, M., S. Atrian, V. de Lorenzo, and L. A. Fernández. 2000. Engineering a mouse metallothionein on the cell surface of Ralstonia eutropha CH34 for immobilization of heavy metals in soil. Nat. Biotechnol. 18:661-665. [DOI] [PubMed] [Google Scholar]
- 48.Veiga, E., V. de Lorenzo, and L. A. Fernández. 1999. Probing secretion and translocation of a beta-autotransporter using a reporter single-chain Fv as a cognate passenger domain. Mol. Microbiol. 33:1232-1243. [DOI] [PubMed] [Google Scholar]
- 49.Veiga, E., E. Sugawara, H. Nikaido, V. de Lorenzo, and L. A. Fernández. 2002. Export of autotransported proteins proceeds through an oligomeric ring shaped by C-terminal domains. EMBO J. 21:2122-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]