Abstract
The complete 2,343,479-bp genome sequence of the gram-negative, pathogenic oral bacterium Porphyromonas gingivalis strain W83, a major contributor to periodontal disease, was determined. Whole-genome comparative analysis with other available complete genome sequences confirms the close relationship between the Cytophaga-Flavobacteria-Bacteroides (CFB) phylum and the green-sulfur bacteria. Within the CFB phyla, the genomes most similar to that of P. gingivalis are those of Bacteroides thetaiotaomicron and B. fragilis. Outside of the CFB phyla the most similar genome to P. gingivalis is that of Chlorobium tepidum, supporting the previous phylogenetic studies that indicated that the Chlorobia and CFB phyla are related, albeit distantly. Genome analysis of strain W83 reveals a range of pathways and virulence determinants that relate to the novel biology of this oral pathogen. Among these determinants are at least six putative hemagglutinin-like genes and 36 previously unidentified peptidases. Genome analysis also reveals that P. gingivalis can metabolize a range of amino acids and generate a number of metabolic end products that are toxic to the human host or human gingival tissue and contribute to the development of periodontal disease.
Periodontal diseases are a group of infections that affect the structures surrounding teeth. If allowed to progress, periodontal disease can cause the destruction of supporting connective tissue and bone, ultimately resulting in tooth loss. Initiation and progression of periodontal diseases is the result of a complex interaction between the bacteria colonizing the gingival crevice and the hosts' immune and inflammatory responses. Since the species most strongly implicated in periodontal disease pathogenesis are also usually present in low numbers in healthy people, the distinction between pathogenic and commensal bacteria in the human host is not clearly defined.
The gram-negative anaerobe Porphyromonas gingivalis belongs to the family Porphyromonadaceae, order Bacteroidales in the phylum Bacteroidetes, previously known as the Cytophaga-Flavobacteria-Bacteroides (CFB) group (6). The bacterium is a major causative agent in the initiation and progression of severe forms of periodontal disease. P. gingivalis is a late or secondary colonizer of the oral cavity, a process that is facilitated by other microbial species that provide attachment sites, as well as supply growth substrates, and reduce oxygen tension to levels optimal for growth of P. gingivalis. Among the early plaque organisms that P. gingivalis adheres to are the oral streptococci (35, 36) and Actinomyces naeslundii (19). Adherence is facilitated by a variety of bacterial surface proteins, including fimbriae, hemagglutinins, and proteinases. P. gingivalis also binds to late colonizers such as Fusobacterium nucleatum, Treponema denticola, and Bacteroides forsythus (now renamed Tanerella forsythensis) (20, 30, 72). The use of a variety of metabolic strategies appears to enable the success of this microbial community. Once established, P. gingivalis cells participate in intercellular communication networks with other oral prokaryotic cells, as well as with eukaryotic cells (37).
P. gingivalis is the third oral pathogen (1, 28) and the second member of the CFB group to be sequenced. The genome of P. gingivalis strain W83 (also known as strain HG66) is presented here. Strain W83 was isolated in the 1950s by H. Werner (Bonn, Germany) from an undocumented human oral infection (41, 46) and was brought to The Pasteur Institute by Madeleine Sebald during the 1960s. The strain was subsequently obtained by Christian Mouton (Quebec, Canada) during the late 1970s. It is anticipated that the availability of the complete genome sequence from this oral pathogen will give tremendous insight into the mechanisms that result in disease progression in an ecological niche deep within the oral cavity.
MATERIALS AND METHODS
Library preparation and random sequencing of P. gingivalis.
P. gingivalis strain W83 was obtained from Christian Mouton, Laval University, Quebec City, Quebec, Canada. Genomic DNA was extracted twice with buffered phenol and once with 25:24:1 phenol-chloroform-isoamyl alcohol and precipitated with alcohol. Cloning, sequencing, and assembly were as described previously for genomes sequenced by The Institute for Genomic Research (TIGR) (13). One small-insert plasmid library (1.5 to 2.5 kb) was generated by random mechanical shearing of genomic DNA. One large-insert library was generated by partial Tsp5091 digestion and ligation to the λ-DASHII/EcoRI vector (Stratagene). In the initial random sequencing phase, ∼8-fold sequence coverage was achieved with 39,623 sequences (average read length, 534 bases). The plasmid and λ sequences were jointly assembled by using TIGR Assembler. Sequences from both ends of 506 λ clones served as a genome scaffold, verifying the orientation, order, and integrity of the contigs. Sequence gaps were closed by editing the ends of sequence traces and/or primer walking on plasmid clones. Physical gaps were closed by direct sequencing of genomic DNA or combinatorial PCR, followed by sequencing of the PCR product. The final molecule has 8.35× sequence redundancy.
ORF prediction and gene family identification.
An initial set of open reading frames (ORFs) that likely encode proteins was identified with GLIMMER (57), and those shorter than 90 bp, as well as some of those with overlaps, were eliminated. (For more details on the annotation process that was used to identify all of the ORFs in the P. gingivalis genome, see reference 70.) A region containing the likely origin of replication was identified, and bp 1 was designated adjacent to the dnaA gene that is in this region. ORFs were searched against a nonredundant protein database as previously described. Frameshifts and point mutations were detected and corrected where appropriate as described previously (51). Remaining frameshifts and point mutations are considered authentic, and corresponding regions were annotated as an “authentic frameshift” or an “authentic point mutation,” respectively. ORF prediction and gene family identification was completed by using the methodology described previously (49). Two sets of hidden Markov models (HMMs) were used to determine ORF membership in families and superfamilies. These included 721 HMMs from Pfam v2.0 and 631 HMMs from TIGR ortholog resource. TMHMM (33) was used to identify membrane-spanning domains in proteins.
Comparative genomics.
All genes and predicted proteins from this genome, as well as from all other completed genomes, were compared by using the basic local alignment search tool (BLAST). For the identification of recent gene duplications, all genes from the P. gingivalis genome were compared to each other. A gene was considered recently duplicated if the most similar gene (as measured by P value) was another gene within the same genome (relative to genes from the two other genomes).
Database submission.
The nucleotide sequence of the whole genome of P. gingivalis was submitted to GenBank under accession number AE015924.
RESULTS
Description of the genome.
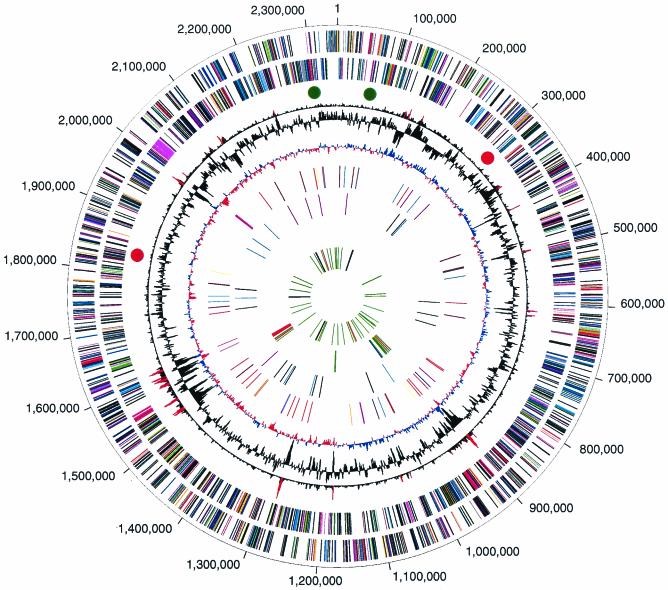
The genome of P. gingivalis is 2,343,479 bp, with an average G+C content of 48.3% (Fig. 1). There are four ribosomal operons (5S-23S-tRNAAla-tRNAIle-16S) and 2 structural RNA genes, as well as 53 tRNA genes with specificity for all 20 amino acids. A total of 1,990 ORFs could be identified in the genome (13). Of these, 1,075 (54%) could be assigned to biological role categories (54), 184 (9.2%) were conserved hypothetical proteins or conserved domain proteins, 208 (10.5%) were of unknown function, and 523 (26.3%) encoded hypothetical proteins. More than 85% of the genome encodes ORFs.
FIG. 1.
Circular representation of the P. gingivalis genome. The outer circle shows the predicted coding regions on the plus strand color-coded by role categories as follows: violet, amino acid biosynthesis; light blue, biosynthesis of cofactors, prosthetic groups, and carriers; light green, cell envelope; red, cellular processes; brown, central intermediary metabolism; yellow, DNA metabolism; light gray, energy metabolism; magenta, fatty acid and phospholipid metabolism; pink, protein synthesis and fate; orange, purines, pyrimidines, nucleosides, and nucleotides; olive, regulatory functions and signal transduction; dark green, transcription; teal, transport and binding proteins; gray, unknown function; salmon, other categories; blue, hypothetical proteins. The second circle shows the predicted coding regions on the minus strand. The third circle presents the χ2 analysis of atypical nucleotide composition; χ2 values of >600 are indicated in red. The fourth circle shows the %G+C. The fifth circle shows atypical nucleotide composition (GC skew). The sixth circle shows the IS elements, indicated by color as follows: orange, ISPg1; light green, ISPg2; magenta, ISPg3; cyan, ISPg4; brown, ISPg5; gold, ISPg6; blue-green, ISPg7; pink, ISPg8; and violet, ISPg9; salmon, ISPg10; olive, ISPg11. The seventh circle shows MITE239 (magenta), MITE700 (cyan), and MITE464 (black). The eighth circle shows Tn4555 (blue), CTn (red), and other transposable elements (green). The ninth circle shows tRNA (green), rRNA (black), and sRNA (red).
Repetitive elements occupy ca. 6% of the P. gingivalis genome and fall into two major classes: DNA repeats and transposable elements. The DNA repeats include uninterrupted direct repeats (Table 1), and a subclass of dispersed repeats known as clustered regularly interspaced short palindromic repeats (CRISPRs) (Table 2). Strain W83 does not appear to contain other classes of dispersed repetitive DNA sequence elements such as ERIC and REP elements. The transposable elements include insertion sequence (IS) elements and miniature inverted-repeat transposable elements (MITEs), which are summarized in Table 3, and large stretches of genes that resemble remnants of conjugable and mobilizable transposons based on sequence similarity to elements previously described in Bacteroides species (11). The locations of transposon-associated genes are shown in Fig. 1. Although there are 96 complete or partial copies of IS elements and MITEs present in strain W83 that occupy more than 94 kb of the genome, the transposable elements are rarely found in a functional gene. Instead, these elements have inserted almost exclusively into intergenic regions and other copies of transposable elements, except for one insertion into a putative outer membrane protein (PG0176/PG0178) that is intact in at least four other strains of P. gingivalis (accession numbers AB069977 to AB069980). Analysis of the IS elements reveals two possible chromosomal inversions that most likely arose by homologous recombination between identical copies of elements at widely separated insertion sites. These potential DNA rearrangements are revealed by inspection of both the duplicated target site sequences (direct repeats) that flank some of the IS elements and a transposon gene disrupted by one IS insertion. The sites of these putative inversions are shown in Fig. 1. In one case, 821 kb (35%) of the chromosome appears to be inverted between one copy of ISPg2 (PG1746) 512 kb before the origin and another copy of ISPg2 (PG0277) 309 kb after the origin (red dots). In the second case, 103 kb (4.4%) of the chromosome appears to be inverted between two copies of ISPg4, which are located on opposite sides of the origin (PG2194 and PG0050) (green dots). Since inversions about the origin do not invert the direction of transcription relative to replication of genes on the segment, such inversions may be selectively neutral. It will be interesting to determine whether strain W83 and other strains of P. gingivalis share a common genetic structure, or whether the proposed chromosomal inversions are relatively recent events.
TABLE 1.
Major families of contiguous repeats and their copy numbers in the genome
| Designationa | No. of copies | No. of direct repeats (mean no.) | Sequence of direct repeatb |
|---|---|---|---|
| CR-41-A | 12 | 2-16 (8) | TGCAGGACGCGATTGTCAA·CTGATTCTTGCTTCCTGCACGA |
| CR-41-B | 1 | 4 (4) | GCTTGCAGGAAGCGATTTCG·GCAAAAAAATTCCTGTTTGCA |
| CR-41-C | 1 | 4 (4) | TGCAGGAAGCCGTTGTCAC·CTGACAATCACTCCCTGCACGA |
| CR-23-A | 14 | 3-14 (9) | TATCCTCTCAT·ATGAGACCATAG |
| CR-23-B | 5 | 5 (5) | ACTAAACTAT·ATACTTTGGTNNN |
| CR-22-A | 32 | 2-8 (5) | CGATTTATA·TATAAATCGAAAA |
| CR-22-B | 4 | 4-6 (5) | TATTTCCGTAT·ATACGGAAATA |
| CR-22-C | 2 | 4-5 (5) | AAACGTCT·AGACGTTTCGAGCA |
The number indicates the length of the repetitive element; the letter indicates the family designation. Not included in table are CR-23-C and -D and CR-22-D, -E, -F, -G, and -H.
A dot indicates the location of dyad symmetry; underlined bases indicate palindromic match(es).
TABLE 2.
CRISPRs
| Designation | Associated gene(s) | No. of direct repeats | Sequence of direct repeatb |
|---|---|---|---|
| CRISPR-30-36a | PG2013 cas2, PG2014 cas1, PG2015 cas4, PG2016 cas3 | 23 | GTTTTAATTCCTGTA·TGGTGCAATTGAAAT |
| CRISPR-36-30-A | PG0338 | 8 | GTTGGATCTACCCTCTAT·TCGAAGGGTACACACAAC |
| CRISPR-36-30-B | PG1164 | 8 | GTTGGGAATACCCTTAGT·TAGAAGGGTGGAGACAAC |
| CRISPR-37-35 | PG1981, PG1982 | 8 | GTCTTAATAGCCTTACGGACTGTGTATGTATAGTGAG |
TABLE 3.
IS elements and MITEs
| Elementa | Family | No. of copies (no. of fragments)b | Inverted repeat (length [bp])c | Direct repeat lengthd | Length (bp) |
|---|---|---|---|---|---|
| ISPg1 | IS5 | 25 (7) | GAGACCTTTGCA (12) | 5 | 1,334 |
| ISPg2 | ISAs1 | 5 (11) | CAGGGCTGACGCATTAAA (18) | 9 | 1,207 |
| ISPg3 | IS5 | 6 (1) | ACGTCAGTTCGA (12) | 7 | 1,070 |
| ISPg4 | IS5 | 10 (0) | CATTAGTGTCCTAAAAG (17) | 9 | 1,394 |
| ISPg5 | IS3 | 11 (1) | TTGACGTGTACTGAAAAAAGTTGACAGTTGG (31) | 3 | 1,514 |
| ISPg6 | ISAs1 | 1 (2) | CAGGGCAACCGCATCAAA (18) | NDe | 1,139 |
| ISPg7 | ISRm7 | 1 (3) | GTCGTCCCTTAAgAAtCCTtTT (22) | 6 | 1,428 |
| ISPg8 | IS5 | 0 (1) | Unknown | ND | ND |
| ISPg9 | IS5 | 2 (2) | GAGACCATTGCA (12) | 5 | 1,326 |
| ISPg10 | IS256 | 1 | TGaCatTtTtAAAAAAaGAcATGG (24) | ND | ∼1,283 |
| ISPg11 | IS256 | (1) | Unknown | ND | ND |
| MITE239 | ISPg3 | 5 | ACGysAGTTCGATmTAArsGAA | 6-8 | 239 |
| MITE464 | ISPg1 | 13 | GAGACTGTTGCA | 5 | 464-1,071 |
| MITE700 | ISPg3 | 8 | ACGTCAGTTCGA | 7 | 455-717 |
ISPg1, ISPg2, and ISPg3 were originally described as IS1126 (44), PGIS2 (69), and IS195 (40).
That is, the number of intact copies with the number of fragments in parentheses.
Lowercase lettering indicates sequence at a position which is not part of the perfect inverted repeat.
Direct repeat or duplicated target site length.
ND, not determined.
There are 21 areas of the genome that display an atypical nucleotide composition identified by χ2 analysis (67) and that also correspond to regions of higher or lower G+C content than the rest of the genome. The areas range in size from 11 to 68 kb and range in G+C content from 29.4 to 61.6%. A variety of genes that could possibly have been acquired by this bacterium through lateral gene transfer are encoded in these regions. The genes include three restriction system proteins (PG0971, most similar to Anabaena sp. strain PCC 7120; PG0968, most similar to Anabaena sp. strain PCC 7120; and PG1469 most similar to Agrobacterium tumefaciens); hemagglutinin proteins B and C (HagB, PG1972, P. gingivalis specific; and HagC, PG1975, P. gingivalis specific); many capsular biosynthesis proteins, 20 transposase genes, two large mobile elements (PG1473 to PG1480, resembles only a conjugative element of Bacteroides thetaiotaomicron; and PG0868 to PG0875, whose sequence and gene organization most closely resembles the antibiotic-resistant mobilizable transposon Tn4555 from Bacteroides fragilis (68); and a thiamine biosynthesis operon (PG2107 to PG2111, which is most similar to the thiamine biosynthesis operon of Escherichia coli). These atypical regions in the P. gingivalis genome also encode many hypothetical and conserved hypothetical proteins, which undoubtedly contribute to the unique biology of this organism.
Comparative genomics.
Comparison of the predicted proteome of P. gingivalis with that of other completely sequenced genomes confirms the close relationship of P. gingivalis to other members of the CFB, including B. fragilis and B. thetaiotaomicron. Outside of the CFB phyla, the genome most similar to that of P. gingivalis is the Chlorobium tepidum genome, supporting previous phylogenetic studies that indicated the chlorobia and CFB phyla are related, albeit distantly. The proteomes most similar to that of P. gingivalis (in terms of the number of proteins with the best scoring matches) were those of B. thetaiotaomicron and B. fragilis with 572 and 437 best-scoring matches (P < 10−5), respectively.
A total of 332 genes were identified as being putatively duplicated in the P. gingivalis lineage. These duplicated genes are likely an indication that there is some selective evolutionary advantage to retaining these genes in the genome. Among these genes are 10 that encode DNA-binding histone-like proteins that have a distinctive domain architecture compared to HU and related histone-like proteins. These DNA-binding proteins have been designated a superfamily (i.e., a set of proteins that share a given domain architecture; TIGRFAMs family TIGR01201). Outside of P. gingivalis, the single known example of a DNA-binding histone-like protein is found in the gut bacterium B. fragilis. All members of this superfamily are distantly related to the bacterial DNA-binding protein HU family (Pfam family PF00216, five of which are also found in the P. gingivalis genome) but differ in architecture, sharing both an N-terminal extension and a glycine-rich C terminus. HU has been shown, among other DNA-binding functions, to assist the unwinding of oriC DNA by the DNA replication initiation protein DnaA (4). Interestingly, all 10 members of the TIGR01201 family in P. gingivalis have direct repeats upstream of their genes that may act as binding sites for the DNA-binding proteins that are encoded by the nearby gene and perhaps regulate their own expression. Alternatively, the repeats may also coordinate expression of the other chromosomal genes that they flank.
Metabolism and transport.
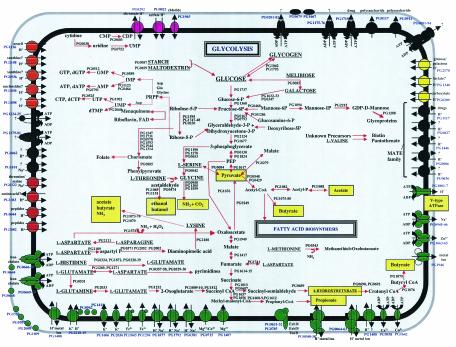
The microbial species that exist in supragingival plaque of the oral cavity are exposed to the host's dietary intake, and many of these bacteria, including the oral streptococci, ferment carbohydrates to acidic end products such as lactic acid for the purpose of energy production. On the other hand, anaerobic species in the subgingival plaque are exposed to crevicular fluid and to the host tissue proteins (61). The availability of the complete genome sequence of P. gingivalis W83 allowed for an analysis of the physiological potential of this species. Based on this analysis, the range of transport capabilities and metabolic pathways that could be identified is presented in Fig. 2.
FIG.2.
Overview of metabolism and transport in P. gingivalis. Primary substrates for energy metabolism are capitalized and underlined. End products of fermentation are highlighted by yellow boxes. Transporters are grouped by substrate specificity and indicated by color as follows: inorganic cations (green), inorganic anions (magenta), organic nutrients (yellow), and drug efflux and other (black). Arrows indicate direction of transport for substrates (and coupling ions, where appropriate).
Genome analysis suggests that P. gingivalis possesses a limited capacity for the uptake and metabolism of organic nutrients. Glucose utilization by P. gingivalis is known to be very poor, and carbohydrates in general do not appear to readily support growth (61). Strain W83 does, however, contain putative ORFs for all enzymes of the glycolytic pathway, as well as ORFs for a putative glucose/galactose transporter and glucose kinase. Sequence analysis shows that the glucose kinase is encoded in a split ORF generated by a missense mutation, and this is a likely explanation for the poor utilization of glucose to support growth. Four putative ORFs for the pentose phosphate pathway were identified, and it is likely that this pathway plays a role in the generation of precursor metabolites during anaerobic growth (Fig. 2).
Whole-genome analysis suggests that P. gingivalis can metabolize several sugars, including melibiose, galactose, starch, and maltodextrin. The bacterium also possesses enzymes for the degradation of complex amino sugars in the form of hexose aminidases. It is still unclear whether these complex sugars are metabolized, but one possibility is that the removal of amino sugars from host glycoproteins likely renders these proteins more susceptible to degradation by bacterial proteinases. In addition, at least 11 amino acids may serve as substrates for energy production (Fig. 2). These amino acids are most likely derived from the degradation of host tissues (see virulence section below) or from the breakdown of other bacterial cells in the oral cavity. Pathways for glutamate and aspartate utilization have been characterized by enzyme assays (65), and ORFs coding for all of these activities were found in the W83 genome. Intracellular glutamate is deaminated to 2-oxoglutatarate by glutamate dehydrogenase and then decarboxylated to succinyl coenzyme A (succinyl-CoA) by a CoA-dependent 2-oxoglutarate oxidoreductase. The possession of this activity is somewhat unusual in bacterial species (23, 25). It has been established that two-thirds of the succinyl-CoA produced in this reaction is converted to butyryl-CoA and then to butyrate. The remaining third may be converted to propionate by a pathway that involves the enzymes methylmalonyl-CoA mutase and acyl-CoA:acetate-CoA transferase, as reported for other propionate-producing bacteria (18). This pathway appears to be unique to P. gingivalis since other anaerobes catabolize glutamate through the hydroxyglutarate, methylaspartate, and/or the aminobutyrate pathways (5, 17, 18). P. gingivalis did not possesss activities for three key enzymes of these pathways: hydroxyglutarate dehydrogenase, 3-methylaspartate ammonia lyase, and 4-aminobutyrate aminotransferase (65). Peptide-derived aspartate is deaminated to fumarate by aspartate ammonia lyase and then either oxidized to acetate or reduced to propionate and butyrate (65).
Results from Takahashi et al. (65) suggest that P. gingivalis prefers to utilize arginine and lysine as free amino acids rather than in peptide form; thus, carboxy-terminal arginine and lysine residues could be released from proteins by carboxypeptidase activities. Masuda et al. (45) found such an activity in culture supernatants, and an ORF coding for an unspecified carboxypeptidase (PG0232) was identified in the genome. A report that P. gingivalis produces citrulline and ornithine from denatured protein (14) implies that the bacterium degrades arginine through the arginine deiminase pathway. Indeed, a gene with homology to arginine deiminase from Bacillus licheniformis (43) was identified. In addition, two genes—pyrB and pyrI (PG0357 and PG0358)—were contiguous in the genome and shared homology with aspartate/ornithine transcarbamylase catalytic and regulatory chains from Vibrio sp. strain 2693 and Pyrococcus abyssi, respectively.
The lysine catabolic pathways appear to be very similar to those found in Clostridium sp. ORFs were identified for the first steps of both l- and d-lysine catabolism; thus, the isomers are apparently degraded by two different pathways that yield butyric acid, acetic acid, and ammonia. Lysine 2,3-aminomutase (KamA) catalyzes the interconversion of l-lysine and l-β-lysine, the first step in the lysine degradation pathway in Clostridium subterminale SB4 (56). In P. gingivalis W83, kamA was found clustered with the genes kamD and kamE (PG1070, PG1073, and PG1074) that encode subunits of d-lysine 5,6-aminomutase, the first enzyme of the d-lysine degradative pathway. Genes encoding enzymes for the subsequent conversion of lysine to butyrate and acetate were located 3′ to kamE. It is not yet known whether these genes are transcribed as an operon.
Little is known about serine and threonine catabolism in P. gingivalis; however, an ORF was detected with homology to serine dehydratase (PG0084) that hydrolyzes serine to pyruvate, ammonia, and water. Threonine may be split to glycine and acetaldehyde by the activity of threonine aldolase, for which an ORF was detected (PG0474). In summary, P. gingivalis appears to catabolize amino acids through pathways that generate ammonia. The organism has a growth pH optimum of >7.5, and ammonia generation may have evolved as a strategy to shift the local pH to the favored alkaline range.
Several studies have shown that P. gingivalis preferentially uses peptides as sources of carbon and nitrogen (60, 65, 71) and, in addition to the previously described proteinases that are known to degrade host proteins, a number of peptidases that may be involved in the further digestion of protein fragments to smaller peptides and amino acids could be identified from the genome.
There are two carboxylate transporters possibly for lactate and formate, and no sugar transporters other than the aforementioned glucose/galactose importer. Although P. gingivalis possesses a broad assortment of secreted peptidases and pathways for the metabolism of amino acids, the bacterium appears to rely on two predicted peptide uptake systems and has only one amino acid transporter, the characterized sodium ion-driven serine/threonine uptake protein SstT (12). A LysE-type amino acid efflux protein is present that may protect the organism from toxic concentrations of amino acids.
The major fermentation products that can be produced based on whole-genome analysis and in vitro end product analyses are propionate, butyrate, isobutyrate, isovalerate, acetate, ethanol, and butanol (27). Many of these end products are probably toxic to human host tissues (see virulence section below).
Nucleosides and nucleobases may represent a hitherto-unsuspected important nutrient source for P. gingivalis and might be used either as building blocks for nucleic acid biosynthesis or may be catabolized as carbon and energy sources. There are three predicted purine uptake systems, a NupG nucleoside uptake system, and a homolog of the Salmonella enterica serovar Typhimurium nicotinamide mononucleotide transporter PnuC. In addition, there are four homologs of E. coli DinF, a DNA damage-induced protein related to sodium ion-driven drug efflux transporters, that are hypothesized to play a role in nucleoside and/or nucleotide efflux (8).
Common to most human pathogens, iron acquisition appears to be an important priority in P. gingivalis, and there are two iron chelate ABC uptake systems, two TonB-dependent iron receptors, and two FeoB ferrous iron uptake systems. There is an array of metal ion homeostasis transporters, including three sodium ion/proton exchangers, which may be important since a significant number of P. gingivalis transporters are predicted to be sodium ion driven.
Virulence and P. gingivalis.
The availability of the complete genome sequence of P. gingivalis facilitates the identification of putative virulence factors associated with the establishment and survival of the bacterium in the gingival crevice and subsequent penetration into host cells (Table 4). Initially, the bacterium must navigate the oral cavity where, as an obligate anaerobe, it is exposed to limited amounts of oxygen before it establishes itself in an anaerobic environment. A cluster of genes (PG1582 to PG1586) was identified with high levels of similarity to the recently described aerotolerance operon of B. fragilis (66). These functions promote the survival of B. fragilis upon exposure to oxygen, and their presence in P. gingivalis suggests that this system may also ensure tolerance to oxygen in the oral cavity. The genome also encodes a superoxide dismutase (PG1545), genes for an alkyl hydroperoxide reductase (PG0618 and PG0619) (55), a thiol peroxidase (PG1729), and a Dps homolog (PG0090) that is involved in the repair of oxidatively damaged nucleic acids (33).
TABLE 4.
Previously characterized and newly identified putative virulence agents from the P. gingivalis genome sequencea
| Function and virulence agent | Description (reference) | Identification |
|---|---|---|
| Acquisition of iron | ||
| PG1875 | Hemolysin | New identification |
| Adhesion | ||
| PG0411 | Hemagglutinin, putative | New identification |
| PG0443 | Hemagglutinin-related protein | New identification |
| PG1326 | Hemagglutinin, putative | New identification |
| PG1427 | Thiol protease/hemagglutinin PrtT precursor, putative | New identification |
| PG1548 | Thiol protease/hemagglutinin PrtT precursor | New identification |
| PG1674 | Hemagglutinin protein HagB, degenerate | New identification |
| PG1837 | Hemagglutinin protein HagA (21) | Previously known |
| PG1844 | Hemagglutinin protein HagD (64) | Previously known |
| PG1972 | Hemagglutinin protein HagB | Previously sequenced |
| PG1975 | Hemagglutinin protein HagC (39) | Previously known |
| PG2024 | Hemagglutinin protein HagE (64) | Previously known |
| PG2132 | Fimbrilin (16) | Previously known |
| PG2198 | Hemagglutinin protein, truncation | New identification |
| Evasion | ||
| PG0011 | Glycosyl hydrolase, family 3 | New identification |
| PG0032 | Beta-mannosidase, putative | New identification |
| PG0043 | Beta-hexosaminidase | New identification |
| PG0070 | Acyl-(acyl-carrier-protein)-UDP-N-acetylglucosamine acyltransferase | New identification |
| PG0071 | UDP-3-O-acyl-GlcNAc deacetylase | New identification |
| PG0072 | UDP-3-O-3-hydroxymyristoyl glucosamine N-acyltransferase | New identification |
| PG0076 | N-acetylmuramoyl-l-alanine amidase, family 4 | New identification |
| PG0106 | Glycosyl transferase, group 4 family protein | New identification |
| PG0108 | UDP-N-acetyl-d-mannosaminuronic acid dehydrogenase | New identification |
| PG0110 | Glycosyl transferase, group 1 family protein | New identification |
| PG0111 | Capsular polysacharride biosynthesis gene, putative | New identification |
| PG0112 | Conserved hypothetical protein, authentic frameshift | New identification |
| PG0113 | Conserved domain protein | New identification |
| PG0115 | Hexapeptide transferase family protein | New identification |
| PG0117 | Polysaccharide transport protein, putative | New identification |
| PG0118 | Glycosyl transferase, group 2 family protein | New identification |
| PG0119 | Glycosyl transferase, WecB/TagA/CpsF family | New identification |
| PG0120 | UDP-N-acetylglucosamine 2-epimerase | New identification |
| PG0129 | Mannosyltransferase | New identification |
| PG0139 | Membrane-bound lytic murein transglycosylase D, putative | New identification |
| PG0264 | Glycosyl transferase, group 2 family protein | New identification |
| PG0294 | Glycosyl transferase, group 2 family protein | New identification |
| PG0311 | Glycosyl transferase, group 2 family protein | New identification |
| PG0334 | Glycosyl transferase, group 2 family protein | New identification |
| PG0352 | Sialidase, putative | New identification |
| PG0369 | Phosphopantetheine adenylyltransferase | New identification |
| PG0435 | Capsular polysaccharide biosythesis protein, putative | New identification |
| PG0436 | Capsular polysaccharide transport protein, putative | New identification |
| PG0437 | Polysaccharide export protein, BexD/CtrA/VexA family | New identification |
| PG0638 | Tetraacyldisaccharide 4′-kinase | New identification |
| PG0750 | Glycosyl transferase, group 2 family protein | New identification |
| PG0902 | Alpha-1,2-mannosidase family protein | New identification |
| PG0920 | Glycosyl transferase, group 2 family protein | New identification |
| PG0973 | Alpha-1,2-mannosidase family protein | New identification |
| PG1048 | N-acetylmuramoyl-l-alanine amidase, family 3 | New identification |
| PG1058 | Outer membrane protein | New identification |
| PG1135 | Bacterial sugar transferase | New identification |
| PG1140 | Glycosyl transferase, group 2 family protein | New identification |
| PG1141 | Glycosyl transferase, group 1 family protein | New identification |
| PG1143 | Sugar dehydrogenase, UDP-glucose/GDP-mannose dehydrogenase family | New identification |
| PG1149 | Glycosyl transferase, group 1 family protein | New identification |
| PG1155 | ADP-heptose-LPS heptosyltransferase, putative | New identification |
| PG1184 | Alginate O-acetyltransferase, putative | New identification |
| PG1277 | UDP-glucose 6-dehydrogenase, putative | New identification |
| PG1288 | GDP-mannose 4,6-dehydratase | New identification |
| PG1289 | GDP-fucose synthetase | New identification |
| PG1345 | Glycosyl transferase, group 1 family protein | New identification |
| PG1346 | Glycosyl transferase, group 1 family protein | New identification |
| PG1560 | dTDP-glucose 4,6-dehydratase | New identification |
| PG1561 | dTDP-4-dehydrorhamnose reductase | New identification |
| PG1562 | dTDP-4-dehydrorhamnose 3,5-epimerase | New identification |
| PG1563 | Glucose-1-phosphate thymidylyltransferase | New identification |
| PG1565 | 3-Deoxy-d-manno-octulosonic-acid transferase, putative | New identification |
| PG1682 | Glycosyl transferase, group 1 family protein | New identification |
| PG1711 | Alpha-1,2-mannosidase family protein | New identification |
| PG1712 | Alpha-1,2-mannosidase family protein | New identification |
| PG1743 | 2-Dehydro-3-deoxyphosphooctonate aldolase | New identification |
| PG1750 | Alpha-1,3/4-fucosidase, putative | New identification |
| PG1783 | Glycosyl transferase, group 2 family protein | New identification |
| PG1815 | 3-Deoxy-d-manno-octulosonate cytidylyltransferase | New identification |
| PG1880 | Glycosyl transferase, group 2 family protein | New identification |
| PG1884 | Alpha-l-fucosidase precursor, putative | New identification |
| PG1964 | Bacterial sugar transferase | New identification |
| PG2038 | N-acetylmuramoyl-l-alanine amidase, putative | New identification/PICK> |
| PG2162 | Lipid A disaccharide synthase | New identification |
| PG2215 | Mannose-1-phosphate guanylyltransferase | New identification |
| PG2223 | Glycosyl transferase, group 2 family protein | New identification |
| Invasion | ||
| PG0151 | Signal recognition particle-docking protein FtsY | New identification |
| PG0293 | Secretion activator protein, putative | New identification |
| PG0485 | Preprotein translocase, YajC subunit | New identification |
| PG0514 | Preprotein translocase, SecA subunit | New identification |
| PG1115 | Signal recognition particle protein | New identification |
| PG1598 | Lipoprotein signal peptidase, putative | New identification |
| PG1762 | Protein-export membrane protein SecD/protein-export membrane protein SecF | New identification |
| PG1918 | Preprotein translocase, SecY subunit | New identification |
| PG2000 | Signal peptidase-related protein | New identification |
| PG2001 | Signal peptidase I | New identification |
| Stress response | ||
| PG0090 | Dps family protein | New identification |
| PG0245 | Universal stress protein family | New identification |
| PG0618 | Alkyl hydroperoxide reductase, C subunit | New identification |
| PG0619 | Alkyl hydroperoxide reductase, F subunit | New identification |
| PG0876 | Thiophene and furan oxidation protein ThdF | New identification |
| PG1330 | Large conductance mechanosensitive channel protein | New identification |
| PG1545 | Superoxide dismutase, Fe-Mn | New identification |
| PG1582 | BatA protein | New identification |
| PG1583 | BatB protein | New identification |
| PG1584 | BatC protein | New identification |
| PG1585 | BatD protein | New identification |
| PG1586 | BatE protein | New identification |
| PG1648 | RelA/SpoT family protein | New identification |
| PG1729 | Thiol peroxidase | New identification |
| PG1808 | Guanosine-3′,5′-bis(diphosphate) 3′-pyrophosphohydrolase | New identification |
| PG2163 | Stationary-phase survival protein SurE | New identification |
| Antibiotic resistance | ||
| PG0135 | Dimethyladenosine transferase | New identification |
| PG0540 | AcrB/AcrD/AcrF family protein | New identification |
| PG0604 | Penicillin tolerance protein LytB | New identification |
| PG1407 | Nitroimidazole resistance protein, putative | New identification |
| PG1538 | Undecaprenol kinase, putative | New identification |
| Virulence and acquisition of peptides | ||
| PG1754 | Conserved domain protein | New identification |
| PG2115 | Protease PrtT, degenerate | New identification |
| PG0010 | ATP-dependent Clp protease, ATP-binding subunit ClpC | New identification |
| PG0088 | Peptidase, M16 family | New identification |
| PG0137 | Aminoacyl-histidine dipeptidase | New identification |
| PG0159 | Endopeptidase PepO | New identification |
| PG0196 | Peptidase, M16 family | Previously sequenced |
| PG0232 | Zinc carboxypeptidase, putative | New identification |
| PG0235 | Carboxyl-terminal protease | New identification |
| PG0317 | Peptidase, M49 family | New identification |
| PG0383 | Membrane-associated zinc metalloprotease, putative | New identification |
| PG0417 | ATP-dependent Clp protease, ATP-binding subunit ClpX | New identification |
| PG0418 | ATP-dependent Clp protease, proteolytic subunit | New identification |
| PG0445 | Peptidase T | New identification |
| PG0503 | Dipeptidyl aminopeptidase IV(31) | Previously known |
| PG0506 | Arginine-specific cysteine proteinase(63) | Previously known |
| PG0537 | Aminoacyl-histidine dipeptidase | New identification |
| PG0553 | Extracellular protease, putative | New identification |
| PG0561 | Peptidase, M20/M25/M40 family | New identification |
| PG0593 | HtrA protein | New identification |
| PG0620 | ATP-dependent protease La | New identification |
| PG0639 | Signal peptide peptidase SppA, 67K type | New identification |
| PG0724 | Prolyl oligopeptidase family protein | New identification |
| PG0753 | Protease | Previously sequenced |
| PG0758 | Peptidyl-dipeptidase Dcp | New identification |
| PG0889 | Peptidase, M24 family protein | New identification |
| PG0956 | Peptidase, M23/M37 family, putative | New identification |
| PG1004 | Prolyl oligopeptidase family protein | New identification |
| PG1055 | Thiol protease (7) | Previously known |
| PG1060 | Carboxyl-terminal protease | New identification |
| PG1118 | ClpB protein | New identification |
| PG1210 | Peptidase, M24 family protein | New identification |
| PG1361 | Dipeptidyl aminopeptidase IV, putative | New identification |
| PG1605 | Aminopeptidase C | New identification |
| PG1654 | d-alanyl-d-alanine dipeptidase | New identification |
| PG1701 | trp-G type glutamine amidotransferase/dipeptidase | New identification |
| PG1724 | O-sialoglycoprotein endopeptidase | New identification |
| PG1788 | Cysteine peptidase, putative | New identification |
| PG1789 | Peptidyl-dipeptidase Dcp | New identification |
| PG1855 | Carboxyl-terminal protease | New identification |
| PG2192 | Peptidase, M23/M37 family | New identification |
The newly identified virulence agents were selected based on the presence of signal peptide sequences or transmembrane domains or by homology to previously described virulence agents in P. gingivalis.
The bacterium uses fimbriae to adhere to other bacterial species and host tissues. Hemagglutinins and various proteases (gingipains) are also involved in tissue colonization through adhesion to extracellular matrix proteins (38, 53, 59). Hemagglutinins in particular may mediate the binding of bacteria to receptors on human cells (21), and the gene sequences for six newly identified putative hemagglutinin-like proteins (PG0411, PG1326, PG1674, PG1427, PG1548, and PG2198) could be identified. Four of these are recent duplications in the genome of HagA and HagD adhesin domain-related sequences. A total of 42 proteinases were identified in the genome sequence that may enable adherence of the bacterium to host tissues, as well as to other bacterial cells, and that may also degrade host proteins (as discussed above). In vitro experiments have demonstrated that proteases attack a range of host proteins, including extracellular matrix proteins (32, 38, 53, 59) and cell adhesion molecules (29), the destruction of which leads to a loss of cell surface receptors (59) and tissue integrity (29). Protease destruction of cytokines (15, 42, 74) and gamma interferon (73) can result in disruption of polymorphonuclear leukocyte function (48) and ultimately affect the host immune response.
A single hemolysin for the release of iron and protophoryn IX (PG1875) was identified. This sequence has full-length homology only to the characterized hemolysin gene of another periodontal pathogen, Prevotella melaninogenica (3). These two hemolysins show absolutely no homology to any other biochemically characterized hemolysin and have weak homology to a conserved hypothetical protein/putative hemolysin fusion protein sequence from Vibrio cholerae.
In P. gingivalis, metabolic end products from the catabolism of various substrates include short-chained carboxylic acids that can affect the host defense system in a variety of ways. When applied directly to healthy human gingiva tissue, short-chain carboxylic acids have been shown to stimulate a gingival inflammatory response and inflammatory cytokine release (50). Short-chain carboxylic acids have also been shown to alter cell function and gene expression and may also contribute to the initiation and prolongation of gingival inflammation (50).
The capsule of P. gingivalis is most likely involved in the evasion of the host response and has been shown to be one of the important virulence determinants in this bacterium (34). Whole-genome analysis reveals at least four capsular biosynthesis gene clusters (PG0106 to PGPG0120, PG0435 to PG0437, PG1140 to PG1149, and PG1560 to PG1565) that are located across the genome. Closer investigation of these gene clusters suggests that mannose, glucose, and rhamnose may be some of the sugars that are present in the capsule of P. gingivalis strain W83. In several pathogens the secretion of virulence factors targeted to the host cells is mediated by type III protein secretion systems. The complete genome of P. gingivalis was searched for the presence of a cluster of nine genes Sct (Hrc/Ysc) that are known to be components of type III protein secretion systems (24). No BLAST matches with these motifs were found. Although several sec gene homologs are present in the genome, including SecA, SecY, SecD, and SecF, the main terminal branch of the general secretory pathway (type II) could not be identified, suggesting that this pathway is not functional in this bacterium.
DISCUSSION
It is estimated that 35% of the U.S. population has some form of periodontitis (2). Traditional methods for the prevention or treatment of periodontal disease include the mechanical removal of plaque and the use of antimicrobial agents. The availability of the genome sequences of at least three oral pathogens and recent investigations into the microbial composition of the human oral microbiome will afford new opportunities to investigate ways to alter the composition of the subgingival biofilm. Potential strategies include those that would decrease opportunities for biofilm formation, reduce attachment among species in the biofilm, or limit the availability of required nutrients. Inhibition of the primary colonizers could prevent the successful establishment of late colonizers such as P. gingivalis.
Recently, specific bacteria have been found to be associated with systemic diseases, e.g., Helicobacter pylori, as etiological agent in gastric ulcers, and the tentative association between the presence of Chlamydia pneumoniae in atherosclerotic plaques and cardiovascular disease (9). Stimulated by these studies and the detection of P. gingivalis and other oral pathogens in atherosclerotic plaques (10, 62), new research is assessing associations between periodontal infection and cardiovascular disease (22, 47, 52, 58). That the pathogenicity of oral bacteria may extend beyond their known ecological niche to other organ systems introduces an important and exciting new dimension to defining the genetic complement of these organisms. Although the genome sequence has not revealed many of the classical virulence factors that are associated with pathogens, it is anticipated that the newly described putative virulence factors of this bacterium, as well as from the others that have been recently sequenced or whose sequencing is nearing completion, will enable the development of antimicrobial agents that can be used against one of the major causative agents of periodontal disease. Ultimately, the genome sequence of P. gingivalis will facilitate an increased understanding of the virulence of this periodontal pathogen and will enable the development of improved diagnostics and therapeutics.
Acknowledgments
We thank Michael Heaney, Michael Holmes, Vadim Sapiro, and Emmanuel Mongodin for informatics support at TIGR. We thank J. F. Tomb for assistance in the early stages of the project.
This work was supported by the National Institute of Dental and Craniofacial Research at the National Institutes of Health (R01DE-13914).
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albandar, J. M., J. A. Brunelle, and A. Kingman. 1999. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988-1994. J. Periodontol. 70:13-29. [DOI] [PubMed] [Google Scholar]
- 3.Allison, H. E., and J. D. Hillman. 1997. Cloning and characterization of a Prevotella melaninogenica hemolysin. Infect. Immun. 65:2765-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahloul, A., F. Boubrik, and J. Rouviere-Yaniv. 2001. Roles of Escherichia coli histone-like protein HU in DNA replication: HU-beta suppresses the thermosensitivity of dnaA46ts. Biochimie 83:219-229. [DOI] [PubMed] [Google Scholar]
- 5.Barker, H. A. 1981. Amino acid degradation by anaerobic bacteria. Annu. Rev. Biochem. 50:23-40. [DOI] [PubMed]
- 6.Boone, D. R., and R. W. Castenholtz (ed.). 2001. Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y.
- 7.Bourgeau, G., H. Lapointe, P. Peloquin, and D. Mayrand. 1992. Cloning, expression, and sequencing of a protease gene (tpr) from Porphyromonas gingivalis W83 in Escherichia coli. Infect. Immun. 60:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, M. H., I. T. Paulsen, and R. A. Skurray. 1999. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31:394-395. [DOI] [PubMed] [Google Scholar]
- 9.Byrne, G. I., S. I. Skarlotos, C. Grunfeld, M. V. Kalayoglu, P. Libby, P. Saikku, J. T. Summersgill, and P. Wyrick. 2000. Collaborative multidisciplinary workshop report: interface of lipid metabolism, atherosclerosis, and Chlamydia infection. J. Infect. Dis. 181(Suppl. 3):S490-S491. [DOI] [PubMed] [Google Scholar]
- 10.Chiu, B. 1999. Multiple infections in carotid atherosclerotic plaques. Am. Heart J. 138:S534-S536. [DOI] [PubMed] [Google Scholar]
- 11.Churchward, G. 2002. Conjugative transposons and related elements, p. 177-191. In N. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 12.Dashper, S. G., L. Brownfield, N. Slakeski, P. S. Zilm, A. H. Rogers, and E. C. Reynolds. 2001. Sodium ion-driven serine/threonine transport in Porphyromonas gingivalis. J. Bacteriol. 183:4142-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endo, J., M. Otsuka, E. Ohara, M. Sato, and R. Nakamura. 1989. Cleavage action of a trypsin-like protease from Bacteroides gingivalis 381 on reduced egg-white lysozyme. Arch. Oral Biol. 34:911-916. [DOI] [PubMed] [Google Scholar]
- 15.Fletcher, J., K. Reddi, S. Poole, S. Nair, B. Henderson, P. Tabona, and M. Wilson. 1997. Interactions between periodontopathogenic bacteria and cytokines. J. Periodontal Res. 32:200-205. [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara, T., S. Morishima, I. Takahashi, and S. Hamada. 1993. Molecular cloning and sequencing of the fimbrilin gene of Porphyromonas gingivalis strains and characterization of recombinant proteins. Biochem. Biophys. Res. Commun. 197:241-247. [DOI] [PubMed] [Google Scholar]
- 17.Gharbia, S. E., and H. N. Shah. 1991. Pathways of glutamate catabolism among Fusobacterium species. J. Gen. Microbiol. 137(Pt. 5):1201-1206. [DOI] [PubMed] [Google Scholar]
- 18.Gottschalk, G. 1986. Bacterial fermentations, p. 208-282. In G. Gottschalk (ed.), Bacterial metabolism, 2nd ed. Springer-Verlag, New York, N.Y.
- 19.Goulbourne, P. A., and R. P. Ellen. 1991. Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J. Bacteriol. 173:5266-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grenier, D. 1992. Demonstration of a bimodal coaggregation reaction between Porphyromonas gingivalis and Treponema denticola. Oral Microbiol. Immunol. 7:280-284. [DOI] [PubMed] [Google Scholar]
- 21.Han, N., J. Whitlock, and A. Progulske-Fox. 1996. The hemagglutinin gene A (hagA) of Porphyromonas gingivalis 381 contains four large, contiguous, direct repeats. Infect. Immun. 64:4000-4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herzberg, M. C., and M. W. Meyer. 1998. Dental plaque, platelets, and cardiovascular diseases. Ann. Periodontol. 3:151-160. [DOI] [PubMed] [Google Scholar]
- 23.Hoffman, P. S., A. Goodwin, J. Johnsen, K. Magee, and S. J. Veldhuyzen van Zanten. 1996. Metabolic activities of metronidazole-sensitive and -resistant strains of Helicobacter pylori: repression of pyruvate oxidoreductase and expression of isocitrate lyase activity correlate with resistance. J. Bacteriol. 178:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes, N. J., C. L. Clayton, P. A. Chalk, and D. J. Kelly. 1998. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J. Bacteriol. 180:1119-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen, R., J. D. Embden, W. Gaastra, and L. M. Schouls. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565-1575. [DOI] [PubMed] [Google Scholar]
- 27.Kaczmarek, F. S., and A. L. Coykendall. 1980. Production of phenylacetic acid by strains of Bacteroides asaccharolyticus and Bacteroides gingivalis (sp. nov.). J. Clin. Microbiol. 12:288-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapatral, V., I. Anderson, N. Ivanova, G. Reznik, T. Los, A. Lykidis, A. Bhattacharyya, A. Bartman, W. Gardner, G. Grechkin, L. Zhu, O. Vasieva, L. Chu, Y. Kogan, O. Chaga, E. Goltsman, A. Bernal, N. Larsen, M. D'Souza, T. Walunas, G. Pusch, R. Haselkorn, M. Fonstein, N. Kyrpides, and R. Overbeek. 2002. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J. Bacteriol. 184:2005-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katz, J., V. Sambandam, J. H. Wu, S. M. Michalek, and D. F. Balkovetz. 2000. Characterization of Porphyromonas gingivalis-induced degradation of epithelial cell junctional complexes. Infect. Immun. 68:1441-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinder, S. A., and S. C. Holt. 1989. Characterization of coaggregation between Bacteroides gingivalis T22 and Fusobacterium nucleatum T18. Infect. Immun. 57:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiyama, M., M. Hayakawa, T. Shiroza, S. Nakamura, A. Takeuchi, Y. Masamoto, and Y. Abiko. 1998. Sequence analysis of the Porphyromonas gingivalis dipeptidyl peptidase IV gene. Biochim. Biophys. Acta 1396:39-46. [DOI] [PubMed] [Google Scholar]
- 32.Kontani, M., H. Ono, H. Shibata, Y. Okamura, T. Tanaka, T. Fujiwara, S. Kimura, and S. Hamada. 1996. Cysteine protease of Porphyromonas gingivalis 381 enhances binding of fimbriae to cultured human fibroblasts and matrix proteins. Infect. Immun. 64:756-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 34.Laine, M. L., and A. J. van Winkelhoff. 1998. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol. Immunol. 13:322-325. [DOI] [PubMed] [Google Scholar]
- 35.Lamont, R. J., S. Gil, D. R. Demuth, D. Malamud, and B. Rosan. 1994. Molecules of Streptococcus gordonii that bind to Porphyromonas gingivalis. Microbiology 140(Pt. 4):867-872. [DOI] [PubMed] [Google Scholar]
- 36.Lamont, R. J., G. W. Hsiao, and S. Gil. 1994. Identification of a molecule of Porphyromonas gingivalis that binds to Streptococcus gordonii. Microb. Pathog. 17:355-360. [DOI] [PubMed] [Google Scholar]
- 37.Lamont, R. J., and H. F. Jenkinson. 2000. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol. Immunol. 15:341-349. [DOI] [PubMed] [Google Scholar]
- 38.Lantz, M. S., R. D. Allen, L. W. Duck, J. L. Blume, L. M. Switalski, and M. Hook. 1991. Identification of Porphyromonas gingivalis components that mediate its interactions with fibronectin. J. Bacteriol. 173:4263-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lepine, G., and A. Progulske-Fox. 1996. Duplication and differential expression of hemagglutinin genes in Porphyromonas gingivalis. Oral Microbiol. Immunol. 11:65-78. [DOI] [PubMed] [Google Scholar]
- 40.Lewis, J. P., and F. L. Macrina. 1998. IS195, an insertion sequence-like element associated with protease genes in Porphyromonas gingivalis. Infect. Immun. 66:3035-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loos, B. G., D. W. Dyer, T. S. Whittam, and R. K. Selander. 1993. Genetic structure of populations of Porphyromonas gingivalis associated with periodontitis and other oral infections. Infect. Immun. 61:204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madianos, P. N., P. N. Papapanou, and J. Sandros. 1997. Porphyromonas gingivalis infection of oral epithelium inhibits neutrophil transepithelial migration. Infect. Immun. 65:3983-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maghnouj, A., T. F. de Sousa Cabral, V. Stalon, and C. Vander Wauven. 1998. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor argR. J. Bacteriol. 180:6468-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maley, J., and I. S. Roberts. 1994. Characterisation of IS1126 from Porphyromonas gingivalis W83: a new member of the IS4 family of insertion sequence elements. FEMS Microbiol. Lett. 123:219-224. [DOI] [PubMed] [Google Scholar]
- 45.Masuda, K., H. Hayashi, M. Yoshioka, and R. Nakamura. 1995. Presence of an arginine carboxypeptidase in culture supernatants of Porphyromonas gingivalis. J. Dent. Res. 74:974. [Google Scholar]
- 46.Menard, C., and C. Mouton. 1995. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect. Immun. 63:2522-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer, D. H., and P. M. Fives-Taylor. 1998. Oral pathogens: from dental plaque to cardiac disease. Curr. Opin. Microbiol. 1:88-95. [DOI] [PubMed] [Google Scholar]
- 48.Nakayama, K., T. Kadowaki, K. Okamoto, and K. Yamamoto. 1995. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis: evidence for significant contribution of Arg-gingipain to virulence. J. Biol. Chem. 270:23619-23626. [DOI] [PubMed] [Google Scholar]
- 49.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, C. M. Fraser, et al. 1999. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 50.Niederman, R., J. Zhang, and S. Kashket. 1997. Short-chain carboxylic-acid-stimulated, PMN-mediated gingival inflammation. Crit. Rev. Oral Biol. Med. 8:269-290. [DOI] [PubMed] [Google Scholar]
- 51.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Offenbacher, S., P. N. Madianos, C. M. Champagne, J. H. Southerland, D. W. Paquette, R. C. Williams, G. Slade, and J. D. Beck. 1999. Periodontitis-atherosclerosis syndrome: an expanded model of pathogenesis. J. Periodontal Res. 34:346-352. [DOI] [PubMed] [Google Scholar]
- 53.Pike, R. N., J. Potempa, W. McGraw, T. H. Coetzer, and J. Travis. 1996. Characterization of the binding activities of proteinase-adhesin complexes from Porphyromonas gingivalis. J. Bacteriol. 178:2876-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riley, M. 1993. Functions of the gene products of Escherichia coli. Microbiol. Rev. 57:862-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rocha, E. R., and C. J. Smith. 1999. Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 181:5701-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruzicka, F. J., K. W. Lieder, and P. A. Frey. 2000. Lysine 2,3-aminomutase from Clostridium subterminale SB4: mass spectral characterization of cyanogen bromide-treated peptides and cloning, sequencing, and expression of the gene kamA in Escherichia coli. J. Bacteriol. 182:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scannapieco, F. A., and R. J. Genco. 1999. Association of periodontal infections with atherosclerotic and pulmonary diseases. J. Periodontal Res. 34:340-345. [DOI] [PubMed] [Google Scholar]
- 59.Scragg, M. A., S. J. Cannon, M. Rangarajan, D. M. Williams, and M. A. Curtis. 1999. Targeted disruption of fibronectin-integrin interactions in human gingival fibroblasts by the RI protease of Porphyromonas gingivalis W50. Infect. Immun. 67:1837-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seddon, S. V., H. N. Shah, J. M. Hardie, and J. P. Robinson. 1988. Chemically defined and minimal media for Bacteroides gingivalis. Curr. Microbiol. 17:147-149. [Google Scholar]
- 61.Shah, H. N., and R. A. D. Williams. 1987. Utilization of glucose and amino acids by Bacteroides intermedius and Bacteroides gingivalis. Curr. Microbiol. 15:241-246. [Google Scholar]
- 62.Shelburne, C. E., N. P. VanPoperin, R. M. Gleason, A. Prabhu, and D. E. Lopatin. 2000. Assay of bacterial DNA in atherosclerotic lesions by quantitative PCR. J. Dent. Res. 79:226.
- 63.Slakeski, N., P. S. Bhogal, N. M. O'Brien-Simpson, and E. C. Reynolds. 1998. Characterization of a second cell-associated Arg-specific cysteine proteinase of Porphyromonas gingivalis and identification of an adhesin-binding motif involved in association of the prtR and prtK proteinases and adhesins into large complexes. Microbiology 144(Pt. 6):1583-1592. [DOI] [PubMed] [Google Scholar]
- 64.Slakeski, N., S. M. Cleal, P. S. Bhogal, and E. C. Reynolds. 1999. Characterization of a Porphyromonas gingivalis gene prtK that encodes a lysine-specific cysteine proteinase and three sequence-related adhesins. Oral Microbiol. Immunol. 14:92-97. [DOI] [PubMed] [Google Scholar]
- 65.Takahashi, N., T. Sato, and T. Yamada. 2000. Metabolic pathways for cytotoxic end product formation from glutamate- and aspartate-containing peptides by Porphyromonas gingivalis. J. Bacteriol. 182:4704-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang, Y. P., M. M. Dallas, and M. H. Malamy. 1999. Characterization of the Batl (Bacteroides aerotolerance) operon in Bacteroides fragilis: isolation of a B. fragilis mutant with reduced aerotolerance and impaired growth in in vivo model systems. Mol. Microbiol. 32:139-149. [DOI] [PubMed] [Google Scholar]
- 67.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 68.Tribble, G. D., A. C. Parker, and C. J. Smith. 1999. Genetic structure and transcriptional analysis of a mobilizable, antibiotic resistance transposon from Bacteroides. Plasmid 42:1-12. [DOI] [PubMed] [Google Scholar]
- 69.Wang, C. Y., V. C. Bond, and C. A. Genco. 1997. Identification of a second endogenous Porphyromonas gingivalis insertion element. J. Bacteriol. 179:3808-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.White, O. Bacterial genome annotation at The Institute for Genomic Research. In C. M. Fraser, T. D. Read, and K. E. Nelson (ed.), Microbial genomics, in press. Humana Press, Inc., New York, N.Y.
- 71.Wyss, C. 1992. Growth of Porphyromonas gingivalis, Treponema denticola, T. pectinovorum, T. socranskii, and T. vincentii in a chemically defined medium. J. Clin. Microbiol. 30:2225-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao, E. S., R. J. Lamont, S. P. Leu, and A. Weinberg. 1996. Interbacterial binding among strains of pathogenic and commensal oral bacterial species. Oral Microbiol. Immunol. 11:35-41. [DOI] [PubMed] [Google Scholar]
- 73.Yun, P. L., A. A. DeCarlo, and N. Hunter. 1999. Modulation of major histocompatibility complex protein expression by human gamma interferon mediated by cysteine proteinase-adhesin polyproteins of Porphyromonas gingivalis. Infect. Immun. 67:2986-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, J., H. Dong, S. Kashket, and M. J. Duncan. 1999. IL-8 degradation by Porphyromonas gingivalis proteases. Microb. Pathog. 26:275-280. [DOI] [PubMed] [Google Scholar]