Abstract
A 254-nucleotide model mRNA, designated ΔermC mRNA, was used to study the effects of translational signals and ribosome transit on mRNA decay in Bacillus subtilis. ΔermC mRNA features a strong ribosome-binding site (RBS) and a 62-amino-acid-encoding open reading frame, followed by a transcription terminator structure. Inactivation of the RBS or the start codon resulted in a fourfold decrease in the mRNA half-life, demonstrating the importance of ternary complex formation for mRNA stability. Data for the decay of ΔermC mRNAs with stop codons at positions increasingly proximal to the translational start site showed that actual translation—even the formation of the first peptide bond—was not important for stability. The half-life of an untranslated 3.2-kb ΔermC-lacZ fusion RNA was similar to that of a translated ΔermC-lacZ mRNA, indicating that the translation of even a longer RNA was not required for wild-type stability. The data are consistent with a model in which ribosome binding and the formation of the ternary complex interfere with a 5′-end-dependent activity, possibly a 5′-binding endonuclease, which is required for the initiation of mRNA decay. This model is supported by the finding that increasing the distance from the 5′ end to the start codon resulted in a 2.5-fold decrease in the mRNA half-life. These results underscore the importance of the 5′ end to mRNA stability in B. subtilis.
mRNA decay is known to play an important role in the posttranscriptional regulation of gene expression in bacteria. Numerous studies on mRNA decay in Escherichia coli have led to the following, still controversial (26, 28), model: RNase E, an endoribonuclease, is the major decay-initiating enzyme. Internal cleavage of an mRNA by RNase E follows obligatory binding to the 5′ end (8, 20, 28). RNase II and/or polynucleotide phosphorylase, both 3′-to-5′ exonucleases, rapidly degrade the mRNA fragments generated by RNase E cleavage (12, 35). Oligoribonuclease is required for the final turnover of RNA oligonucleotides to mononucleotides (17).
The accessibility of the 5′ end of an mRNA to RNase E has been shown to be an important mediator of stability in E. coli. The presence of stable secondary structures at the 5′ end of an mRNA interferes with RNase E-initiated decay (1, 6). Recently, it was shown in vivo that the circularization of an mRNA resulted in a four- to sixfold increase in the half-life (30). Whether the translation of an mRNA is important for stability in E. coli is not clear. The stability of some, but not all, mRNAs is affected by the introduction of premature stop codons. Uncoupling transcription and translation decreases mRNA half-life, while the effect of introducing translational inhibitors may depend on the mode of action of these inhibitors. (See references 20, 24, and 34 for a comprehensive listing of references addressing these points.)
Much less is known about mRNA decay in Bacillus subtilis. The question of how mRNA decay occurs in this organism is of particular interest, since the B. subtilis genome reveals a lack of homologues to RNase E, RNase II, and oligoribonuclease, all of which are major RNase activities in E. coli (9). Previous experiments with B. subtilis showed that, as in E. coli, the 5′ end seems to be an important mediator of mRNA stability. The presence of a strong ribosome-binding site (RBS) in the gsiB mRNA was critical for its long half-life (25). A secondary structure present in the 5′ untranslated region was also shown to be important for the stability of the aprE mRNA (22). The binding of a regulatory protein to the 5′-terminal region of glpD mRNA was found to be important for mRNA stability (18).
Our studies of mRNA decay in B. subtilis have concentrated on the message encoded by the ermC gene, a plasmid-borne erythromycin resistance (Emr) gene. The addition of erythromycin to a strain containing the ermC gene results in 15- to 20-fold stabilization of ermC mRNA, which is due to erythromycin-induced ribosome stalling near the 5′ end of the message (3, 4). In earlier work, it was shown that stabilization was the result only of ribosome stalling and was not dependent on the translation of the body of the message or on a particular structure at the 5′ end. Furthermore, ribosome stalling at the 5′ end conferred stability to diverse downstream sequences, thus defining the ermC leader RNA as a “5′ stabilizer” (2, 11). The stalled ribosome was proposed to confer stability by protecting the 5′ end from the binding of a 5′-end-dependent endoribonuclease. Similar results were obtained by others studying erythromycin-induced ribosome stalling on ermA mRNA (36) and chloramphenicol-induced ribosome stalling on cat mRNA (13).
A deletion version of the ermC gene has been constructed, providing a model mRNA—designated ΔermC mRNA—that is 254 nucleotides (nt) long (14). The small size of ΔermC mRNA makes it a convenient substrate for studies of in vivo decay, including the observation of endonucleolytic cleavage at the site of ribosome stalling (14). Since the previous ermC mRNA studies involved the addition of erythromycin and ribosome stalling, which is not a normal physiological state for this mRNA, we sought to gain a better understanding of mRNA decay processes in B. subtilis by studying the decay of ΔermC mRNA in the absence of erythromycin addition. Here we focus on the effects of translational signals, ribosome transit, and 5′-end accessibility.
MATERIALS AND METHODS
Bacterial strains.
The B. subtilis host was BG1, which is trpC2 thr-5. E. coli DH5α (19) was the host for plasmid constructions. Hosts for M13 phage derivatives were JM109 (42) and CJ236 (27).
Standard procedures.
The preparation and transformation of B. subtilis competent cell cultures (15), β-galactosidase assays (7), and site-directed mutagenesis (27) were as described previously.
Northern blot analysis.
RNA was isolated from B. subtilis cultures grown to mid-logarithmic phase in minimal medium containing Spizizen salts with 0.5% glucose, 0.1% Casamino Acids, 0.001% yeast extract, 0.1% tryptophan, 0.1% threonine, and 1 mM MgSO4 as described previously (3). Northern blot analysis of RNA on 6% denaturing polyacrylmide gels was done as previously described (40). For ermC-lacZ fusions, total RNA was electrophoresed through a 1% formaldehyde-morpholinepropanesulfonic acid (MOPS)-agarose gel and transferred to a nylon membrane. Quantitation of radioactivity in bands on Northern blots was done with a PhosphorImager instrument (Molecular Dynamics). The riboprobe used was complementary to nt 13 to 114 of ΔermC mRNA. The riboprobe was synthesized by T7 RNA polymerase transcription in the presence of [α-32P]UTP with an isolated PCR fragment as a template. The PCR fragment was generated with a downstream primer that contained, at its 5′ end, the T7 RNA polymerase promoter sequence, allowing transcription of the PCR product. After being probed for ΔermC mRNA, the membrane was stripped by being placed in a metal pan containing boiling 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% sodium dodecyl sulfate. The pan was removed from the flame and allowed to cool for 15 min. This procedure was repeated, and then the stripped membrane was probed for 5S rRNA to normalize RNA levels loaded in each lane (31). Probing for 5S RNA was done with a 5′-end-labeled DNA oligonucleotide that was complementary to nt 61 to 80 of 5S RNA. 5S prehybridization and hybridization conditions were as described previously (29).
Plasmids.
ΔermC mRNA was expressed from plasmid pYH196, an E. coli-B. subtilis shuttle plasmid that has been described elsewhere (14). To facilitate the cloning of mutated DNA fragments, an EcoRI site was introduced downstream of the ΔermC transcription terminator, giving plasmid pYH231. NcoI and HindIII sites are present at the beginning of the ΔermC transcription unit (23). These sites were used in conjunction with the downstream EcoRI site to insert fragments of M13 replicative-form DNAs that had been mutated as described by Kunkel et al. (27) or PCR fragments that were generated with mutagenic 5′ primers containing NcoI or HindIII sites. The 27- and 73-nt insertions at the 5′ end were described previously (14). For integration of ΔermC-lacZ fusions at the amyE site, ΔermC DNA fragments were inserted into integration vector pAC5, a chloramphenicol-resistant version of pAC7 (41). Following digestion of the pAC5 construct with NruI, BG1 was transformed to chloramphenicol resistance, and disruption of the amyE locus was tested on starch plates.
Data analysis.
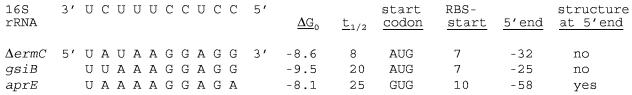
Half-lives were determined by linear regression analysis plots of the percentage of RNA remaining versus time. Half-life data reported here are based on at least three independent experiments in which the R2 value was greater than 0.9. A two-sample t test comparing wild-type and mutant mRNA half-lives was used to calculate P values. A P value of less than 0.05 was considered significant. The free-energy values for 16S rRNA binding (see Fig. 6) were determined at the Zuker RNA website (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form6.cgi) by using a temperature of 37°C, a 0.1 mM RNA concentration, and a 10 mM Na+ concentration.
FIG. 6.
Comparison of translation initiation regions of ΔermC, gsiB, and aprE. At the top left is the 10-nt sequence at the 3′ terminus of 16S rRNA, with the RBS regions of mRNAs lined up below this sequence. Shown at the right are the free energy of the RBS-16S rRNA interaction (ΔG0), the mRNA half-life (t1/2) (in minutes), the start codon sequence, the distance between the RBS and the start codon, the distance between the 5′ end and the start codon, and the presence or absence of a 5′ structure.
RESULTS
Effects of RBS and start codon mutations on ΔermC mRNA stability.
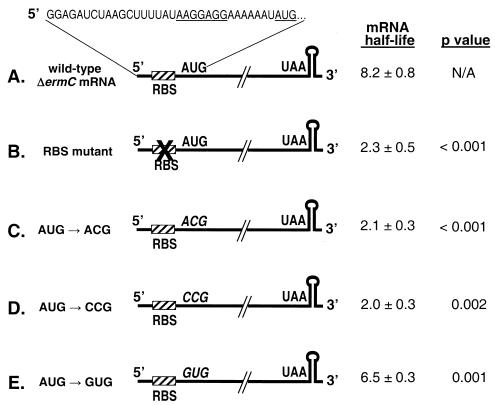
ΔermC mRNA, shown schematically in Fig. 1A, is a 254-nt mRNA that contains a strong RBS, located 19 nt from the 5′ end, followed by a 62-amino-acid-encoding open reading frame (ORF) (14). The ORF consists of the first 16 codons of the ermC leader peptide fused in frame with the last 48 codons of the ermC methylase-encoding sequence. Immediately downstream of the ORF stop codon is a strong transcription terminator structure, with a predicted stability of −42 kcal/mol. From Northern blot analysis of ΔermC mRNA decay after the addition of rifampin, a half-life of 8.2 min was measured (Fig. 2A).
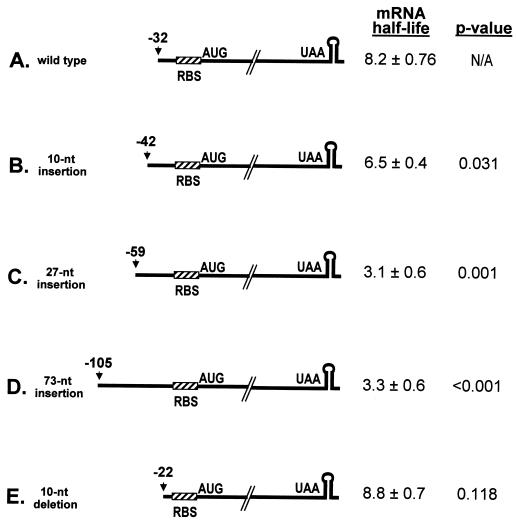
FIG. 1.
Schematic diagrams and half-lives of wild-type ΔermC mRNA and derivatives with a mutated RBS or a mutated start codon. The nucleotide sequence from the transcription start site to the translational start codon is shown at the top, with the RBS and start codon underlined. Half-lives (in minutes) are reported as means and standard deviations. N/A, not applicable.
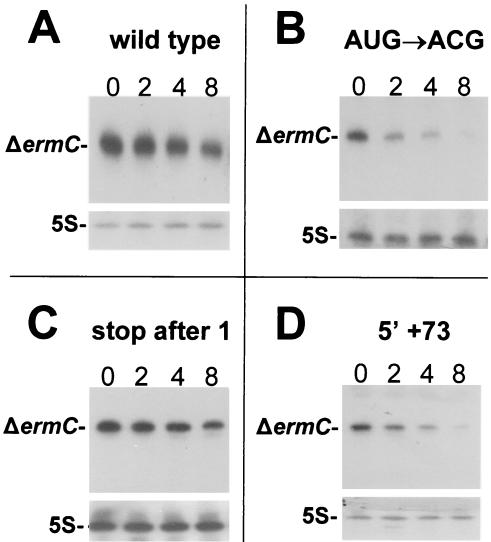
FIG. 2.
Representative Northern blot analyses of ΔermC mRNAs. Total RNA was isolated from a wild-type strain (A), a start codon mutant (B), a mutant with a stop codon inserted immediately after the start codon (C), and a mutant with a 73-nt insertion at the 5′ end (D). RNA was isolated at 0, 2, 4, and 8 min after rifampin addition (times are indicated at top of panels). Also shown are blots reprobed for 5S rRNA, for normalization of the RNA quantity.
To examine the effects of ribosome binding on the stability of ΔermC mRNA, constructs that contained a mutated RBS or a mutated start codon were made. When the RBS sequence was mutated from 5′-AAGGAGG-3′ to 5′-AAGATCT-3′, the half-life of ΔermC mRNA was decreased about fourfold (Fig. 1B). A similar fourfold decrease in the ΔermC mRNA half-life was observed when the AUG start codon was mutated to the nonfunctional ACG or CCG start codon (Fig. 1C and D and 2B). These results suggested that ribosome binding and the formation of the ribosome-mRNA-charged tRNA ternary complex were important for mRNA stability. It was of interest to determine whether a functional but weaker start codon would also affect ΔermC mRNA stability. B. subtilis uses three start codons, AUG, UUG, and GUG, with protein synthesis being about three- to fivefold less efficient with GUG than with AUG as the start codon (39). Mutating the start codon of ΔermC mRNA from AUG to GUG resulted in a modest but significant decrease in the mRNA half-life, from 8.2 to 6.5 min (Fig. 1E).
Effect of ribosome transit on ΔermC mRNA stability.
We next examined whether ribosome transit along ΔermC mRNA was critical for its stability. UAA stop codons were inserted in frame at various positions along the ΔermC mRNA ORF (Fig. 3). Initially, we made insertion constructs with the stop codon located more distal from the start codon (Fig. 3A to C). However, as these resulted in wild-type half-lives, the site of insertion was moved ever closer to the start codon (Fig. 3D to F). Surprisingly, even when a stop codon was inserted directly after the start codon, such that no peptide bond formation could take place, the mRNA was as stable as the wild type (Fig. 2C and 3G).
FIG. 3.
Schematic diagrams and half-lives of ΔermC mRNAs with stop codon insertions. Half-lives (in minutes) are reported as means and standard deviations.
The efficiency of translation termination can vary depending on the sequence of the stop codon encountered and the surrounding context of the stop codon (32, 33, 38). To confirm that translation was terminating efficiently at the inserted stop codons, translational fusions that placed the E. coli lacZ coding sequence after codon 47 of ΔermC mRNA were created. ΔermC-lacZ fusions were made for the wild type and for derivatives with stop codons after codons 1, 2, 5, and 17. These constructs were integrated into the amyE locus of B. subtilis, and β-galactosidase assays were performed to assess stop codon efficiency. In all instances, the amounts of β-galactosidase activity from the constructs containing a premature stop codon were less than 0.6% that from the wild type (data not shown). Translation was therefore shown to be efficiently terminated in the RNAs with inserted stop codons, yet there was no decrease in half-life. These results indicated that, although ribosome binding and, by inference, the formation of a ternary complex were critical for the stability of this mRNA, actual translation was not required.
Translation and stability of a larger mRNA.
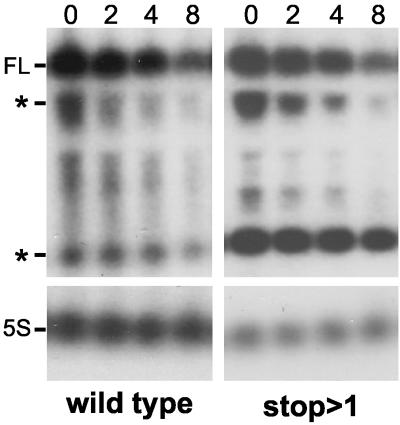
It could be argued that the small size of ΔermC mRNA provided fewer target sites for the initiation of decay and that a longer transcript would be susceptible to rapid decay in the absence of translation. The ermC-lacZ mRNAs described above were suitable for addressing this issue. The ΔermC-lacZ transcript was approximately 3.2 kb long, about 12 times longer than ΔermC mRNA. Northern blot analysis was performed with wild-type ΔermC-lacZ mRNA and ΔermC-lacZ RNA containing a stop codon immediately after the start codon (Fig. 4). The wild-type ΔermC-lacZ mRNA had a half-life of 6.3 ± 0.73 (mean and standard deviation) min, while the half-life of the untranslated ΔermC-lacZ RNA was 6.8 ± 0.84 min (P = 0.414) (Fig. 4). We conclude from these data that ribosome transit did not play a role in determining stability, even for a longer RNA.
FIG. 4.
Northern blot analysis of ΔermC-lacZ mRNA. The migration of full-length ΔermC-lacZ mRNA (FL) and two prominent intermediates (marked by asterisks) is indicated. Similar decay intermediates from other ermC-lacZ fusions have been observed elsewhere (11). The greater prominence of the lower decay intermediate in the mutant with the stop after position 1 may be due to increased processing by a 3′ exonuclease in the absence of competing ribosome flow. Values at top of panels indicate times in minutes.
Half-lives of ΔermC mRNAs with 5′ additions or deletions.
The results so far suggested that ternary complex formation was required for wild-type stability. It was reasonable to assume that the presence of the ternary complex near the 5′ end offered protection against the initiation of decay, possibly by interfering with the binding of a 5′-end-dependent endonuclease. The 5′ end of ΔermC mRNA is located at −32 (i.e., 32 nt upstream of the A in the start codon). We hypothesized that increasing the distance from the 5′ end to the start codon would result in less protection of the 5′ end and a decreased mRNA half-life, whereas decreasing this distance might result in an increased mRNA half-life (assuming that the efficiency of ribosome binding would not be affected by changes in the sequence and distance from the 5′ end). To test this hypothesis, the distance from the 5′ end was increased by adding 10, 27, and 73 nt between the transcriptional and translational start sites (Fig. 5). These nucleotides were selected randomly from the E. coli lacZ coding sequence and were not predicted to form any significant secondary structure when inserted near the 5′ end of ΔermC mRNA. Interestingly, the mRNA with the 10-nt insert showed a small decrease in half-life (6.5 min), and the mRNAs with inserts of 27 and 73 nt showed even shorter half-lives (3.1 and 3.3 min, respectively) (Fig. 2D and 5B to D). Thus, increasing the distance from the 5′ end to the start codon, at least up to 59 nt, resulted in a corresponding decrease in the half-life. In another experiment, the distance from the 5′ end to the start codon was decreased by 10 nt (Fig. 5E). In this experiment, the mRNA half-life was not significantly different from that of the wild type.
FIG. 5.
Schematic diagrams and half-lives of ΔermC mRNAs with 5′ insertions and a deletion. The distance from the 5′ end to the start codon is indicated above the arrows. Half-lives (in minutes) are reported as means and standard deviations. N/A, not applicable.
DISCUSSION
In this work, we have analyzed several translation parameters and their relevance to the decay of ΔermC mRNA, a constitutively transcribed, 254-nt mRNA. The factors that were considered were the RBS, start codon, extent of translation of the ΔermC ORF, and location of the RBS or start codon relative to the 5′ end. Although it would be difficult to extrapolate from a study of ΔermC mRNA alone to B. subtilis mRNAs in general, we believe that intensive study of a representative mRNA species can be informative as to the primary mechanism of decay initiation.
The 8.2-min half-life of ΔermC mRNA is at the high end expected for bacterial mRNAs. In a recent global analysis of the half-lives of mRNAs from E. coli grown in minimal medium, it was found that 80% of individual mRNA half-lives were between 2 and 8 min, with a mean half-life of 5.7 min (5). The ΔermC mRNA RBS consists of a 7-nt sequence complementary to the 3′-end sequence of 16S rRNA (Fig. 6) and is surrounded by AU-rich sequences on both sides (Fig. 1, upper portion). The absence of G and C residues around the RBS is thought to be optimal for ribosome binding (21). In addition, the small size of ΔermC mRNA may offer fewer internal decay initiation sites. Although we believe that the initiation of decay in B. subtilis is controlled primarily by access to the 5′ end (see below), alternative mechanisms operating through direct internal entry are also likely to occur. Such alternative mechanisms would explain the decreased half-life of ΔermC-lacZ mRNA (6.3 min, as opposed to 8.2 min for ΔermC mRNA). Nevertheless, the fact that ΔermC-lacZ mRNA is about 12 times the size of ΔermC mRNA but showed only a 23% decrease in half-life demonstrates the overriding importance of the 5′ end. Furthermore, the fact that there was no difference in the half-lives of translated and untranslated ΔermC mRNA and ΔermC-lacZ mRNA suggests that any minor degradation pathway is not affected by ribosome flow.
Our study goes well beyond two earlier reports about the stability of B. subtilis mRNAs with regard to translational signals. In one report (25), the 20-min half-life of gsiB mRNA, encoding a stress response protein, was shown to be dependent on a strong RBS. Changing 3 nt of the gsiB RBS resulted in a fourfold decrease in the half-life. The 5′ end of gsiB is at −25; thus, the results obtained with gsiB mRNA are very similar to our results, in which the RBS mutation resulted in a fourfold decrease in the half-life. Other translational effects on stability were not determined for gsiB mRNA. Figure 6 shows that the gsiB RBS is predicted to have a higher free energy for binding 16S rRNA than the ΔermC RBS. Whether the difference in the free energy of RBS interactions between ΔermC and gsiB (−8.6 and −9.5 Kcal/mol, respectively) accounts for the large difference in the mRNA half-lives (8 and 20 min) is a matter of speculation. This notion could be tested by substituting the gsiB RBS sequence with that of ΔermC and vice versa.
The other report (22), which appeared as our studies of ΔermC mRNA were in progress, looked at the stability of aprE mRNA, which has a half-life of ≥25 min. aprE mRNA also showed a fourfold decrease in stability when one or two residues of its RBS were mutated. Another result similar to ours was that the insertion of a stop codon after aprE codon 4 did not affect stability, demonstrating that translation of the coding sequence was not needed for wild-type stability. On the other hand, mutating the aprE start codon resulted in only a small decrease in half-life, from ≥25 min to 18 min, a finding which is very different from our finding that mutating the ΔermC mRNA start codon had as great a negative effect on the half-life as mutating the RBS. This difference can be explained by the fact that the aprE start codon is GUG, which we have shown gives less stability than the AUG start codon when present in ΔermC mRNA (Fig. 1E). Thus, the aprE start codon may not contribute as much to the relative stability of aprE mRNA as the AUG start codon does to ΔermC mRNA. More importantly, the aprE mRNA has a 5′-terminal secondary structure, which is very important for stability; weakening of this structure resulted in a fivefold decrease in stability (22). It is likely that the stability of aprE mRNA is determined primarily by this structure, since the strength of the RBS interaction, the GUG start codon, and the suboptimal spacing between the RBS and the start codon (Fig. 6) all suggest weak ribosome binding. Thus, the negative effect of interfering with ternary complex formation on aprE mRNA may be mitigated by the presence of a 5′-terminal secondary structure. Analysis of the 5′ untranslated region of ΔermC mRNA with the GCG FOLD program (Wisconsin Package version 10.0; Genetics Computer Group, Madison, Wis.) predicted no secondary structure for this sequence.
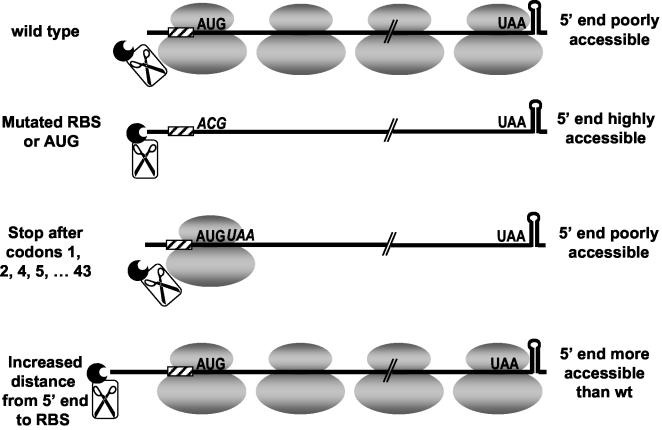
The recent report of Yusupova et al. on the path of mRNA through the ribosome (43) is germane to thinking about mRNA protection achieved by ribosome binding. The ribosome was shown to protect a region of mRNA from −15 to +16 (the A residue of the AUG start codon is at +1), similar to much earlier estimates of −16 to +16, which were based on the protection of ribosome-bound mRNA from endonuclease digestion (37). According to these data, we can estimate that the binding of a ribosome in a ternary complex on ΔermC mRNA would leave about 16 or 17 nt unprotected at the 5′ end. Based on earlier studies of RNase E-dependent decay in E. coli, one might expect that a 5′-terminal, single-stranded stretch of 16 or 17 nt would render the mRNA susceptible to rapid decay. In E. coli studies, it was demonstrated that, although 5′-terminal secondary structures could function as 5′ stabilizers, single-stranded extensions of 4 nt or more upstream of the structure decreased RNA stability (6, 16). To explain why the predicted 5′-terminal, single-stranded sequence of ΔermC mRNA does not lead to instability, one could propose that, although the 5′-terminal 16 or 17 nt may not be in the mRNA tunnel of the ribosome, they could be in contact with the surface of the ribosome, thus limiting accessibility to a decay-initiating RNase. A model for our results, based on this view of ribosome binding and mRNA stability, is presented in Fig. 7. According to this model, the mRNA half-life is determined primarily by access of the 5′ end to a 5′-end-dependent endoribonuclease. A flow of ribosomes that bind near the 5′ end protects the mRNA from RNase binding. Formation of the ternary complex alone is sufficient for the ribosome to compete with the RNase; the protective effect of a bound ternary complex would be evident even in the complete absence of downstream translation. Earlier conclusions regarding the ability of erythromycin-induced ribosome stalling to stabilize downstream RNA sequences (4, 11) can thus be viewed as a special case of the protective effect of ribosome binding.
FIG. 7.
Model for interference with binding of the decay-initiating, 5′-end-dependent RNase. The RNase is depicted as a 5′-binding pocket attached to scissors representing the endonuclease activity.
An alternative hypothesis for explaining why an unbound 5′-terminal stretch might not render the mRNA susceptible to rapid decay has to do with the nature of the putative RNase E-like endoribonuclease that is thought to exist in B. subtilis (10). From comparisons of 5′-end-dependent decay of mRNAs in E. coli and B. subtilis, it has been proposed that the RNase E-like RNase of B. subtilis may have a different mechanism for translocating from the 5′ end to an internal cleavage site (9, 24). While E. coli RNase E can loop around a 5′-proximal bound ribosome or an RNA secondary structure to cleave the mRNA at a downstream site, the putative, functionally equivalent B. subtilis RNase may track along the mRNA to its cleavage site. Thus, the initiation of decay is hindered by a 5′-proximal ribosome to a greater degree in B. subtilis. Testing of such models awaits the identification of a decay-initiating enzyme in B. subtilis.
Acknowledgments
This work was supported by Public Health Service grant GM-48804 from the National Institutes of Health.
We thank Jeanne DiChiara for making the AUG→ACG and AUG→CCG mutants as well as the mutants with stop codons after codons 2 and 5.
REFERENCES
- 1.Arnold, T. E., J. Yu, and J. G. Belasco. 1998. mRNA stabilization by the ompA 5′ untranslated region: two protective elements hinder distinct pathways for mRNA degradation. RNA 4:319-330. [PMC free article] [PubMed] [Google Scholar]
- 2.Bechhofer, D. H. 1993. 5′ mRNA stabilizers, p. 31-52. In J. G. Belasco and G. Brawerman (ed.), Control of mRNA stability. Academic Press, Inc., San Diego, Calif.
- 3.Bechhofer, D. H., and D. Dubnau. 1987. Induced mRNA stability in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 84:498-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bechhofer, D. H., and K. H. Zen. 1989. Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J. Bacteriol. 171:5803-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouvet, P., and J. G. Belasco. 1992. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature 360:488-491. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, J., A. A. Guffanti, W. Wang, T. A. Krulwich, and D. H. Bechhofer. 1996. Chromosomal tetA(L) gene of Bacillus subtilis: regulation of expression and physiology of a tetA(L) deletion strain. J. Bacteriol. 178:2853-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coburn, G. A., and G. A. Mackie. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acids Res. Mol. Biol. 62:55-108. [DOI] [PubMed] [Google Scholar]
- 9.Condon, C. 2003. RNA processing and degradation in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 67:157-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condon, C., H. Putzer, D. Luo, and M. Grunberg-Manago. 1997. Processing of the Bacillus subtilis thrS leader mRNA is RNase E-dependent in Escherichia coli. J. Mol. Biol. 268:235-242. [DOI] [PubMed] [Google Scholar]
- 11.DiMari, J. F., and D. H. Bechhofer. 1993. Initiation of mRNA decay in Bacillus subtilis. Mol. Microbiol. 7:705-717. [DOI] [PubMed] [Google Scholar]
- 12.Donovan, W. P., and S. R. Kushner. 1986. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 83:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreher, J., and H. Matzura. 1991. Chloramphenicol-induced stabilization of cat messenger RNA in Bacillus subtilis. Mol. Microbiol. 5:3025-3034. [DOI] [PubMed] [Google Scholar]
- 14.Drider, D., J. M. DiChiara, J. Wei, J. S. Sharp, and D. H. Bechhofer. 2002. Endonuclease cleavage of messenger RNA in Bacillus subtilis. Mol. Microbiol. 43:1319-1329. [DOI] [PubMed] [Google Scholar]
- 15.Dubnau, D., and R. Davidoff-Abelson. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J. Mol. Biol. 56:209-221. [DOI] [PubMed] [Google Scholar]
- 16.Emory, S. A., P. Bouvet, and J. G. Belasco. 1992. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 6:135-148. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh, S., and M. P. Deutscher. 1999. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc. Natl. Acad. Sci. USA 96:4372-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glatz, E., M. Persson, and B. Rutberg. 1998. Antiterminator protein GlpP of Bacillus subtilis binds to glpD leader mRNA. Microbiology 144:449-456. [DOI] [PubMed] [Google Scholar]
- 19.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193-227. [DOI] [PubMed] [Google Scholar]
- 21.Hager, P. W., and J. C. Rabinowitz. 1985. Translational specificity in B. subtilis, p. 1-31. In D. Dubnau (ed.), The molecular biology of the bacilli. Academic Press, Inc., San Diego, Calif.
- 22.Hambraeus, G., K. Karhumaa, and B. Rutberg. 2002. A 5′ stem-loop and ribosome binding but not translation are important for the stability of Bacillus subtilis aprE leader mRNA. Microbiology 148:1795-1803. [DOI] [PubMed] [Google Scholar]
- 23.Hue, K. K., S. D. Cohen, and D. H. Bechhofer. 1995. A polypurine sequence that acts as a 5′ mRNA stabilizer in Bacillus subtilis. J. Bacteriol. 177:3465-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joyce, S. A., and M. Dreyfus. 1998. In the absence of translation, RNase E can bypass 5′ mRNA stabilizers in Escherichia coli. J. Mol. Biol. 282:241-254. [DOI] [PubMed] [Google Scholar]
- 25.Jurgen, B., T. Schweder, and M. Hecker. 1998. The stability of mRNA from the gsiB gene of Bacillus subtilis is dependent on the presence of a strong ribosome binding site. Mol. Gen. Genet. 258:538-545. [DOI] [PubMed] [Google Scholar]
- 26.Kennell, D. 2002. Processing endoribonucleases and mRNA degradation in bacteria. J. Bacteriol. 184:4645-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 28.Kushner, S. R. 2002. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 184:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Z., and M. P. Deutscher. 1995. The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. Proc. Natl. Acad. Sci. USA 92:6883-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackie, G. A. 2000. Stabilization of circular rpsT mRNA demonstrates the 5′-end dependence of RNase E action in vivo. J. Biol. Chem. 275:25069-25072. [DOI] [PubMed] [Google Scholar]
- 31.Makarova, O. V., E. M. Makarov, R. Sousa, and M. Dreyfus. 1995. Transcribing of Escherichia coli genes with mutant T7 RNA polymerases: stability of lacZ mRNA inversely correlates with polymerase speed. Proc. Natl. Acad. Sci. USA 92:12250-12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mottagui-Tabar, S., M. F. Tuite, and L. A. Isaksson. 1998. The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem. 257:249-254. [DOI] [PubMed] [Google Scholar]
- 33.Pavlov, M. Y., D. V. Freistroffer, V. Dincbas, J. MacDougall, R. H. Buckingham, and M. Ehrenberg. 1998. A direct estimation of the context effect on the efficiency of termination. J. Mol. Biol. 284:579-590. [DOI] [PubMed] [Google Scholar]
- 34.Petersen, C. 1993. Translation and mRNA stability in bacteria: a complex relationship, p. 117-147. In J. G. Belasco and G. Brawerman (ed.), Control of mRNA stability. Academic Press, Inc., San Diego, Calif.
- 35.Regnier, P., and C. M. Arraiano. 2000. Degradation of mRNA in bacteria: emergence of ubiquitous features. Bioessays 22:235-244. [DOI] [PubMed] [Google Scholar]
- 36.Sandler, P., and B. Weisblum. 1989. Erythromycin-induced ribosome stall in the ermA leader: a barricade to 5′-to-3′ nucleolytic cleavage of the ermA transcript. J. Bacteriol. 171:6680-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steitz, J. A. 1969. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature 224:957-964. [DOI] [PubMed] [Google Scholar]
- 38.Tate, W. P., E. S. Poole, J. A. Horsfield, S. A. Mannering, C. M. Brown, J. G. Moffat, M. E. Dalphin, K. K. McCaughan, L. L. Major, and D. N. Wilson. 1995. Translational termination efficiency in both bacteria and mammals is regulated by the base following the stop codon. Biochem. Cell Biol. 73:1095-1103. [DOI] [PubMed] [Google Scholar]
- 39.Vellanoweth, R. L., and J. C. Rabinowitz. 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 6:1105-1114. [DOI] [PubMed] [Google Scholar]
- 40.Wang, W., and D. H. Bechhofer. 1997. Bacillus subtilis RNase III gene: cloning, function of the gene in Escherichia coli, and construction of Bacillus subtilis strains with altered rnc loci. J. Bacteriol. 179:7379-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinrauch, Y. M., T. Msadek, F. Kunst, and D. Dubnau. 1991. Sequence and properties of comQ, a new competence regulatory gene of Bacillus subtilis. J. Bacteriol. 173:5685-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 43.Yusupova, G. Z., M. M. Yusupov, J. H. Cate, and H. F. Noller. 2001. The path of messenger RNA through the ribosome. Cell 106:233-241. [DOI] [PubMed] [Google Scholar]