Abstract
Respiratory syncytial virus (RSV) has recently been recognized as a serious pathogen in elderly and immunocompromised adults. Diagnosis of acute infection in adults is often difficult due to the insensitivity of viral culture, and reverse transcription-PCR (RT-PCR) is a more sensitive alternative. The relationship of quantitative RT-PCR to viable virus has never been studied for RSV. Therefore, we compared a quantitative real-time RT-PCR with viral culture to assess viral load in adult volunteers challenged with the RSV A2 strain. Twelve of 13 volunteers were infected, and there was a high correlation (r = 0.84) between quantitative RT-PCR and viral titer by cell culture. However, RT-PCR was more sensitive, with 73 of 169 (43%) samples positive compared to 58 (34%) samples positive by culture. The correlation between the two tests was highest early in the course of viral shedding (r = 0.91, days 0 to 6), whereas during days 7 to 13, there was more variability (r = 0.70). All subjects were culture negative by day 11, whereas one subject remained RT-PCR positive on day 12. All subjects were RT-PCR negative at day 28 postinfection. Quantitative RT-PCR has an excellent correlation with viral titers, as measured by culture, and should be a useful tool for future studies addressing viral load and disease pathogenesis.
Respiratory syncytial virus (RSV) is a frequent cause of serious lower respiratory tract disease in young children (13). Reinfections occur throughout life and are generally mild in healthy adults. However, elderly or immunocompromised adults may develop severe RSV infection (4, 5, 23). In contrast to children, diagnosis in adults is often difficult due to the insensitivity of viral culture (7). This is likely due to thermolability of the virus and low titers of virus shed by adults and may also be due to the presence of preexisting nasal antibody in clinical specimens resulting in in vitro neutralization. Thus, investigators have increasingly relied on new sensitive molecular techniques such as reverse transcription PCR (RT-PCR) for the immediate diagnosis of RSV infection in adults (6, 9-11, 14, 30). In addition, the recent development of real-time RT-PCR, in which the concentration of amplification products is monitored as they accumulate during thermal cycling, allows quantification of RNA in specimens (18). The ability to assess the relationship of viral load to severity of illness and determination of the location of viral replication in the respiratory tract would improve the understanding of disease pathogenesis in adults as well as children. Quantitative RT-PCR has been used successfully to monitor disease progression and assess response to therapy for malignant diseases and other viral infections that are difficult to cultivate, such as human immunodeficiency virus and hepatitis C virus (2, 12, 15, 20, 29). However, the relationship of RNA copy to viable organisms and clinical illness is less well defined for respiratory pathogens, including RSV (9, 16, 17, 19, 22). Therefore, we developed a quantitative real-time RT-PCR and compared it to viral culture to assess viral load in adult volunteers challenged with the RSV A2 strain.
MATERIALS AND METHODS
Subjects.
Thirteen healthy adults, ages 21 to 50 years, were inoculated with the RSV A2 challenge pool of virus developed by the National Institute of Allergy and Infectious Diseases. Infection was established by nasal inoculation of virus at a dose of 104.7 50% tissue culture infective doses (TCID50). Subjects underwent nasal washes on day 0 (prior to challenge), days 1 through 12, and day 28 postinfection. Nasal washes were performed by instilling 5 ml of sterile saline in each nostril. Recovered nasal wash was diluted 1:5 with 5× viral transport medium. Samples were immediately transported on wet ice to the laboratory for viral culture. This study was approved by the University of Rochester Institutional Review Board, and informed consent was obtained from all subjects prior to enrollment.
Virus culture.
Within 2 h of collection, samples were inoculated onto cell culture. Four hundred microliters of nasal sample was placed onto HEp-2 cells in roller tubes. Tubes were incubated at 35°C on a rotating wheel and examined daily for cytopathic effect (CPE). CPE characteristic of RSV was confirmed by immunofluorescent staining with RSV-specific monoclonal antibodies (Bartel's Diagnostics, Issaquah, Wash.). Simultaneously, nasal wash samples were inoculated onto 96-well tissue culture plates to determine the viral titer by standard methods (24). Briefly, 100 μl of five 10-fold dilutions of each sample was added in duplicate to HEp-2 cell monolayers and observed daily for CPE. The viral titer was calculated as the TCID50 per milliliter of nasal sample (21). Following cultures, the remaining sample was aliquoted, fast frozen, and at stored at −70°C for RT-PCR testing at a later date.
RT-PCR.
Quantitative RT-PCR was performed on all samples and nested nonquantitative RT-PCR was done on a subset of samples that were negative by the quantitative assay. The quantitative RT-PCR was developed to be group A or B specific, whereas the nested RT-PCR method detects group A and B RSV. Since the challenge study utilized the A2 strain of RSV, the quantitative RT-PCR specific for group A virus was performed.
Sample extraction.
Forty micrograms of tRNA (Gibco BRL, Gaithersburg, Md.) was added to each 250-μl sample, which was then extracted with 750 μl of LS Stat (Tel-Test, Inc., Friendswood, Tex.) according to the manufacturer's instructions. The RNA was precipitated with 100% isopropanol for 15 min at −70°C and centrifuged, and the pellet was washed with 75% ethanol. After drying, the RNA pellet was dissolved in 10.5 μl of water for RT-PCR.
Primers and probes.
The primers and probes used in this study are based on nucleotide sequences from the F gene of RSV group A and B viruses (3). Probes utilized the fluorescent dyes Texas red and FAM with black hole quenchers (Integrated DNA Technologies, Coralville, Iowa).
Quantitative RT-PCR.
For the RT step, 5.25 μl of RNA was combined with 10 U of avian myeloblastosis virus reverse transcriptase, 10 U of RNasin, 200 μM deoxynucleoside triphosphates (dNTP), and the forward inner primer for group A (FA; 5′ CACCCTGTTGGAAAC) at a final concentration of 200 nM. The mixture was incubated at 42°C for 1 h. Five microliters of the resulting cDNA product was used in the PCR.
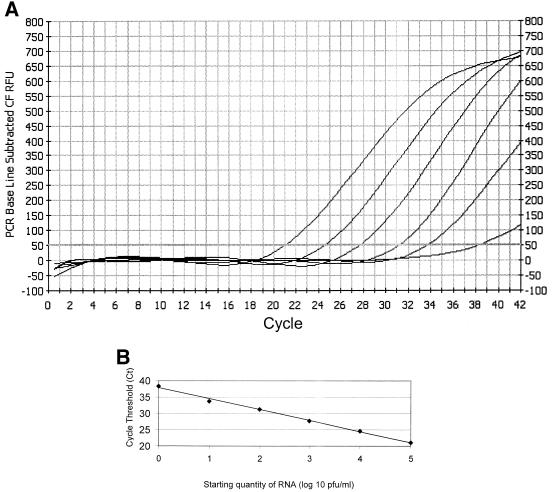
The amplification mixture contained 200 nM FA, the inner reverse common group A and B primer (IRC; 5′ CTCTGTCAGTTCTTG 3′), and the group A probe (FAM; 5′ ATGTTGGACCCTTCTTTTGTGTTGGTTGTA 3′). The mixture also contained 5 μl of RT product, 5 mM MgCl2, 400 μM dNTP (with dUTP replacing dTTP), 5 U of Taq polymerase, and 1 U of uracyl DNA-glycosylase (UNG). This mixture was placed at room temperature for 10 min. The UNG was then inactivated at 95°C for 2 min. Amplification was performed on a Bio-Rad iCycler for 40 cycles of 95°C for 5 s, 42°C for 40 s, and 68°C for 10 s. Relative fluorescent units (RFU) for the FAM probe were monitored during each cycle at 490 nM. The cycle threshold (Ct), which is the cycle number at which a positive amplification reaction was identified, was defined when the RFU exceeded 10 times the standard deviation of baseline RFU values for all samples. The lower limit of detection for the assay was 10 PFU/ml. A standard curve was developed with serial 10-fold dilutions of stock RSV A2 strain (106 PFU/ml). (Fig. 1) The Ct values were plotted against virus quantity in log10 PFU per milliliter. This curve was used to convert the Ct values for nasal wash specimens to PFU-per-milliliter equivalents.
FIG. 1.
Quantitative RT-PCR. (A) The PCR uses the Taqman principle with RFU monitored on an iCycler thermocycler. RFU were plotted, and the Ct for each reaction was determined. Six 10-fold dilutions of stock virus (106 PFU/ml) were used to generate a standard curve. Shown from left to right are the curves of decreasing concentrations of stock virus from dilutions of 10−1 to 10−6. The Ct is defined as the point at which the curve crosses the horizontal threshold line at 60 RFU. The limit of detection for the assay was defined as the 10−5 dilution (10 PFU/ml), because the 10−6 dilution was not reliably positive. (B) The log10 titer of virus in a specimen was plotted against the Ct value, and a best fit line was constructed. The linear range of the assay is from 1 to 105 PFU/ml, with a correlation coefficient of 0.99. RSV quantity in unknown clinical samples, expressed as log10 PFU-per-milliliter equivalents, is derived by plotting the Ct of an unknown on the standard curve.
Real-time single-tube nested RT-PCR.
The real-time single-tube nested RT-PCR nonquantitative assay is a modification of a previously developed “hanging droplet” PCR method that incorporates a Taqman reaction (27). The quantitative reaction described above serves as the nested component; however, additional primers are utilized to amplify an outer larger segment of the gene and to detect both group A and B viruses. Briefly, extracted RNA was converted to cDNA by RT using the outer forward common (OFC) primer (5′ ATGCAGGTGTAACAACACCTTTAAGCACTTACATGTTAAC 3′) and dNTPs. The cDNA is placed in a PCR tube with the outer reverse common (ORC) primer (5′ GTAATGTTAAACTGTTCATAGTGTCACAAAATACTCGATT 3′) and additional OFC primer, dNTPs with dUTP replacing dTTP, Taq polymerase, and N-glycosylase (UNG). A 9-μl droplet containing the FA primer and forward group B primer (FB; 5′ CACCTTGCTGGAAAT 3′) and the IRC primer with the group A probe and group B probe (Texas red; 5′ ATATTTGATCCTTCTTTGATGTTGGTGGTG 3′) was placed in the underside of the PCR tube lid. Additional Taq polymerase and 21% glycerol were also placed in the droplet. After the first round of PCR (30 cycles of 95°C for 5 s, 42°C for 40 s, and 68°C for 10 s), the tubes were inverted to incorporate the hanging droplet containing the inner primers and probes, and the second round of PCR was run for 40 cycles using the same parameters as in the first round. RFU are measured at 490 nm (FAM) and 580 nm (Texas red). The lower limit of detection for the assay is 1.0 PFU/ml for RSV group A or B.
Nasal IgA to RSV.
Titers of immunoglobulin A (IgA) to RSV fusion protein were performed by enzyme immunoassay (EIA) according to published methods (26). Briefly, the RSV group A fusion protein was purified by affinity chromatography as previously described (25). EIA plates (96 wells each) were coated with antigen, and 100 μl of nasal wash was added at serial twofold dilutions from 1:2 to 1:16 in duplicate. The plates were incubated overnight, and bound IgA was detected by alkaline phosphatase-conjugated goat anti-human IgA, followed by substrate. Titers are expressed as the log2 of the nasal dilution in which the optical density is 0.2 and at least twice the background value of wells without antigen. The nasal titer of RSV IgA was standardized with a control nasal wash containing a defined quantity of RSV-specific IgA.
RESULTS
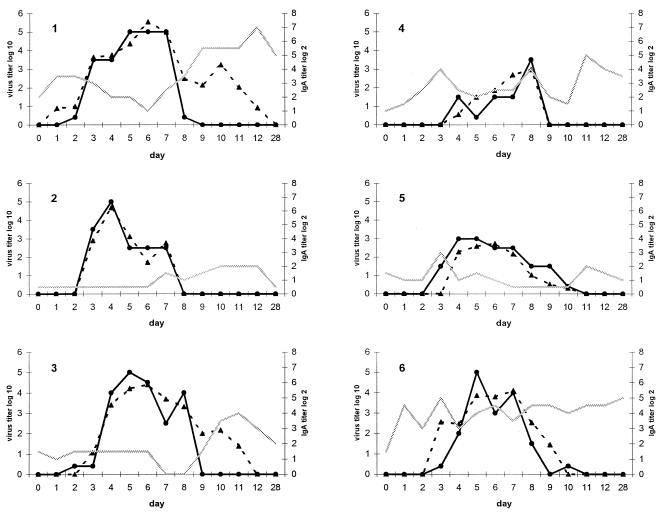
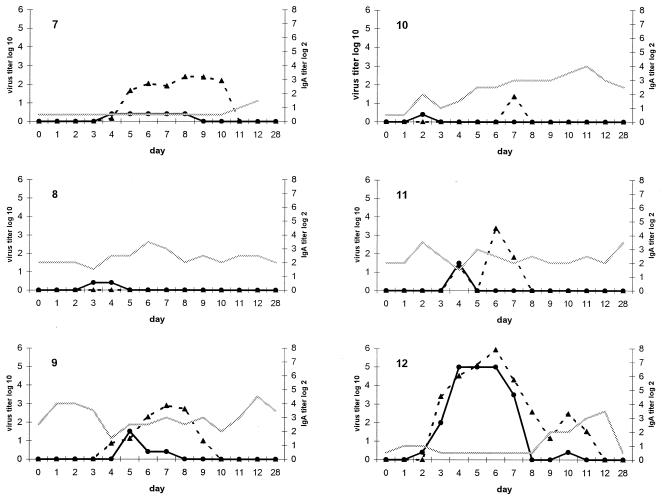
Twelve of 13 subjects were infected, as judged by at least one nasal wash positive by viral culture. One subject was culture and RT-PCR negative in all samples. Overall, there was good correlation (r = 0.84) between quantitative RT-PCR and viral titer by cell culture. However, RT-PCR was more sensitive, with 73 of 169 (43%) samples positive by RT-PCR, while 58 (34%) samples were positive by culture. One of 12 subjects who shed virus as detected in two undiluted specimens cultured in roller tubes had negative quantitative and nested RT-PCR on all days. The patterns of shedding and titers of RSV over time were very similar for both methods (Fig. 2). The correlation between the two tests was best early in the course of viral shedding (r = 0.91, days 0 to 6), whereas during days 7 to 13, there was more variability (r = 0.70). Half of the subjects had RT-PCR titers that were higher and remained positive after virus could be cultured (subjects 1, 3, 7, 10, 11, and 12). The average time to virus shedding was approximately 3 days by either method. (RT-PCR, 3.4 ± 1.6 days; viral culture, 3.1 ± 1.0 days). On day 1 postinoculation, only two subjects were positive by quantitative RT-PCR, and four additional subjects were detected by the more sensitive nested RT-PCR. The average last day subjects were RT-PCR positive was day 9.2 ± 1.8, compared to day 7.2 ± 2.6 by viral culture (P = 0.05). Titers peaked slightly earlier by viral culture than by RT-PCR (4.6 ± 1.5 versus 6.5 ± 1.1 days), but this difference was not significant. All persons were culture negative by day 11. At day 12 postchallenge, 1 of 12 viral shedders was positive by quantitative RT-PCR, whereas 4 were positive on day 12 by nested RT-PCR. All samples were negative by quantitative and nested RT-PCR on day 28.
FIG. 2.
Graphic representation of the relationship of RSV viral load in nasal specimens (left y axis) and nasal IgA titer (right y axis and shaded line) in 12 infected volunteers challenged with the RSV A2 strain. The virus titer is reported as TCID50 per milliliter for culture (solid line) and as PFU-per-milliliter equivalents for RT-PCR (dotted line) as described in Materials and Methods and the legend to Fig. 1. Each graph represents 1 of the 12 subjects infected with RSV.
Nasal IgA titers were determined in the nasal wash specimens from days 0 to 12 and day 28. Although titers varied from day to day, in 5 of 12 infected persons, the titers of nasal IgA to the F protein rose at approximately the time viral shedding by culture ceased (day 8). There did not appear to be a consistent relationship between nasal IgA titers and how well RT-PCR and viral culture correlated, as illustrated by patients 6 and 7. Subject 7 had very low IgA and minimal cultivatable virus with relatively high RT-PCR titers. In contrast, subject 6 had high IgA titers as well as high titers of virus detected by both RT-PCR and culture.
DISCUSSION
The development of molecular techniques for the diagnosis and quantification of pathogens that cannot be cultured by traditional techniques has revolutionized the field of microbiology and infectious diseases. Certain organisms, such as RSV, can be grown in cell culture, yet the virus is relatively labile, and many investigators have begun to use RT-PCR to identify infection (30). This is particularly true for adults, for whom viral culture is relatively insensitive (7, 8). Nested RT-PCR has been successfully used in epidemiologic studies of adults, with a sensitivity of 73% and specificity of 99% (6). However, concern has arisen that new molecular techniques that detect minute quantities of viral RNA may not be associated with clinically meaningful illnesses and that residual nucleic acid may persist in the nasopharynx of infected individuals for long periods of time. Only one previous study has compared quantitative RT-PCR to viral culture for assessment of a respiratory virus over time (22). In this investigation of six subjects with symptomatic influenza virus infection, virus could be detected for up to 7 days with PCR compared to 1 to 2 days with viral culture. The present study helps to define the meaning of quantitative RT-PCR in relation to viral culture for RSV and addresses the issue of low-level RNA persistence following infection.
The overall correlation of quantitative RT-PCR and viral titers was excellent. The pattern of viral shedding was quite similar by either method, although subjects tended to remain RT-PCR positive several days after virus was no longer isolated by culture. However, we did not find that subjects had detectable viral RNA for a prolonged period after becoming culture negative. Only one subject was RT-PCR positive on day 12 by quantitative PCR (limit of detection, 1.0 PFU). Even using the more sensitive method of nested RT-PCR (limit of detection, 0.1 PFU), only 4 of 12 subjects were positive on day 12, and all were negative on day 28. These results indicate that a low level of RNA does not persist in normal healthy adults. We conclude that the detection of viral RNA reflects active viral replication, although it will be important to study elderly and immunocompromised adults to determine if the same is true for these groups as well. Interestingly, 10 of 13 subjects were RT-PCR negative on day 1 by quantitative methodology, and 7 were negative by nested RT-PCR. These samples were collected only 24 h after inoculation of 104.7 TCID50, indicating the viral RNA can be rapidly cleared in the absence of virus replication.
Evaluation of subjects in a challenge study is very different from the evaluation of ill patients. In the setting of natural infection, specimens are rarely handled with precision, and transit times and conditions can be quite variable. In a previous study of patients infected with RSV who were ill for an average of 5 days at the time of evaluation, RT-PCR sensitivity was 73%, compared to 39% for culture (6). Assuming a 3-day incubation period, subjects were typically cultured approximately 8 days after exposure. This time period corresponds to the days when the correlation between RT-PCR and culture declined in the present challenge study. Thus, it seems most likely that the discrepancy between culture and RT-PCR in natural infection is the result of both variable specimen processing and the time point during the course of illness when people seek medical attention. It is also possible that wild-type RSV isolates are more difficult to grow in cell culture than RSV A2, a tissue culture-adapted strain.
In vitro neutralization of virus by preexisting nasal antibody could be a factor in the difficulty in isolating virus in adults or children with RSV reinfection. Notwithstanding some variability in nasal IgA titer due to collection methods, it did not appear that high nasal IgA titers were not consistently associated with a discrepancy between culture and RT-PCR positivity. Correction of nasal IgA titers by total protein or urea nitrogen was not possible, because the nasal wash specimens were placed into viral transport medium, which contains protein.
Many questions about the pathophysiology of RSV disease in both children and adults exist. There are few studies of infants and none of adults that have addressed the issue of severity of illness and viral load, and those that do offer conflicting conclusions (1, 28). In infants, hospitalization for severe illness typically occurs at the point at which titers of culturable virus are declining, raising the possibility that disease is mediated by the immune response rather than viral cytotoxicity. Little data exist regarding the presence and quantity of RSV in the lower airways in adults or older children with severe disease. In the elderly, it is not clear whether severe disease is due to associated comorbid conditions or whether immunosenescence leads to prolonged and greater viral replication. Until now, many of these questions could not be addressed due to the difficulties of viral quantification by culture. Quantitative RT-PCR appears to be an excellent tool for future studies to address these important questions.
In summary, quantitative RT-PCR has an excellent correlation with viral titers, as measured by culture when specimens are obtained under optimal circumstances. Detection of RSV by RT-PCR indicates recent viral replication, and viral RNA does not persist for long periods following infection.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (NIAID AI-45248) and Aventis-Pasteur, Ltd.
REFERENCES
- 1.Buckingham, S. C., A. J. Bush, and J. P. DeVincenzo. 2000. Nasal quantity of respiratory syncytial virus correlates with disease severity in hospitalized infants. Pediatr. Infect. Dis. J. 19:113-117. [DOI] [PubMed] [Google Scholar]
- 2.Cilloni, D., E. Gottardi, D. De Micheli, A. Serra, G. Volpe, F. Messa, G. Rege-Cambrin, A. Guerrasio, M. Divona, F. Lo Coco, and G. Saglio. 2002. Quantitative assessment of WT1 expression by real-time quantitative PCR may be a useful tool for monitoring minimal residual disease in acute leukemia patients. Leukemia 16:2115-2121. [DOI] [PubMed] [Google Scholar]
- 3.Collins, P. L., Y. T. Huang, and G. W. Wertz. 1984. Nucleotide sequence of the gene encoding the fusion (F) glycoprotein of human respiratory syncytial virus. Proc. Natl. Acad. Sci. USA 81:7683-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Englund, J. A., C. J. Sullivan, M. C. Jordan, L. P. Dehner, G. M. Vercellotti, and H. H. Balfour. 1988. Respiratory syncytial virus infection in immunocompromised adults. Ann. Intern. Med. 109:203-208. [DOI] [PubMed] [Google Scholar]
- 5.Falsey, A. R., C. K. Cunningham, W. H. Barker, R. W. Kouides, J. B. Yuen, M. Menegus, L. B. Weiner, C. A. Bonville, and R. F. Betts. 1995. Respiratory syncytial virus and influenza A infections in the hospitalized elderly. J. Infect. Dis. 172:389-394. [DOI] [PubMed] [Google Scholar]
- 6.Falsey, A. R., M. A. Formica, and E. E. Walsh. 2002. Diagnosis of respiratory syncytial virus infection: comparison of reverse transcription-PCR to viral culture and serology in adults with respiratory illness. J. Clin. Microbiol. 40:817-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falsey, A. R., R. M. McCann, W. J. Hall, and M. M. Criddle. 1996. Evaluation of four methods for the diagnosis of respiratory syncytial virus infection in older adults. J. Am. Geriatr. Soc. 44:71-73. [DOI] [PubMed] [Google Scholar]
- 8.Falsey, A. R., J. J. Treanor, R. F. Betts, and E. E. Walsh. 1992. Viral respiratory infections in the institutionalized elderly: clinical and epidemiologic findings. J. Am. Geriatr. Soc. 40:115-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan, J., K. J. Henrickson, and L. L. Savatski. 1998. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization (hexaplex) assay. Clin. Infect. Dis. 26:1-6. [DOI] [PubMed] [Google Scholar]
- 10.Freymuth, F., G. Eugene, A. Vabret, J. Petitjean, E. Gennetay, J. Brouard, J. F. Duhamel, and B. Guillois. 1995. Detection of respiratory syncytial virus by reverse transcription-PCR and hybridization with a DNA enzyme immunoassay. J. Clin. Microbiol. 33:3352-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert, L. L., A. Dakhama, B. M. Bone, E. E. Thomas, and R. G. Hegele. 1996. Diagnosis of viral respiratory tract infections in children by using a reverse transcription-PCR panel. J. Clin. Microbiol. 34:140-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginzinger, D. G. 2002. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp. Hematol. 30:503-512. [DOI] [PubMed] [Google Scholar]
- 13.Hall, C. B. 2001. Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 334:1917-1928. [DOI] [PubMed] [Google Scholar]
- 14.Henkel, J. H., S. W. Aberle, M. Kundi, and T. Popow-Kraupp. 1997. Improved detection of respiratory syncytial virus in nasal aspirates by semi-nested RT-PCR. J. Med. Virol. 53:366-371. [PubMed] [Google Scholar]
- 15.Jung, R., K. Soondrum, and M. Neumaier. 2000. Quantitative PCR. Clin. Chem. Lab. Med. 38:833-836. [DOI] [PubMed] [Google Scholar]
- 16.Kuoppa, Y., J. Boman, L. Scott, U. Kumlin, I. Eriksson, and A. Allard. 2002. Quantitative detection of respiratory Chlamydia pneumoniae infection by real-time PCR. J. Clin. Microbiol. 40:2273-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen, H. H., J. A. Kovacs, F. Stock, V. H. Vestereng, B. Lundgren, S. H. Fischer, and V. J. Gill. 2002. Development of a rapid real-time PCR assay for quantitation of Pneumocystis carinii f. sp. carinii. J. Clin. Microbiol. 40:2989-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak, K. J., S. J. Flood, L. Marmaro, W. Giusti, and K. Deete. 1995. Oligo nucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 19.Mygind, T., S. Birkelund, E. Falk, and G. Christiansen. 2001. Evaluation of real-time quantitative PCR for identification and quantification of Chlamydia pneumoniae by comparison with immunohistochemistry. J. Microbiol. Methods 46:241-251. [DOI] [PubMed] [Google Scholar]
- 20.Saag, M. S., M. Holodniy, D. R. Kuritzkes, W. A. O'Brien, R. Coombs, M. E. Poscher, D. M. Jacobsen, G. M. Shaw, D. D. Richman, and P. A. Volberding. 1996. HIV viral load markers in clinical practice. Nat. Med. 2:625-629. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt, N. J., and R. W. Emmons (ed.). 1989. General principles of laboratory diagnostic methods for viral, rickettsial and chlamydial infections, p. 1-36. In Diagnostic procedures for viral, rickettsial and chlamydial infections. American Public Health Association,Washington, D.C.
- 22.van Elden, L. J. R., M. Nijhuis, P. Schipper, R. Schuurman, and A. M. van Loon. 2001. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol. 39:196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh, E. E., A. R. Falsey, and P. A. Hennessey. 1999. Respiratory syncytial virus and other infections in persons with chronic cardiopulmonary disease. Am. J. Respir. Crit. Care Med. 160:791-795. [DOI] [PubMed] [Google Scholar]
- 24.Walsh, E. E. and C. B. Hall. 1989. Respiratory syncytial virus in diagnostic procedures for viral rickettsial and chlamydial infections, p. 693-712. In N. J. Schmidt and R. W. Emmons (ed.), Diagnostic procedures for viral, rickettsial and chlamydial infections. American Public Health Association, Washington, D.C.
- 25.Walsh, E. E., M. W. Brandriss, and J. J. Schlesinger. 1985. Purification and characterization of the respiratory syncytial virus fusion protein. J. Gen. Virol. 66:409-415. [DOI] [PubMed] [Google Scholar]
- 26.Walsh, E. E., and A. R. Falsey. 1999. A simple and reproducible method for collecting nasal secretions from frail elderly adults for measurement of virus-specific IgA. J. Infect. Dis. 179:1268-1273. [DOI] [PubMed] [Google Scholar]
- 27.Walsh, E. E., A. R. Falsey, I. A. Swinburne, and M. A. Formica. 2001. Reverse transcription polymerase chain reaction (RT-PCR) for diagnosis of respiratory syncytial virus infection in adults: use of a single-tube “hanging droplet” nested PCR. J. Med. Virol. 63:259-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright, P. F., W. C. Gruber, M. Peters, G. Reed, Y. Zhu, F. Robinson, S. Coleman-Dockery, and B. S. Graham. 2002. Illness severity, viral shedding, and antibody responses in infants hospitalized with bronchiolitis caused by respiratory syncytial virus. J. Infect. Dis. 185:1011-1018. [DOI] [PubMed] [Google Scholar]
- 29.Yousef, G. M., A. Scorilas, L. G. Kyriakopoulou, L. Rendl, M. Diamandis, R. Ponzone, N. Biglia, M. Giai, R. Roagna, P. Sismondi, and E. P. Diamandis. 2002. Human kallikrein gene 5 (KLK5) expression by quantitative PCR: an independent indicator of poor prognosis in breast cancer. Clin. Chem. 48:1241-1250. [PubMed] [Google Scholar]
- 30.Zambon, M. C., J. D. Stockton, J. P. Clewley, and D. M. Fleming. 2001. Contribution of influenza and respiratory syncytial virus to community cases of influenza-like illness: an observational study. Lancet 358:1410-1416. [DOI] [PubMed] [Google Scholar]