Abstract
We compared the transcriptome, proteome, and nucleotide sequences between the parent strain Escherichia coli W3110 and the l-threonine-overproducing mutant E. coli TF5015. DNA macroarrays were used to measure mRNA levels for all of the genes of E. coli, and two-dimensional gel electrophoresis was used to compare protein levels. It was observed that only 54 of 4,290 genes (1.3%) exhibited differential expression profiles. Typically, genes such as aceA, aceB, icdA, gltA, glnA, leu operon, proA, thrA, thrC, and yigJ, which are involved in the glyoxylate shunt, the tricarboxylic acid cycle, and amino acid biosynthesis (l-glutamine, l-leucine, proline, and l-threonine), were significantly upregulated, whereas the genes dadAX, hdeA, hdeB, ompF, oppA, oppB, oppF, yfiD, and many ribosomal protein genes were downregulated in TF5015 compared to W3110. The differential expression such as upregulation of thr operon and expression of yigJ would result in an accumulation of l-threonine in TF5015. Furthermore, two significant mutations, thrA345 and ilvA97, which are essential for overproduction of l-threonine, were identified in TF5015 by the sequence analysis. In particular, expression of the mutated thrABC (pATF92) in W3110 resulted in a significant incremental effect on l-threonine production. Upregulation of aceBA and downregulation of b1795, hdeAB, oppA, and yfiD seem to be linked to a low accumulation of acetate in TF5015. Such comprehensive analyses provide information regarding the regulatory mechanism of l-threonine production and the physiological consequences in the mutant stain.
In recent years, the completion of the genome project on numerous organisms has accelerated the development of very powerful tools for functional genomics such as DNA arrays (6) and two-dimensional gel electrophoresis (31). Comparative analysis of the gene expression profiles has provided extensive biological information on a genome scale regarding response to stress and/or environmental change, dissection of regulatory circuitry, drug target characterization or identification, cellular response to bacterial infection, and other information for many organisms, including Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, and human cells (1, 6, 22, 25, 38). In addition to studies of transcription levels, proteome analysis is important in the understanding of global regulatory processes in living organisms (13, 14, 17, 19, 31) since the gene expression profiles often do not directly relate to protein expression levels (28). In this sense, functional genomic techniques, along with genomic information, may enable us to unravel the global regulatory processes or complex metabolic networks in living organisms (18), consequently offering a comprehensive blueprint of the physiology of the bacterium (17, 19, 22, 38).
Amino acids have been the prominent target metabolites from microorganisms in bioindustry due to large commercial demands for flavor enhancers, animal feed, sweeteners, and therapeutic agents. Of them, l-threonine, one of the essential amino acids, is widely used as a feed and food additive, and various industrial strains that more efficiently produce l-threonine have been successfully developed by traditional approaches, including the deregulation of enzymes, the elimination of competitive pathways, and the amplification of genes (7, 20, 26, 36). In a previous study, we developed the l-threonine-producing Escherichia coli strain TF5015 via recursive mutations (24). This strain requires both l-methionine and l-isoleucine for growth and shows resistance to various chemical analogues. It was suggested that the mechanism of l-threonine production of TF5015 probably resulted from releasing the feedback regulation and blocking the carbon flow into undesirable by-products. Regulation mechanism involved in l-threonine biosynthesis in E. coli has been relatively well characterized (30). However, to understand the overall regulatory mechanism and the physiological events in response to the accumulation of l-threonine in TF5015, detailed information, including the cellular regulations, entire metabolic fluxes, and dynamic responses of the complex metabolic networks is crucial.
In the present study, to gain further insight into the global regulatory mechanism for l-threonine biosynthesis, we carried out the comparative analyses of transcriptome, proteome, and nucleotide sequences between the prototrophic E. coli W3110 and the l-threonine-producing E. coli TF5015. Expression patterns of the genes and proteins were investigated for both strains by using DNA macroarrays containing virtually every gene of E. coli and two-dimensional gel electrophoresis. The profiles were analyzed in terms of the accumulation of l-threonine and physiological consequences in the mutant strain. The details of these analyses are presented below.
MATERIALS AND METHODS
Bacterial strains.
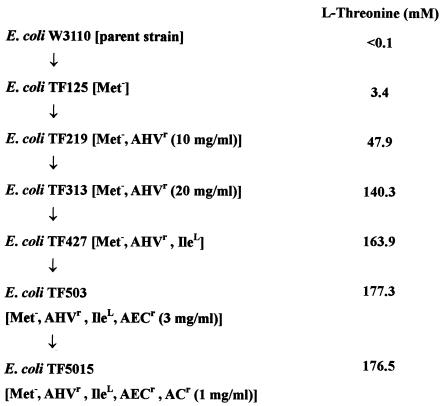
E. coli W3110 [F− IN(rrnD-rrnE) rpoS+], which produces a negligible level of threonine (<0.1 mM), was used as a parent strain for comparative analysis (24). The threonine-producing mutant E. coli TF5015 (Met− IleL AHVr AECr ACr rpoS+) was obtained from a previously constructed threonine-producing E. coli strain, TF427, by mutations produced by using N-methyl-N′-nitro-N-nitrosoguanidine and shows resistance to 20 mg of α-amino-β-hydroxyvaleric acid (AHV; threonine analogue), 3 mg of 2-aminoethyl-l-cysteine (AEC; lysine analogue), and 1 mg of l-azetidine-2-carboxylic acid (AC; proline analogue)/ml (Fig. 1) (24). This strain requires both methionine and isoleucine for growth. E. coli Hfr 3000 YA73 (thrB relA1 spoT1 thi-1) was used for cloning of the thr operon (39).
FIG. 1.
Genealogy of E. coli mutants producing l-threonine from E. coli W3110. Abbreviations: Met−, l-methionine auxotroph; AHVr, resistant to α-amino-β-hydroxyvaleric acid; AECr, resistant to 2-aminoethyl-l-cysteine; IleL, l-isoleucine leaky character; ACr, resistant to l-azetidine-2-carboxylic acid. The production of l-threonine was conducted in a 250-ml flask (24).
Media and culture conditions.
Strains were routinely grown in Luria-Bertani medium or M9 medium (34). If necessary, ampicillin was used at a final concentration of 100 μg/ml. For extraction of total RNA and protein, strains W3110 and TF5015 were grown in a 5-liter jar fermentor containing 1.5 liter of fermentation medium [70 g of glucose, 10 g of (NH4)2SO4, 2 g of KH2PO4, 0.5 g of MgSO4·7H2O, 5 mg of FeSO4·7H2O, 5 mg of MnSO4·4H2O, 3 g of yeast extract, and 800 mg of methionine per liter of water at pH 6.0] (24). A seed culture was grown at 33°C for 4 h in a 500-ml flask containing 75 ml of modified Luria-Bertani medium and then inoculated into a 5-liter jar fermentor. During the cultivation, a mixture of glucose and phosphate at final concentrations of 60 g and 0.5 g/liter, respectively, was fed two times when the glucose level was <5 g/liter. The culture conditions were as follows: the pH was kept at 6.0 with NH4OH, the temperature was kept at 31°C, the aeration rate was 1 vvm (air volume/working volume/minute), and the agitation speed was 800 rpm. Production of threonine by a recombinant strain was conducted in flasks under optimized conditions as described in our previous work (24).
RNA purification.
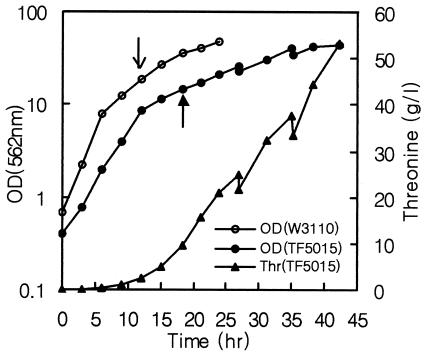
For analysis of transcription levels between the prototrophic strain and threonine-producing mutant at the threonine production stage, both strains were cultivated to an early stationary phase in the fermentation medium (Fig. 2), and culture broth was taken for extraction and purification of total RNA by using RNeasy kits (Qiagen) according to the procedures reported elsewhere (1, 17, 32, 38). Briefly, 0.2 ml of culture broth was lysed in boiling lysis buffer (1), and the resulting solution was extracted twice by using acidic hot phenol (pH 4.3) at 65°C with vigorous vortex and incubation for 5 min, followed by acid phenol-chloroform (5:1) extraction. The RNA was precipitated with ethanol, redissolved in water, treated with DNase I (Ambion), and applied to RNeasy column. The purified RNA was redissolved in RNase-free water and stored at −72°C. The absence of genomic DNA was confirmed according to the reported procedure (32). The RNA concentration was determined by using the spectrophotometer.
FIG. 2.
Growth curve of E. coli W3110 and TF5015 in the fermentation medium. Cells were harvested at the time indicated in arrows.
cDNA labeling, hybridization, and scanning.
RNA was labeled according to the general procedure described elsewhere (17, 32, 38). One microgram of total RNA was mixed with E. coli cDNA labeling primers (Sigma-GenoSys, Inc.) in a final volume of 15 μl. The reaction mixture was heated to 90°C for 2 min and then cooled to 42°C over a period of 20 min. Fifty units of avian myeloblastosis virus reverse transcriptase, together with 20 μCi of [α-33P]dCTP (≥2,500 Ci/mmol; Amersham-Pharmacia Biotech) was added to the cDNA labeling mixture, followed by incubation for 2 h 30 min at 42°C. Unincorporated-radiolabeled nucleotides were removed by applying the reaction mixture to a Sephadex G-25 gel filtration spin column.
Hybridizations were performed in roller bottles (3.5 by 30 cm) in a hybridization oven. After the blots were rinsed in 50 ml of 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) for 5 min, the Panorama gene arrays (Sigma-GenoSys) were prehybridized for 2 h at 65°C in 5 ml of hybridization solution (17, 32, 38). The radiolabeled cDNA mixture was added to the hybridization solution and preincubated at 90 to 95°C for 10 min in a water bath. The denatured labeled cDNA in hybridization solution was added to the blots in the roller bottles and hybridized at 65°C for 18 h. After the hybridization solution was decanted, the array membranes were first washed with 50 ml of wash solution three times for 5 min each time at room temperature, followed by a three 20-min washes with 100 ml of prewarmed wash solution at 65°C. The membranes were then wrapped in clear wrap and exposed to a phosphorimager screen (BAS-SR2025 imaging plate [20 by 40 cm]; Fuji Photo Film Co., Ltd.) for 24 to 72 h. The exposed imaging screens were scanned with a pixel size of 50 μm for greater resolution of spots on an FLA 3000 phosphorimager (Fuji).
Analysis of DNA arrays.
The spot intensities were analyzed by using ArrageGauge software from Fuji. Background values were automatically subtracted by using the global subtraction method. The density values for the duplicate spots were averaged and then normalized by the global normalization method described in the software. In this case, the sums of total spot densities, including data and controls, were the same in the two arrays. Identification of each gene and its functional grouping was carried out by using the manufacturer's E. coli array information file. The correlation coefficients of spot intensities were calculated from the duplicate spots on the same membrane and from the duplicate experiments with total RNA purified from TF5015, and the coefficients ranged from 0.997 to 0.999 and from 0.910 to 0.980, respectively. The expression ratio (relative intensity of genes in TF5015 to those in W3110) for genes showing confidence levels higher than 99.9% in two sets of experiments was 1.78; thus, the genes showing a level greater than this value were considered to be upregulated. The genes with an expression ratio less than 0.56 were regarded as downregulated. In addition to above criteria, genes whose signal intensities were lower than the mean background intensity plus three standard deviations in both W3110 and TF5015 samples were excluded (32).
Two-dimensional gel electrophoresis and mass spectrometric analysis.
Two-dimensional gel electrophoresis was carried out according to a procedure described elsewhere (13, 14, 17) with a slight modification. For protein extraction, cells grown on fermentation medium were harvested at the same growth phase as for total RNA preparation. A 2-ml portion of culture broth was centrifuged at 2,500 × g and 4°C for 5 min and then washed four times with a wash solution (14). The resulting pellet was resuspended in 10 mM Tris buffer (pH 8.0) containing 1 mM phenylmethylsulfonyl fluoride, sonicated for 2 min on ice, and centrifuged at 10,000 × g for 30 min. The supernatant was treated with 1 U of DNase I at 37°C for 15 min, centrifuged for 30 min at 10,000 × g and 4°C, and dialyzed in 5 mM Tris buffer overnight at 4°C. After protein quantification, 30-, 50-, and 100-μg portions of protein extracts were dried by vacuum centrifugation and stored at −72°C until use.
Extracted protein dissolved in 400 μl of rehydration solution (8 M urea; CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}; 18 mM dithiothreitol; 0.5% IPG buffer at pH 4 to 7 L or pH 3 to 10 NL [Amersham-Pharmacia Biotech]) was loaded onto 24-cm IPG strips (pH 4 to 7 L or pH 3 to 10 NL type [Amersham-Pharmacia Biotech]). After rehydration of strips for 12 h, proteins were focused by using a series of voltage increases at 500 V for 1 h, 1,000 V for 1 h, and 8,000 V for 12 h. The second dimension was carried out in a 12.5% polyacrylamide gel (Ettan DALT gel, 255 by 196 by 1 mm [Amersham-Pharmacia Biotech]). After fixation and silver staining, the wet gel was scanned with an ImageScanner and quantified with ImageMaster 2D Elite software (Amersham Pharmacia Biotech). Spot densities were determined on three gel images from duplicate cultures and then normalized to the total spot volume of the protein spots on each gel.
Protein spots showing distinctly different expression patterns were excised and destained by a silver stain-destain protocol (Scripps Center for Mass Spectrometry [http://masspec.scripps.edu/services/protein/]) and subjected to gel digestion with 20 ng of sequencing-grade trypsin (Boehringer Mannheim)/μl in 8 μl of 2.5 mM Tris-HCl at pH 8.5 (13, 14, 35). The resulting samples were analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (PE Biosystems Voyager System 4095 [PerSeptive Biosystems, Inc.]). Peptide fragment peaks (at 805.416 and 2,163.056 m/z) produced by autodigestion of trypsin were used as an internal calibration. Proteins were identified by using the ProteinProspector server (http://prospector.ucsf.edu/) and by comparison with the E. coli reference gel in the SWISS-2DPAGE database (http://kr.expasy.org/ch2d/).
Recombinant DNA techniques.
DNA manipulations and PCR were carried out by standard procedures (34). Chromosomal DNA and PCR fragments were purified by using a Wizard genomic DNA purification kit (Promega) and a QIAquick gel extraction kit (Qiagen), respectively. Fifteen genes were amplified by PCR with Pfu DNA polymerase (Strategene) from genomic DNA of E. coli W3110 and TF5015. The nucleotide sequence was determined by the dideoxy chain termination method (34).
Cloning of thrABC operon.
The wild-type and mutated thrABC operons were cloned in a pBR322 plasmid (New England Biolabs) by complementation of E. coli mutant. Genomic DNAs of E. coli W3110 and TF5015 were completely digested with HindIII and BamHI, and DNA fragments ranging from 5 to 7 kb were isolated, respectively. The digested fragments were ligated with 4.3-kb pBR322/HindIII/BamHI and transformed into thrB auxotrophic E. coli Hfr 3000 YA73 (39). The plasmids, pAW88 (10.5kb) and pATF92 (10.5kb) containing wild-type and mutated thr operons, respectively, were obtained from thr+ transformants. The presence of thrABC genes and its regulatory region in pAW88 and pATF92 was confirmed by DNA sequencing. The constructed plasmids were introduced into W3110 by electroporation (34).
Analysis.
Cell growth was monitored by measuring the optical density at 562 nm by using a spectrophotometer (Beckman DU650). Amino acids were analyzed by high-pressure liquid chromatography (Waters). Acetic acid was determined by using a DX-600 ion chromatograph (Dionex).
RESULTS
Gene expression profiling of E. coli W3110 and E. coli TF5015.
Gene expression levels were monitored on a genome-wide scale for the parent strain W3110 and the mutant TF5015 at the early stationary phase by using DNA macroarray membranes containing every gene of E. coli. This membrane has duplicated spots for each gene, and each experiment was repeated twice. Therefore, four data sets were used for transcriptome analysis. When we compared the spot intensities measured from duplicate spots on a membrane and two sets of experiments by using total RNA purified from TF5015, the consistency of the array data was very good within the same membrane and between membranes. A genome-wide comparison between W3110 and TF5015 revealed that only 54 genes exhibit differential expression profiles (Table 1). Of these genes, the mRNA levels of 19 genes increased, and 35 genes displayed diminished expression levels in TF5015 compared to W3110. When the whole genome of E. coli was grouped according to their functions, most of the upregulated genes were found to be associated with amino acid biosynthesis or metabolism, central intermediary metabolism, and energy metabolism (Table 2). The genes linked with translation or posttranslational modification, amino acid biosynthesis or metabolism, cell structure, and transport or binding protein were downregulated.
TABLE 1.
Genes showing a significant differential expression
| Expression type and gene | Protein | Relative fold increase |
|---|---|---|
| Increased expression | ||
| aceA | isocitrate lyase | 2.1 |
| aceB | malate synthase A | 5.3 |
| aldA | lactaldehyde dehydrogenase A | 2.1 |
| b0427 | hypothetical protein | 2.3 |
| b2530 | hypothetical protein | 1.8 |
| fimA | type 1 fimbrial subunit | 2.0 |
| glnA | glutamine synthetase | 2.6 |
| gltA | citrate synthase | 2.0 |
| icdA | isocitrate dehydrogenase | 2.2 |
| infB | protein chain initiation factor 2 | 1.8 |
| leuA | 2-isopropylmalate synthase | 2.4 |
| leuB | 3-isopropylmalate dehydrogenase | 2.4 |
| leuC | 3-isopropylmalate dehydratase | 1.8 |
| leuD | 3-isopropylmalate dehydratase | 1.9 |
| mdoB | phosphoglycerol transferase I | 1.8 |
| proA | GSA dehydrogenase | 3.0 |
| thrA | AKI-HDI | 6.0 |
| thrC | threonine synthase | 3.2 |
| yigJ | hypothetical 13.3-kDa protein | 2.0 |
| Decreased expression | ||
| asd | aspartate semialdehyde dehydrogenase | 1.8 |
| b0859 | hypothetical protein | 2.0 |
| b1200 | hypothetical protein | 1.8 |
| b1795 | hypothetical protein | 1.8 |
| dadA | d-amino acid dehydrogenase | 2.0 |
| dadX | alanine racemase | 1.9 |
| dapB | dihydrodipicolinate reductase | 1.8 |
| glyA | serine hydroxymethyltransferase | 1.9 |
| hdeA | protein HdeA precursor | 1.8 |
| hdeB | protein HdeB precursor | 3.1 |
| ilvC | ketol-acid reductoisomerase | 1.8 |
| metE | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | 2.1 |
| narG | respiratory nitrate reductase 1 alpha chain | 2.3 |
| ompA | outer membrane protein A | 2.0 |
| ompF | outer membrane protein F precursor | 1.9 |
| ompX | outer membrane protein X precursor | 1.8 |
| oppA | periplasmic oligopeptide-binding protein precursor | 8.8 |
| oppB | oligopeptide transport system permease protein OppB | 2.3 |
| oppF | oligopeptide transport ATP-binding protein OppF | 3.7 |
| prlA | preprotein translocase secy subunit | 1.8 |
| rfaL | hypothetical protein | 2.1 |
| rplI | 50S ribosomal subunit protein L9 | 2.1 |
| rplJ | 50S ribosomal subunit protein L10 | 1.9 |
| rplL | 50S ribosomal subunit protein L7/L12 | 2.5 |
| rplP | 50S ribosomal subunit protein L16 | 3.1 |
| rplQ | 50S ribosomal subunit protein L17 | 2.6 |
| rplV | 50S ribosomal subunit protein L22 | 2.3 |
| rplY | 50s ribosomal subunit protein L25 | 1.9 |
| rpmA | 50S ribosomal subunit protein L27 | 1.9 |
| rpmG | 50S ribosomal subunit protein L33 | 2.1 |
| rpsD | 30S ribosomal subunit protein S4 | 1.8 |
| rpsS | 30S ribosomal subunit protein S19 | 2.2 |
| slyD | probable fkbP-type peptidyl-prolyl cis-trans isomerase SlyD | 1.8 |
| tufB | elongation factor EF-Tu | 2.0 |
| yfiD | hypothetical 14.3-kDa protein | 2.1 |
TABLE 2.
Functional classification of differentially expressed genes
| Functional group | Differentially expressed gene(s)
|
|
|---|---|---|
| Higher on TF5015 | Lower on TF5015 | |
| Amino acid biosynthesis and metabolism | glnA, leuA, leuB, leuC, leuD, proA, thrA, thrC | asd, dadA, dadX, dapB, glyA, ilvC, metE |
| Cell processes | b0427, mdoB | |
| Cell structure | fimA | ompA, ompF, ompX, rfaL |
| Central intermediary metabolism | aceA, aceB, b2530 | |
| Energy metabolism | aldA, icdA, gltA | narG, yfiD |
| Hypothetical, unclassified, unknown | yigJ | b1795, hdeA, hdeB |
| Putative enzymes | b0859, b1200 | |
| Putative transport proteins | prlA | |
| Translation, posttranslational modification | infB | rplI, rplJ, rplL, rplP, rplQ, rplV, rplY, rpmA, rpmG, rpsD, rpsS, slyD, tufB |
| Transport and binding proteins | oppA, oppB, oppF | |
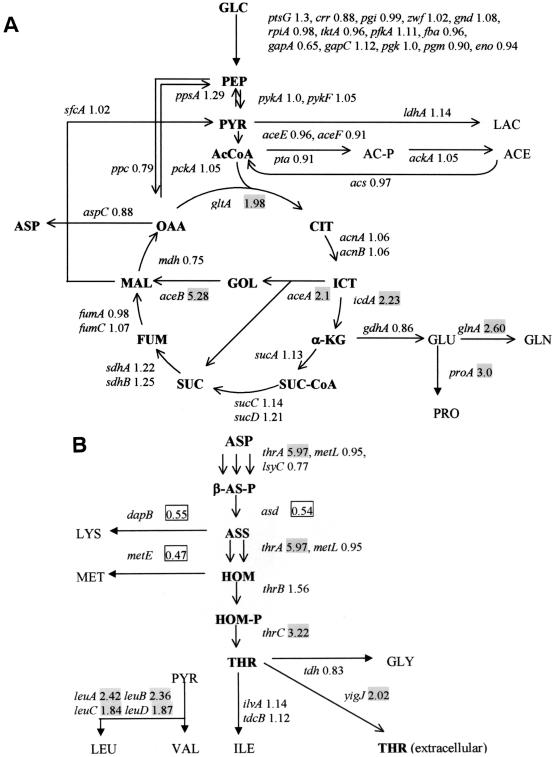
The genes involved in tricarboxylic acid (TCA) cycle and glyoxylate shunt were considerably upregulated in TF5015 (Table 1 and Fig. 3). Interestingly, aceA, aceB, and icdA, which participate in the isocitrate catabolic pathway at the branch point between glyoxylate shunt and TCA cycle, were concomitantly overexpressed in TF5015. The transcript of aceK in contrast to aceBA was similarly expressed in both strains, which seems to be due to premature transcriptional termination of aceK (5). As shown in Fig. 3, ppc, whose gene product not only mediates conversion of phosphoenolpyruvate (PEP) into oxaloacetate but functions as a pivotal anaplerotic reaction (11, 20), was similarly expressed in both W3110 and TF5015. Transcriptome analysis revealed a negligible change in the expression levels of the genes involved in glycolytic and pentose phosphate pathways between W3110 and TF5015 (Fig. 3).
FIG. 3.
Metabolic map showing relative expression ratios (intensity of TF5015 versus that of W3110) of transcript levels in the central metabolic pathway (A) and the threonine biosynthetic pathway (B). Abbreviations: GLC, glucose; PEP, phosphoenolpyruvate; PYR, pyruvate; LAC, lactate; Ac-CoA, acetyl-CoA; AC-P, acetyl-phosphate; ACE, acetate; CIT, citrate; ICT, isocitrate; α-KG, α-ketoglutarate; SUC-CoA, succinyl-CoA; FUM, fumarate; MAL, malate; OAA, oxaloacetate; GOL, glyoxylate; GLU, glutamate; GLN, glutamine; PRO, proline; ASP, aspartate; LYS, lysine; MET, methionine; β-AS-P, β-aspartyl phosphate; ASS, aspartate semialdehyde; HOM, homoserine; HOM-P, homoserine phosphate; THR, threonine; GLY, glycine; ILE, isoleucine; LEU, leucine; VAL, valine. The shaded and boxed numbers in the figure represent up- and downregulated genes, respectively, in TF5015.
The mRNA levels of the thrA and thrC, which encode the key enzymes in the threonine biosynthetic pathway, were found to be upregulated in TF5015 (Table 1 and Fig. 3), even though threonine accumulated at high levels in TF5015 compared to W3110 (Table 3). The lysine level was also significant in TF5015 (Table 3) and increased with fermentation time (data not shown). It was reported that the expressions of asd, dapB, dapE, lysA, and lysC are repressed by lysine in E. coli (30). Transcriptome analysis also confirmed the downregulation of asd and dapB in TF5015 (Table 1 and Fig. 3). In contrast, the expression levels of dapE, lysA, and lysC were not influenced by the accumulation of lysine. No significant differences in the expression levels of the genes associated with the biosynthesis of methionine and isoleucine and the degradation pathway of threonine except for the downregulation of metE were found between W3110 and TF5015 (Fig. 3). The hypothetical protein, yigJ product (recently named as rhtC), involved in the excretion of threonine was recently reported (42). Comparison of expression profiling showed the upregulation of yigJ in TF5015, which well coincides with the overproduction of threonine.
TABLE 3.
Accumulation levels of amino acids and acetic acid in E. coli W3110 and TF5015 at the early stationary phase
| Amino acid or acetic acid | Concn (mM)a in strain:
|
|
|---|---|---|
| W3110 | TF5015 | |
| Glutamate | 0.18 | 1.75 |
| Isoleucine | ND | 0.28 |
| Leucine | ND | 1.24 |
| Lysine | ND | 0.12 |
| Methionine | 2.26 | 3.94 |
| Threonine | ND | 131.1 |
| Valine | ND | 0.39 |
| Acetic acid | 3.17 | 0.88 |
The amino acids and acetic acid concentrations represent the average of triplicate experiments. ND, not detected in culture broth.
The overall transcript expression patterns of the genes involved in the biosynthesis of other amino acids except for aspartate family amino acids were observed to be similar between W3110 and TF5015. Only six genes were expressed at significantly higher levels in TF5015, whereas four genes were downregulated. Those include dadAX, glnA, glyA, ilvC, leuABCD, and proA (Table 1). The transcript levels of hdeA, hdeB, oppA, and yfiD, which are known to be induced either by acetate or at low pH (2, 3, 13, 17, 23), were found to be downregulated in TF5015 (Table 1). This result was also supported by the observation that acetate accumulation level in TF5015 was ∼4-fold lower than in W3110 at the early stationary phase (Table 3). Many ribosomal genes were downregulated in TF5015 (Table 1).
Proteome analysis between W3110 and TF5015.
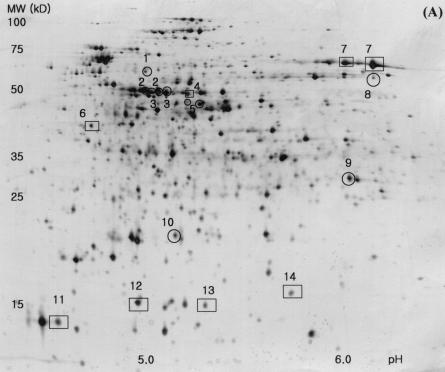
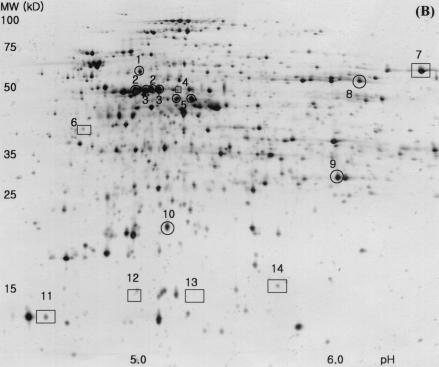
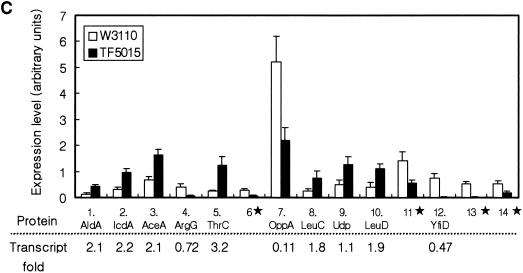
In order to analyze the global gene expression at protein levels and to confirm whether the transcriptome analysis using the DNA arrays would correlate with the protein expression levels, two-dimensional gel electrophoresis was carried out for proteins extracted from W3110 and TF5015 at the same growth stage as for total RNA preparation. As shown in Fig. 4, ca. 500 protein spots were visualized by silver staining. Of them, 18 protein spots displayed significantly different expression patterns; 10 proteins were found to be highly expressed, whereas 8 proteins were downregulated in TF5015 (Fig. 4A, 4B). We identified 14 proteins from 18 spots by using MALDI-TOF (Fig. 4C). Four proteins—AceA, IcdA, OppA, and ThrC—appeared as a doublet spot in two-dimensional gel. Proteins showing a higher expression level in TF5015 were identified as AceA, AldA, IcdA, LeuC, LeuD, ThrC, and Udp, whereas the downregulated proteins were ArgG, OppA, and YfiD. Of the identified 10 proteins, 8 proteins were in good agreement with the expression profiles of the corresponding genes, and these include AceA, AldA, IcdA, LeuC, LeuD, OppA, ThrC, and YfiD. However, the protein levels of ArgG and Udp were not directly correlated with the expression levels of each transcript.
FIG. 4.
Silver-stained two-dimensional gel patterns of E. coli W3110 (A) and E. coli TF5015 (B) at the same growth stage as DNA macroarray and quantification of protein spots showing differential expression patterns (C). In panels A and B, the horizontal axes represent pH, and the vertical axes represent molecular masses in kilodaltons. The transcript fold values (i.e., the relative intensity of transcript in TF5015 versus that of W3110) of corresponding genes are shown below the differently expressed proteins in panel C. 18 proteins (10 spots [○] of higher expression in TF5015 and 8 spots [□] of higher expression in W3110) showing differential expression patterns were selected, and 14 protein spots were identified by MALDI-TOF and shown in (C). Four proteins (IcdA, ThrC, AceA, and OppA) displayed double spots in the two-dimensional gels, respectively. The symbol (★) indicates unidentified protein spots. Spot intensities were measured and normalized as described in Materials and Methods. Error bars represent the standard deviation of the mean intensity. Continued.
In central metabolism, both isocitrate lyase (encoded by aceA) and isocitrate dehydrogenase (encoded by icdA) were overexpressed in TF5015, a finding which confirms the upregulation at the branch point of isocitrate at transcriptional and translational levels. Meanwhile, threonine synthase (encoded by thrC) was observed to be overexpressed, as well as the mRNA level. Induction of lactaldehyde dehydrogenase (encoded by aldA) in both transcriptional (Table 1) and translational levels seems to be due to a higher accumulation of glutamate in TF5015 than in W3110 (Table 3) (34). Overexpression of two 3-isopropylmalate dehydratases (encoded by leuC and leuD) was well correlated with the induction of leu operon and the accumulation of leucine in TF5015 (Table 3) (27). Also, repression of oligopeptide transport periplasmic binding protein (encoded by the oppA) observed by two-dimensional gel analysis well coincided with a significant decrease of oppA in TF5015 (Table 1). A yfiD gene product, a homologue of pyruvate formate lyase (3), was strongly repressed in TF5015, a finding consistent with transcriptome analysis and a low accumulation of acetate in TF5015.
Comparison of nucleotide sequences between W3110 and TF5015.
Analysis of nucleotide sequences between W3110 and TF5015 is expected to provide some insights into regulation mechanism of genes. The analyses of transcriptome and proteome between the two strains revealed that a number of the genes showing the significant expression changes in TF5015 are mainly related to the metabolic pathways affecting directly the biosynthesis and/or metabolism of threonine. Thus, we selected 15 genes in the central metabolic pathway and threonine biosynthetic pathway, and analyzed their nucleotide sequences. As a result, it was found that mutations occurred in the open reading frames (ORFs) of thrA and ilvA (Table 4) in TF5015. The mutation sites of thrA and ilvA genes were S345F (designated thrA345) and S97F (designated ilvA97), respectively. No mutation was observed in other 13 genes.
TABLE 4.
Changes in nucleotide sequences of genes in E. coli TF5015 compared to strain W3110
| Gene | Nucleotide change | Amino acid substitution |
|---|---|---|
| aceBAK | -a | - |
| asd | - | - |
| aspC | - | - |
| iclR | - | - |
| ilvA | C290T | S97F |
| lysC | - | - |
| mdh | - | - |
| metL | - | - |
| ppc | - | - |
| thrA | C1034T | S345F |
| thrB | - | - |
| thrC | - | - |
| yigJ | - | - |
-, No sequence change.
Effect of mutated thrABC operon on l-threonine production.
To evaluate whether the overproduction of threonine by TF5015 is mainly contributed by upregulation of the mutated thrABC operon (thrA345), the wild-type and mutated thrABC operons were cloned and transformed into W3110, respectively. The resulting recombinant strains were subjected to a fermentation experiment in flasks under optimized conditions. As shown in Table 5, production of threonine by W3110/pATF92 harboring the mutated thr operon was significantly increased, whereas W3110/pAW88 containing the wild-type thr operon showed a marginal increase in the level of threonine (ca. 2.8 mM) compared to W3110 (less than 0.1 mM).
TABLE 5.
Production of l-threonine by recombinant E. coli strain
| E. coli strain | Cell growtha (g/liter) | Threonine concn (mM) |
|---|---|---|
| W3110 | 11.9 | <0.1 |
| W3110/pAW88 | 11.5 | 2.8 |
| W3110/pATF92 | 10.3 | 82.4 |
In dry cell weight.
DISCUSSION
In this study, we have demonstrated the regulatory mechanism of threonine production and the physiological events based on the comparative analyses of the nucleotide sequences, transcriptome, and proteome between the parent strain and threonine-producing E. coli mutant. We prepared total RNA and proteins from W3110 and TF5015 at the early stationary phase, the threonine-producing stage of TF5015, and then analyzed the transcriptome and proteome differences between both strains. Interestingly, a number of genes displaying distinctly different expression levels are related to the biosynthesis and/or metabolism of aspartate family amino acids and central intermediary metabolism (Table 2). Two-dimensional gel electrophoresis revealed that differently expressed protein levels are in good agreement with relative mRNA levels of corresponding genes (Fig. 4). Also, two important mutations, thrA345 and ilvA97, were identified by the comparative sequence analysis (Table 4).
Comparison of gene expression profiles between W3110 and TF5015 showed that only 54 of 4,290 genes (1.3%) exhibit differential transcript expression patterns, and this is interesting since TF5015 is the mutant producing a much higher level of threonine compared to the parent strain W3110. From the viewpoint of growth rate, metabolic fluxes, regulation mechanisms, genetic modifications, and accumulation of threonine, although the physiology of TF5015 would greatly differ from that of W3110, global comparisons of the expression profiles suggest that the general trends at the transcriptional level are very similar to each other.
Both transcriptome and proteome analyses revealed the upregulation of glyoxylate shunt and TCA cycle in TF5015 compared to W3110 (Fig. 3). It was reported that the supply of oxaloacetate from PEP via PEP carboxylase and glyoxylate bypass is required for maximal yield of threonine by theoretical stoichiometric analysis (40) and experimental evidence (37). Since the biosynthesis of oxaloacetate via PEP carboxylase is main route from glucose and PEP carboxylase is inhibited by aspartate in E. coli (11), the regulation of ppc and its gene product is the pivotal step for biosynthesis of aspartate family amino acids. When we analyzed the ppc gene at genetic and transcriptional levels between W3110 and TF5015, distinctive differences were not observed (Fig. 3 and Table 4). In this regard, our analyses strongly imply that TF5015 adopts glyoxylate shunt and TCA cycle rather than PEP carboxylase for glucose metabolism at the transcript level.
Pyruvate formation is a main route from glucose metabolism according to stoichiometric analysis of central metabolism in E. coli (15); thus, it seems that the conversion of pyruvate to oxaloacetate without losing carbon as CO2 can be a major factor for improving the production of threonine. Therefore, it seems to be reasonable that upregulation of the ace operon could partly contribute to an increase of acetyl coenzyme A (acetyl-CoA) flux into oxaloacetate and increased production of threonine, even though the precise balanced flow of isocitrate at the branch point is not clear in TF5015. It was known that E. coli could excrete 10 to 30% of carbon flux from glucose as acetate under aerobic conditions (16). Accordingly, we suggest that upregulation of the ace operon can be a way of preventing the accumulation of acetate or accommodating an efficient utilization of acetate (8). This is well supported by a reduced level of acetate in TF5015 compared to W3110 (Table 3). Since glyoxylate shunt is normally repressed during the growth of E. coli on glucose, upregulation of the ace operon is not consistent with the previous observation (5). The expression of aceBAK is known to be significantly influenced by various regulation factors, including arcA, arcB, fadR, fruR, himA, himD, and iclR (5), but expression levels of these factors were almost the same between W3110 and TF5015. In addition, no modifications of DNA sequences in an ORF of aceBAK, an upstream region of aceBAK promoter, and an iclR in both strains were detected by DNA sequencing (Table 4). Thus, some other factors, such as a change of metabolites responding to threonine production or unknown mechanisms, seem to affect the transcriptional control of the ace operon.
Upregulation of the thr operon in TF5015 was confirmed by both transcriptome and proteome analyses (Fig. 3 and 4). In a previous study, we observed that the expression level of aspartokinase I (AKI) in TF427, the parent strain of TF5015, was elevated ca. three to fourfold compared to its parent strain, TF125, in the presence of threonine and isoleucine in culture medium (24). The production level of threonine by TF427 is much higher than TF125. By DNA sequencing of the thr operon, we confirmed the replacement of Ser with Phe at position 345 in the thrA product, AKI-homoserine dehydrogenase I (HDI), of TF5015 (Table 4). Also, the enzyme assay of mutated aspartokinase showed that the activity of AKI was not inhibited by threonine (24). In a previous result about mutant analysis of Serratia marcescens thr operon, threonine-mediated feedback inhibition of both AKI activity and HDI activity was released by single amino acid substitution (the exchange of Gly to Asp at position 330 or Ser to Phe at position 352) in the central region of the AKI-HDI product (29). Similarly, a change at S300Y of Corynebacterium glutamicum AK yielded a threonine- and lysine-insensitive aspartokinase product (21). Genetic analysis revealed that the mutation of thrA results in a release of feedback inhibition of AKI-HDI by threonine in TF5015 rather than a deregulation of feedback repression by threonine plus isoleucine. Meanwhile, amino acid substitution of threonine deaminase (TD; Ser-97→Phe) was detected from the analysis of the nucleotide sequence between W3110 and TF5015 (Table 4). The nutritional requirement of isoleucine and enzyme assay of TD (24) represented that the loss of enzyme activity came from the replacement of Ser by Phe that is located in N2 subdomain of TD (12). Auxotrophic mutants containing the amino acid substitutions (Lys62→Glu, Ala66→Val, or Pro156→Ser) in the N2 subdomain and its nearby loops (12) were reported through genetic experiments (10), which support our interpretation. In this respect, we suggest that the upregulation of thr operon in TF5015 would be caused by limitation of isoleucine due to the inactivation of TD. As shown in Table 5, simple overexpression of the wild-type thr operon resulted in a slight increase in the level of threonine, whereas expression of the mutated thrABC (pATF92) in W3110 gave rise to a significant incremental effect on threonine production. Thus, it is obvious that upregulation of the mutated thr operon in TF5015 led to the overproduction of threonine from the analyses of transcriptome, proteome, and DNA sequences, as well as from the enzyme assay and fermentation experiments. Furthermore, since sequence changes of metL (encodes AKII-HDII) and lysC (encodes AKIII) in contrast to thrA were not detected in ORFs and regulatory regions of both genes (30), we suggest that the biosynthesis of threonine in TF5015 is mainly catalyzed by AKI-HDI rather than by AKII-HDII and AKIII. It is necessary to determine why the expression of thrB was slightly increased in TF5015 in two sets of array experiments. One possibility is that an internal promoter at the 3′ end of thrA allows the formation of thrB transcript in addition to those initiated at the major promoter of thrA upstream (30).
Considering the increased accumulation of lysine in the course of fermentation by TF5015 and feedback regulation of the biosynthetic pathway of aspartate family amino acids by lysine (3), blocking of the lysine pathway is likely to be indispensable for the prevention of lysine accumulation, the derepression of asd, and finally an increase in threonine production in TF5015. The increase of threonine production noted in a previous study was accomplished by auxotrophic mutation of α,ɛ-diaminopimelic acid (26). The asd gene, which is essential for threonine biosynthesis, was downregulated in TF5015 (Fig. 3), and this seems to be due to the accumulation of lysine (Table 3) (30).
We observed drastic changes in transcript or protein levels in some genes in response to the accumulation of other amino acids or difference of growth rate. At some promoters of the Lrp regulon, Lrp action was greatly modified by the presence of leucine (27). The expressions of dadAX, leu operon, glnA, glyA, ompF, oppA, oppB, and oppF seem to directly or indirectly respond to an accumulation of leucine, Lrp, or leucine/Lrp (4, 27). Upregulation of aldA is likely to arise from an accumulation of glutamate (Tables 1 and 3 and Fig. 4) (33). The differential expressions of mdoB, proA, and ompF, which encode phosphoglycerol transferase I, a gamma-glutamyl phosphate reductase, and an outer membrane protein F precursor, respectively, might be involved in the response of osmotic regulation of TF5015 (Table 1) (9, 36, 41). As observed in earlier study (38), the downregulation of many ribosomal genes would result from the slow growth rate of TF5015 compared to W3110 (Fig. 2 and Table 1).
The accumulation of acetate in TF5015 was about fourfold lower than in W3110 at the early stationary phase, although acetate was simultaneously accumulated in both W3110 and TF5015 (Table 3), reaching up to 200 and 66.7 mM, respectively, with fermentation time (data not shown). Previous studies revealed that RpoS-regulated genes, periplasmic transporters for amino acids and peptides, and metabolic enzymes are induced either by acetate or at low pH (2, 3, 13, 17, 23). Of these, we suppose that downregulations of b1795, hdeAB operon, oppA, and yfiD are related to a lower accumulation of acetate in TF5015 compared to W3110. In particular, the induction of YfiD can be a strong indication for internal acidification. A low level of acetate and repression of YfiD in TF5015 imply that this mutant maintains intracellular homeostasis at the early stationary phase, even though the overproduction of threonine was expected to affect to some extent the cellular physiology.
We have demonstrated that a global analysis of expression profiles at mRNA and protein levels between prototrophic strain and threonine-producing mutant provides crucial information for understanding the mechanism of threonine overproduction and the physiological consequences in TF5015. Integrated knowledge regarding the threonine-producing mutant is expected to offer more rational strategies for developing microorganisms with greater potential.
Acknowledgments
We thank Charles Yanofsky for many helpful comments.
This work was supported by the BK21 Program of the Ministry of Education and the National Research Laboratory Program of the Ministry of Science and Technology of Korea.
REFERENCES
- 1.Arfin, S. M., A. D. Long, E. T. Ito, L. Tolleri, M. M. Riehle, E. S. Paegle, and G. W. Hatfield. 2000. Global gene expression profiling in Escherichia coli K-12. J. Biol. Chem. 275:29672-29684. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, C. N., J. Mcelhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic gowth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvo, J. M., and R. G. Mattews. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Eshcerichia coli. Microbiol. Rev. 58:466-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cronan, J. E., Jr., and D. LaPorte. 1996. Tricarboxylic acid cycle and glyoxylate bypass, p. 206-216. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 6.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 7.Eggeling, L., and H. Sahm. 1999. Amino acid production: principles of metabolic engineering, p. 153-176. In S. Y. Lee and E. T. Papoutsakis (ed.), Metabolic engineering. Marcel Dekker, Inc., New York, N.Y.
- 8.Farmer, W. R., and J. C. Liao. 1997. Reduction of aerobic acetate production by Escherichia coli. Appl. Environ. Microbiol. 63:3205-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiedler, W., and H. Rotering. 1985. Characterization of an Escherichia coli mdoB mutant strain unable to transfer sn-1-phosphoglycerol to membrane-derived oligosacchrides. J. Biol. Chem. 260:4799-4806. [PubMed] [Google Scholar]
- 10.Fisher, K. E., and E. Eisenstein. 1993. An efficient approach to identify ilvA mutations reveals an amino-terminal catalytic domain in biosynthetic threonine deaminase from Escherichia coli. J. Bacteriol. 175:6605-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraenkel, D. G. 1996. Glycolysis, p. 189-197. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 12.Gallagher, D. T., G. L. Gilliland, G. Xiao, J. Zondlo, K. E. Fisher, D. Chinchilla, and E. Eisenstein. 1998. Structure and control of pyridoxal phosphate-dependent allosteric threonine deaminase. Structure 6:465-475. [DOI] [PubMed] [Google Scholar]
- 13.Han, M. J., S. S. Yoon, and S. Y. Lee. 2001. Proteome analysis of metabolically engineered Escherichia coli. J. Bacteriol. 183:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatzimanikatis, V., L. H. Choe, and K. H. Lee. 1999. Proteomics: theoretical and experimental considerations. Biotechnol. Prog. 15:312-318. [DOI] [PubMed] [Google Scholar]
- 15.Holms, W. H. 1986. The central metabolic pathways of Escherichia coli: relationship between flux and control at a branch point, efficiency of conversion to biomass, and excretion of acetate. Curr. Top. Cell. Regul. 28:69-105. [DOI] [PubMed] [Google Scholar]
- 16.Holms, W. H. 1996. Flux analysis and control of the central metabolic pathways in Escherichia coli. FEMS Microbiol. Rev. 19:85-116. [DOI] [PubMed] [Google Scholar]
- 17.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. L. Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitering of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated proteins, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 18.Ideker, T., V. Thorsson, J. A. Ranish, R. Christmas, J. Buhler, J. K. Eng, R. Bumgarner, D. R. Goodlett, R. Aebersold, and L. Hood. 2001. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292:929-934. [DOI] [PubMed] [Google Scholar]
- 19.Ireland, M. M. E., J. A. Karty, E. M. Quardokus, J. P. Reilly, and Y. V. Brun. 2002. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol. Microbiol. 45:1029-1041. [DOI] [PubMed] [Google Scholar]
- 20.Jetten, M. S. M., and A. J. Sinskey. 1995. Recent advances in the physiology and genetics of amino acid-producing bacteria. Crit. Rev. Biotechnol. 15:73-103. [DOI] [PubMed] [Google Scholar]
- 21.Kalinowski, J., J. Cremer, B. Bachmann, L. Eggeling, H. Sahm, A. Puhler. 1991. Genetic and biochemical analysis of the aspartokinase from Corynebacterium glutamicum. Mol. Microbiol. 5:1197-1204. [DOI] [PubMed] [Google Scholar]
- 22.Khodursky, A. B., B. J. Peter, N. R. Cozzarelli, D. Botstein, P. O. Brown, and C. Yanofsky. 2000. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:12170-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirkpatrick, C., L. M. Maurer, N. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, J. H., J. W. Oh, H. H. Lee, and H. H. Hyun. 1991. Production of l-threonine by auxotrophs and analogue-resistant mutants of Escherichia coli. Kor. J. Appl. Microbiol. Biotechnol. 19:583-587. [Google Scholar]
- 25.Lucchini, S., A. Thompson, and L. C. D.Hinton. 2001. Microarrays for microbiologists. Microbiology 147:1403-1414. [DOI] [PubMed] [Google Scholar]
- 26.Mizukami, T., M. Yagisawa, S. Kawahara, H. Kase, T. Oka, and A. Furuya. 1986. Essential role of aspartokinase in l-threonine production by Escherichia coli W mutants. Agric. Biol. Chem. 50:1015-1018. [Google Scholar]
- 27.Newman, E. B., R. T. Lin, and R. D'ari. 1996. The leucine/Lrp regulon, p. 1513-1525. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 28.Oh, M. K., and J. C. Liao. 2000. Gene expression profiling by DNA microarrays and meabolic fluxes in Escherichia coli. Biotechnol. Prog. 16:278-286. [DOI] [PubMed] [Google Scholar]
- 29.Omori, K., Y. Imai, S. I. Suzuki, and S. Komatsubara. 1993. Nucleotide sequence of the Serratia marcescens threonine operon and analysis of the threonine operon mutations which alter feedback inhibition of both aspartokinase I and homoserine dehydrogenase I. J. Bacteriol. 175:785-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patte, J. C. 1996. Biosynthesis of threonine and lysine, p. 528-541. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 31.Persidis, A. 1998. Proteomics. Nat. Biotechnol. 16:393-394. [DOI] [PubMed] [Google Scholar]
- 32.Pomposiello, P. J., M. H. J. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quintilla, F. X., L. Baldoma, J. Badia, and J. Aguilar. 1991. Aldehyde dehydrogenase induction by glutamate in Escherichia coli. Eur. J. Biochem. 202:1321-1325. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 36.Song, K. H., H. H. Lee, and H. H. Hyun. 2000. Characterization of salt-tolerant mutant for enhancement of l-threonine production in Escherichia coli. Appl. Microbiol. Biotechnol. 54:647-651. [DOI] [PubMed] [Google Scholar]
- 37.Sugita, T., and S. Komatsubara. 1989. Construction of a threonine-hyperproducing strain of Serratia marcescens by amplifying the phosphoenolpyruvate carboxylase gene. Appl. Microbiol. Biotechnol. 30:290-293. [Google Scholar]
- 38.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theze, J., and I. Saint-Girons. 1974. The threonine locus of Escherichia coli K-12: genetic structure and evidence for an operon. J. Bacteriol. 118:990-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varma, A., B. W. Boesch, and B. O. Palsson. 1993. Biochemical production capabilities of Escherichia coli. Biotechnol. Bioeng. 42:59-73. [DOI] [PubMed] [Google Scholar]
- 41.Wood, L. M. 1999. Osmosensing by bacteria: signal and membrane-based sensors. Microbiol. Mol. Biol. Rev. 63:230-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zakataeva, N. P., V. V. Aleshin, I. L. Tokmakova, P. V. Troshin, and V. A. Livshits. 1999. The novel transmembrane Escherichia coli proteins involved in the amino acid efflux. FEBS Lett. 452:228-232. [DOI] [PubMed] [Google Scholar]