Abstract
Salmonella enterica serotype O1,4,5,12:Hb:1,2, designated according to the current Kauffmann-White scheme as S. enterica serotype Paratyphi B, is a very diverse serotype with respect to its clinical and microbiological properties. PCR and blot techniques, which identify the presence, polymorphism, and expression of various effector protein genes, help to distinguish between strains with systemic and enteric outcomes of disease. All serotype Paratyphi B strains from systemic infections have been found to be somewhat genetically related with respect to the pattern of their virulence genes sopB, sopD, sopE1, avrA, and sptP as well as other molecular properties (multilocus enzyme electrophoresis type, pulsed-field gel electrophoresis [PFGE] type, ribotype, and IS200 type). They have been classified as members of the systemic pathovar (SPV). All these SPV strains possess a new sopE1-carrying bacteriophage (designated ΦSopE309) with high SopE1 protein expression but lack the commonly occurring avrA determinant. They exhibit normal SopB protein expression but lack SopD protein production. In contrast, strains from enteric infections classified as belonging to the enteric pathovar possess various combinations of the respective virulence genes, PFGE pattern, and ribotypes. We propose that the PCR technique for testing for the presence of the virulence genes sopE1 and avrA be used as a diagnostic tool for identifying both pathovars of S. enterica serotype Paratyphi B. This will be of great public health importance, since strains of serotype Paratyphi B have recently reemerged worldwide.
Salmonella enterica is one of the most diverse species in the bacterial kingdom. It is currently subdivided into six subspecies according to fermentative properties and into ca. 2,400 serotypes according to polymorphisms in the lipopolysaccharide (O antigen) and flagellar (H antigen) structures (20). Among S. enterica, two major pathogenic groups causing human infections have been identified: Salmonella strains restricted or adapted to humans (e.g., S. enterica serotype Typhi and S. enterica serotype Paratyphi A, B, and C) cause systemic clinical conditions such as septicemia and organ manifestation (typhoid fever), while the so-called enteritis salmonella strains (e.g., S. enterica serotype Enteritidis) cause local intestinal infections and originate epidemiologically from animal husbandry.
However, human infections due to S. enterica serotype Paratyphi B with the O:H formula O1,4,5,12:Hb:1,2 are not restricted to systemic infections (paratyphoid fever) and human-to-human infection routes (15) but have been associated with gastroenteritis and food-borne infections as well (3, 7, 12). This clinical and epidemiological heterogeneity was regarded as a consequence of fermentative varieties among this serotype. Many such isolates do ferment d-tartrate and have been designated biovar S. enterica serotype Java, in contrast to non-d-tartrate-fermenting strains, designated biovar S. enterica serotype Paratyphi B sensu stricto (3, 12, 13). Moreover, S. enterica serotype Paratyphi B strains have been shown to be highly variable in the presence and polymorphism of several molecular (4, 6, 10, 25) and virulence properties (e.g., effector proteins), which is uncommon among other serotypes (17, 21).
This communication describes molecular properties of S. enterica serotype Paratyphi B strains which underline an unusually great diversity among strains of this serotype. However, patterns of genetic properties allow discrimination between strains from systemic infections and strains from local enteric infections as well as from nonhuman sources. This might have significant public health implications, taking into consideration the recent emergence of such strains (9, 14, 19, 27).
MATERIALS AND METHODS
Bacterial strains.
Ninety-nine strains of S. enterica serotype Paratyphi B were investigated in this study. Sixteen strains belong to Salmonella Reference Collection A (SARA) (Table 1) and have been used for comparative purposes as a set of international S. enterica serotype Paratyphi B reference strains. Seventy-two strains originated from clinical cases (sporadic cases of gastroenteritis and typhoid illnesses among humans), and eleven strains came from poultry (Table 2). They were chosen for this study from our type culture collection according to their various clinical and geographical origins as well as their different years of isolation. The bacteriophage-sensitive tester strains A36 (S. enterica serotype Typhimurium) and 00-08652 (S. enterica serotype Paratyphi B) as well as some other isolates of S. enterica as listed in Table 3 were also derived from the type culture collection of our laboratory. All Salmonella strains were stored as glycerol (20%) cultures at −70°C.
TABLE 1.
Phenotypic properties of serotype S. enterica serotype Paratyphi B strains of SARA
| Designation (classification)a | Phage typeb | DTFc | MLEE typed |
|---|---|---|---|
| SARA 41 (Pb1)e | 3a var 1,2/Q3 | − | 1 |
| SARA 44 (Pb1) | Beccles/M3 | − | 1 |
| SARA 45 (Pb1) | Dundee/BT6 | − | 1 |
| SARA 46 (Pb1a) | Taunton/B7 | − | 1d |
| SARA 47 (Pb2) | 1 var 1/NC | + | 1 |
| SARA 49 (Pb2b) | Beccless/NC | + | 1c |
| SARA 50 (Pb3)f | 1 var 4/NC | + | 2 |
| SARA 51 (Pb3) | 1 var 4/NC | + | 2 |
| SARA 52 (Pb3) | 1 var 4/NC | + | 2a |
| SARA 53 (Pb3) | 1 var 4/NC | + | 2 |
| SARA 54 (Pb3) | 1 var 4/NC | + | 2 |
| SARA 55 (Pb3a) | 1 var 4/NC | + | 2b |
| SARA 56 (Pb4)g | 3b var 2 | + | 1g |
| SARA 57 (Pb5)h | UT/NC | + | 1e |
| SARA 59 (Pb5b) | UT/NC | + | 1c |
| SARA 62 (Pb7)i | UT/UT | + | 13 |
Classification according to Beltran et al. (4).
Phage types according to Rische and Ziesché (23). NC, not characteristic; UT, untypeable.
DTF, d-tartrate fermentation.
MLEE pattern according to Table 3.
Same as SARB43 (Pb1) (6).
Same as SARB44 (Pb3) (6).
Same as SARB45 (Pb4) (6).
Same as SARB46 (Pb5) (6).
Same as SARB47 (Pb7) (6).
TABLE 2.
Serotype S. enterica Serotype Paratyphi B strains isolated between 1996 and 2001 from human clinical cases and nonclinical strains from poultry
| No. of strains tested | Origina | Phage typeb | d-Tartrate fermentation |
|---|---|---|---|
| 15 | Sporadic cases associated with systemic infections, fever, and septicemia | Taunton/B7 | − |
| 1 | Water isolate | Taunton/B7 | − |
| 3 | International outbreak of paratyphoid fever (1999) | Taunton/B7 | − |
| 2 | Septicemic infections | 1 | − |
| 1 | Carrier | 1 | − |
| 1 | Carrier | Jersey/J3 | − |
| 1 | Typhoid infection | 3b/A1 | − |
| 4 | Sporadic cases of systemic infection, fever, and septicemia | 3a1/B6 | − |
| 6 | Sporadic cases of gastroenteritis | 3b var 2 | + |
| 7 | Sporadic cases of gastroenteritis | UT/NC | + |
| 19 | Sporadic cases of gastroenteritis | Dundee/B6 | + |
| 3 | Sporadic cases of gastroenteritis | 1b var 3/1 | + |
| 1 | Gastroenteritis | 1 var 2/nc | + |
| 2 | Sporadic cases of gastroenteritis, diarrhea | Jersey/var 1/nc | + |
| 3 | Sporadic cases of gastroenteritis | 1 var 2 | + |
| 3 | Sporadic cases of gastroenteritis | 3bvar9/1 | + |
| 5 | Poultry, flock B (1999) | Dundee/B6 | + |
| 5 | Poultry, flock A (1999) | UT/NC | + |
| 1 | Poultry, flock C (1999) | 3b/NC | + |
Poultry flocks A, B, and C were from geographically distant locations.
Phage types according to Rische and Ziesché (23). UT, untypeable; NC, not characteristic.
TABLE 3.
sopE1 polymorphism among SPV and EPV strains of S. enterica serotype Paratyphi B and other serotypes and lysogens of S. enterica strains
| Strain | Serotype | Pathovar | sopE1 PCR result | sopE1 RFLP patterna |
sopE1 (I) PCR resultb
|
sopE1 (II) PCR resultc
|
||
|---|---|---|---|---|---|---|---|---|
| Upstream to orfR | Downstream to orf45 | Upstream to orfR | Downstream to orf194 | |||||
| Tester strains (lysogen) | ||||||||
| B309 | Paratyphi B | SPV | + | II | − | − | + | + |
| 00-08652 | Paratyphi B | EPV | − | − | − | − | − | − |
| 00-08652 (ΦSopE309)d | Paratyphi B | EPV | + | II | − | − | + | + |
| A36 | Typhimurium | − | − | − | − | − | − | − |
| A36 (SopEΦ)e | Typhimurium | − | + | I | + | −g | − | − |
| A36 (ΦSopE309)f | Typhimurium | − | + | II | − | − | + | + |
| Clinical strains | ||||||||
| 99-06072 | Paratyphi B | SPV | + | II | − | − | + | + |
| 99-02820 | Paratyphi B | SPV | + | II | − | − | + | + |
| 99-08309 | Paratyphi B | SPV | + | II | − | − | + | + |
| 99-06148 | Paratyphi B | SPV | + | II | − | − | + | + |
| 97-15877 | Paratyphi B | SPV | + | II | − | − | + | + |
| SAR41 | Paratyphi B | SPV | + | II | − | − | + | + |
| 99-08380 | Paratyphi B | SPV | + | I | − | − | + | + |
| 96-01098 | Paratyphi B | EPVh | + | I | + | − | − | + |
| 97-12134 | Paratyphi B | EPVh | + | I | + | + | − | − |
| 99-01096 | Paratyphi B | EPVh | + | I | + | + | − | − |
| 99-01097 | Paratyphi B | EPVh | + | I | + | + | − | − |
| 99-08163 | Paratyphi B | EPVh | + | I | − | − | − | − |
| 99-04814 | Paratyphi B | EPVh | + | I | − | + | + | − |
| 96-08640 | Typhi | − | + | I | + | + | − | − |
| RKI130 | Heidelberg | − | + | I | − | − | + | + |
| 98-11635 | Typhimurium DT68i | + | I | + | + | − | − | |
| 93-00080 | Typhimurium DT204i | + | I | + | + | − | − | |
| 75-01646 | Typhimurium DT175i | + | I | + | + | − | − | |
| 76-E8 | Enteritidis | + | II | − | − | + | + | |
| 63-00647 | Gallinarum | + | II | − | − | + | + | |
| 99-03279 | Hadar | + | II | − | − | + | + | |
| 2229 | Dublin | + | II | − | − | + | + | |
Cluster I sopE1 PCR (17).
Cluster II sopE1 PCR (17).
Lysogenization of 00-08652 (EPV phage-free tester strain) with ΦSopE309 (isolated from SPV S. enterica serotype Paratyphi B strain B 309).
Lysogenization of the S. enterica serotype Typhimurium phage-free tester strain A36 with the sopE1-carrying bacteriophage SopEΦ isolated from clinical S. enterica serotype Typhimurium strains (17).
Lysogenization of the S. enterica serotype Typhimurium phage-free tester strain A36 with the sopE1-carrying bacteriophage ΦSopE309 isolated from systemic pathovar of S. enterica serotype Paratyphi B strain B 309.
The PCR is negative because a kanamycin resistance gene cassette has been introduced (17).
Enteric pathovar carrying the sopE1 determinant.
DT, definitive phage type.
Fermentative tests.
d-Tartrate fermentation was carried out as described by Kauffmann (12) with the modification described by Barker (2).
Phage typing and techniques.
Phage typing was carried out as described by Anderson (1) with the modification of Rische and Ziesché (23). All typing phages applied were obtained from the Laboratory of Enteric Pathogens, Public Health Laboratory Service, London, Colindale, United Kingdom; the typing bacteriophages of the Scholtens system (Bilthoven, The Netherlands) were propagated in our laboratory. The SopEΦ phage is described elsewhere (16) and was used according to the authors' recommendations. The isolation of bacteriophages from and lysogenization of S. enterica serotype Paratyphi B strains were carried out as described by Schmieger (24) using the above-mentioned sensitive tester strains.
Electrotyping.
Multilocus enzyme electrophoresis (MLEE) analysis was carried out as described earlier (26), using 22 enzymes (Table 4). The patterns derived after MLEE were designated arbitrarily by numbering (Table 4). Letters (a, b, and c) associated with numbers designate related patterns exhibiting differences in one or two enzymes with respect to their running positions (Rf [relative to the front] values) (25).
TABLE 4.
Definition of MLEE types and their frequencies among S. enterica serotype Paratyphi B strains of clinical origin and from poultry
| MLEE typea | % | Rf (102)b
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EST | MDH | LDH | G6P | 6PG | ADH | ACP | LAP | ALP | IDH | FUM | ACO | PGM | GOT | PGI | IPO | ME | ADK | GD1 | GD2 | HEX | GP1 | GP2 | CAT | ||
| 1 | 35 | 53 | 58/30 | 58 | 41/38 | 74 | 56 | 22/11 | 83 | 0 | 0 | 58 | 0 | 67 | 95 | 59 | 55 | 42 | 54 | 56 | 55 | 0 | 55 | 60 | 66 |
| 1 | 8 | 53 | 58/30 | 58 | 41/38 | 74 | 56 | 22/11 | 83 | 0 | 72 | 58 | 0 | 67 | 95 | 59 | 55 | 42 | 54 | 56 | 55 | 58 | 55 | 60 | 66 |
| 1a | 1 | 53 | 58/30 | 58 | 41/38 | 74 | 56 | 22/11 | 83 | 0 | 72 | 58 | 0 | 67 | 95/86 | 59 | 55 | 42 | 54 | 56 | 55 | 58 | 55 | 60 | 68 |
| 1b | 1 | 47 | 58/30 | 58 | 41/38 | 74 | 56 | 22/11 | 83 | 0 | 0 | 58 | 0 | 67 | 95/86 | 59 | 55 | 42 | 54 | 56 | 55 | 58 | 55 | 60 | 66 |
| 1c | 5 | 0 | 58/30 | 58 | 41/38 | 74 | 56 | 22/11 | 83 | 0 | 72 | 58 | 0 | 67 | 95 | 59 | 55 | 42 | 54 | 56 | 55 | 58 | 55 | 60 | 68 |
| 2 | 9 | 43 | 58/30 | 58 | 41/38 | 74 | 56 | 22 | 83 | 0 | 76 | 58 | 0 | 67 | 95/86 | 59 | 55 | 42 | 54 | 56 | 55 | 58 | 55 | 60 | 68 |
| 3 | 25 | 56 | 58/30 | 58 | 41/38 | 71 | 56 | 20/16 | 83 | 0 | 72 | 58 | 0 | 67 | 95 | 59 | 55 | 42 | 54 | 56 | 55 | 58 | 55 | 60 | 66 |
| 4 | 3 | 53 | 58/30 | 58 | 41/38 | 71 | 56 | 32/22/11 | 83 | 0 | 76 | 58 | 0 | 67 | 95 | 59 | 55 | 42 | 54 | 56 | 55 | 58 | 55 | 60 | 66 |
| 5 | 1 | 53 | 58/40/30 | 58 | 41/38 | 67 | 56 | 28/22/11 | 83 | 0 | 72 | 58 | 65 | 67 | 95/86 | 59 | 55 | 42 | 54 | 56 | 55 | 58 | 55 | 0 | 68 |
| 6 | 3 | 53 | 58/40/30 | 58 | 41/38 | 71 | 56 | 28/22/11 | 83 | 0 | 72 | 58 | 65 | 71 | 95/86 | 59 | 55 | 42 | 54 | 56 | 55 | 0 | 55 | 0 | 68 |
| 7 | 1 | 53 | 58/30 | 58 | 41/38 | 74 | 56 | 28/22 | 83 | 0 | 0 | 58 | 60 | 71 | 95/86 | 59 | 55 | 42 | 54 | 56 | 55 | 0 | 55 | 0 | 53 |
| 8 | 3 | 53 | 58/40/30 | 58 | 41/38 | 74 | 56 | 26/20 | 0 | 0 | 72 | 58 | 65 | 67 | 95/86 | 59 | 55 | 42 | 54 | 56 | 55 | 0 | 55 | 0 | 68 |
| 9 | 1 | 53 | 58/40/30 | 58 | 41/38 | 74 | 56 | 32/22 | 83 | 0 | 72 | 58 | 65 | 67 | 95/86 | 59 | 55 | 42 | 54 | 56 | 55 | 0 | 55 | 0 | 66 |
| 10 | 1 | 53 | 58/30 | 58 | 41/38 | 74 | 56 | 28/22 | 83 | 76 | 58 | 67 | 95/86 | 59 | 55 | 42 | 54 | 56 | 55 | 58 | 55 | 60 | 53 | ||
| 11 | 1 | 53 | 58/40 | 58 | 41/38 | 74 | 56 | 28/22/11 | 83 | 0 | 76 | 58 | 60 | 67 | 95/86 | 59 | 55 | 42 | 54 | 56 | 55 | 58 | 55 | 0 | 66 |
| 12 | 1 | 47 | 58/30 | 58 | 41/38 | 71 | 56 | 28/22 | 83 | 0 | 72 | 58 | 0 | 71 | 95/86 | 59 | 55 | 42 | 54 | 56 | 0 | 58 | 55 | 60 | 66 |
| 13 | 1 | 53 | 58/30 | 58 | 41/38 | 71 | 56 | 91/32/22 | 83 | 0 | 72 | 58 | 0 | 71 | 95/86 | 59 | 55 | 42 | 54 | 56 | 55 | 58 | 55 | 60 | 0 |
Classes 1d (differing in 6P6), 1f (differing in ACP), and 1g (differing in ACP and CAT) were detected only among SARA strains.
EST, esterases; MDH, malate dehydrogenase; LDH, l-lactate dehydrogenase; G6P, glucose 6-phosphate dehydrogenase; 6PG, 6-phosphogluconase dehydrogenase; ADH, alcohol dehydrogenase; ACP, acid phosphatase; LAP, leucine aminopeptidase; ALP, alkaline phosphatase; IDH, isocitrate dehydrogenase; FUM, fumarase; ACO, aconitase; PGM, phosphoglucomutase; GOT, glutamic-oxalacetic transaminase; PGI, phosphoglucose isomerase; IPO, indophenol oxidase (superoxide dismutase); ME, malic enzyme; ADK, adenylatkinase; GD1, glutamate dehydrogenase (NAD); GD2, glutamate dehydrogenase (NADP); HEX, hexokinase; GP1, glyceraldehyde-phosphate dehydrogenase (NAD); GP2, glyceraldehyde-phosphate dehydrogenase (NADP); CAT, catalase. Values in bold indicate deviations from MLEE type 1.
DNA isolation and PCR.
Chromosomal DNA isolation, DNA cleavage with restriction enzymes, and agarose gel electrophoresis were performed as described by Prager et al. (21). PCRs were carried out with the Gene Amp PCR system 9600 (Perkin-Elmer Applied Biosystems, Weiterstadt, Germany). Amplification was performed with AmpliTaq Gold and Gene Amp 10× PCR buffer II (Applied Biosystems, Weitershausen, Germany) according to the manufacturer's protocol.
The PCR primers and conditions used in this study are summarized in Table 5. In order to identify the vicinity of the various sopE1 determinants, PCR primers were designed which give rise to PCR products overlapping the sopE1 region and its upstream or downstream vicinity (Table 5).
TABLE 5.
PCR primers and conditions for detecting sopB, sopD, and sopE1, as well as avrA and sptP (21)
| Primer | Primer sequence | Target | PCR conditions (denaturing/annealing/ extension; no. of cycles) | Size (bp) of PCR product |
|---|---|---|---|---|
| SopB-RSB1 | 5′-CAA CCG TTC TGG GTA AAC AAG AC-3′ | sopBa | 60 s, 94°C/60 s, 55°C/120 s, 72°C; 30 | 1,348 |
| SopB-RSB2 | 5′-AAG ATT GAG CTC CTC TGG CGA T-3′ | |||
| SopD-SD3 | 5′-TGA TAG TAA ACA GAT CTT GAT GAG C-3′ | sopDa | 60 s, 94°C/60 s, 55°C/60 s, 72°C; 30 | 289 |
| SopD-SD4 | 5′-TTA TGG GAG TCA CTT TAA GAT TCG GTA A-3′ | |||
| SopE1-P4 | 5′-ACA CAC TTT CAC CGA GGA AGC G-3′ | sopE1a | 60 s, 94°C/60 s, 55°C/60 s, 72°C; 30 | 398 |
| SopE1-M2 | 5′-GGA TGC CTT CTG ATG TTG ACT GG-3′ | |||
| SptP-P | 5′-GTT GAG AGG TGG GTT GAT AAA GCC-3′ | sptPb | 60 s, 94°C/60 s, 55°C/60 s, 72°C; 30 | 496 |
| SptP-M | 5′-TGG TAT TGG TCT ATC GCT TCT CCC-3′ | |||
| AvrA-P4 | 5′-GTT ATG GAC GGA ACG ACA TCG G-3′ | avrAb | 60 s, 94°C/60 s, 64°C/60 s, 72°C; 30 | 385 |
| AvrA-M1 | 5′-ATT CTG CTT CCC GCC GCC-3′ | |||
| Up-SopE(I)-Fc | 5′-CTA ACA TCA AAA AGC AAT CC-3′ | orfR | 30 s, 94°C/60 s, 48°C/90 s, 72°C; 25 | 1,009 |
| Up-SopE(I)-R | 5′-TCT GTC ATA ATG ATC TTC TCC-3′ | sopE1 | ||
| Down-SopE(I)-Fc | 5′-ACA CAC TTT CAC CGA GGA AGC G-3′ | sopE1 | 60 s, 94°C/60 s, 57°C/60 s, 72°C; 25 | 821 |
| Down-SopE(I)-R | 5′-ACG GCT GGA AGC ATG GGA ACT TT-3′ | orf45 | ||
| Up-SopE(II)-Fd | 5′-CAT AAA TAA TCG CTA CCT GC-3′ | orfR | 30 s, 94°C/60 s, 48°C/90 s, 72°C; 25 | 1,000 |
| Up-SopE(II)-R | 5′-TCT GTC ATA ATG ATC TTC TCC-3′ | sopE1 | ||
| Down-SopE(II)-Fd | 5′-ACA CAC TTT CAC CGA GGA AGC G-3′ | sopE1 | 30 s, 94°C/60 s, 62°C/60 s, 72°C; 25 | 842 |
| Down-SopE(II)-R | 5′-TCG CAA CAG ATG ATG AGA AAG C-3′ | orf194 |
The nucleotide sequences correspond to the respective genes from S. enterica serotype Dublin (EMBL accession numbers: for sopE1, L78992; for sopB, AF060858; and for sopD, AF030589); the sopE2 determinant present in all S. enterica serotype Paratyphi B strains (data not shown) was not considered throughout this study.
The nucleotide sequences correspond to the respective genes from S. enterica serotype Typhimurium (avrA, AF013573, and sptP, TU63293).
PCR-generated DNA probes were purified by using GFX PCR DNA and a gel band purification kit (Amersham Biosciences, Little Chalfont, United Kingdom).
Restriction fragment length polymorphisms (RFLP) of PCR products were analyzed as described earlier (21).
Southern blots.
Southern blotting techniques and DNA probes were essentially as described earlier (21). DNA fragments were transferred to a positively charged nylon membrane (Roche) by vacuum blotting as recommended by the supplier (Pharmacia, Uppsala, Sweden) and fixed to the membrane by UV cross-linking (GS Gene Linker; Bio-Rad, Munich, Germany). The PCR-generated DNA probes were labeled with digoxigenin-11-dUTP by using a random primed labeling kit (Roche). The labeled probe was hybridized to the membrane-bound nucleic acid and detected with a digoxigenin luminescence detection kit (Roche) by using CSPD {3-(4-methoxyspiro[1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.1(3,7)]decan]-4-yl) phenyl phosphate}. Digoxigenin-labeled bacteriophage lambda DNA digested with EcoRI and HindIII (Boehringer, Mannheim, Germany) served as molecular mass standards.
Harvesting of Sop proteins.
The proteins SopB, SopE1, and SopD have been identified in supernatants of Salmonella cultures. All strains under study were incubated in 20 ml of Luria-Bertani broth containing 0.3 M NaCl using a 100-ml bulb flask with a narrow neck overnight on a longitudinal shaker (100/min). Cultures were transferred to an ice bath for 30 min. The supernatants were harvested by centrifugation (1 h at 18,000 × g) and filtrated through a Millipore filter (0.45 μm). Proteins from the supernatants were precipitated with 10% trichloroacetic acid on ice for 1 h. After centrifugation (1 h at 20,000 rpm), precipitates were transferred to 0.4 ml of 0.1 M NaOH and 2.0 ml of ice-cold acetone (−20°C). After 20 min at −20°C, precipitates were harvested by centrifugation (15 min at 20,000 rpm), washed again with ice-cold acetone, and harvested by centrifugation. The sediments were dried overnight, dissolved in 100 μl of Laemmli buffer, boiled 5 min at 95°C, and subsequently subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
SDS-PAGE and Western blots.
SDS-PAGE was carried out as described by Bollang and Edelstein (5) in a 10% polyacrylamide gel using a Mini-Protean apparatus (Bio-Rad). Western blot analysis was performed with semidry blots using polyvinylidene difluoride membranes and a Fast Blotter (Bio-Rad) for 15 min at 22 V and 150 mA. For detection of Sop proteins, the polyclonal rabbit antibodies α-SopB, α-SopE1, and α-SopD were applied. The antibodies were raised in rabbits against the respective recombinant Sop proteins which were purified by using the pET-Directional TOPO expression kit (Invitrogen BV, Breda, The Netherlands) and the ÄKTAexplorer NT100 (Amersham) according to the manufacturer's instructions (W. Streckel et al., unpublished data). Since SopE1 and SopE2 have both been found to react to α-SopE1, they are distinguished by their different molecular sizes and expression profiles (data not shown; see Fig. 4D).
FIG. 4.
Qualitative and quantitative differences in the presence of SopE1, SopB, and SopD in supernatants of SPV and EPV strains of serotype Paratyphi B. (S. enterica serotype Paratyphi B strains which fail to produce SopE2 under the culture conditions used were selected.) (A) SDS gel; (B) Western blot of SopB; (C) Western blot of SopD; (D) Western blot of SopE1 (EVP strains which belong to variant 4 were selected; see Table 7). c, molecular weight standard.
Clonal analysis by PFGE, IS200 typing, and ribotyping.
Pulsed-field gel electrophoresis (PFGE) and Southern blotting for IS200 and ribotyping were carried out according to Prager et al. (22) using the RFLPScan system (Scanalytics, Fairfax, Va.) for reading and interpretation.
The genetic distances for PFGE patterns (dendrogram) were calculated as described by Claus et al. (8).
Definition of S. enterica serotype Paratyphi B pathovars.
On the basis of the specific pathogenic patterns described below, we propose the designations “systemic pathovar” (SPV) for all strains of S. enterica serotype Paratyphi B which were found to be associated with mainly systemic or paratyphoid infections and “enteric pathovar” (EPV) for all serotype Paratyphi B strains associated with enteric and food-borne infections.
RESULTS
Biological and molecular properties of S. enterica serotype Paratyphi B.
In order to detect pathogenic and molecular differences among S. enterica serotype Paratyphi B strains for the purpose of clinical diagnosis and epidemiology, 16 SARA reference strains (Table 1) and 83 clinical strains of human origin and nonclinical strains from poultry (Table 2) were analyzed. All these strains with the antigenic formula 1,4,5,12:b:1,2 were characterized with regard to various biological and molecular properties, such as d-tartrate fermentation (Tables 1 and 2), phage types (Tables 1 and 2), their clonal relatedness (Tables 1 and 4; Fig. 1 and 2), and their virulence characters (presence, polymorphism, and expression of the effector protein genes sopB, sopE1, sopD, sptP, and avrA [Tables 6, 7, and 8]; sopE2 was not considered [Table 5; see Discussion]).
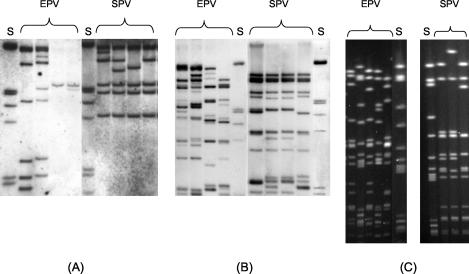
FIG. 1.
PFGE patterns (A), IS200 types (B), and ribotypes (C) of SPV and EPV strains of serotype Paratyphi B. S, molecular standard.
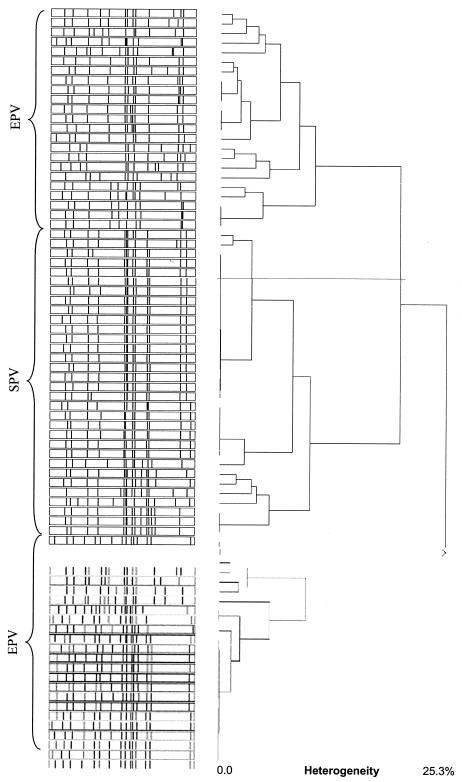
FIG. 2.
Cluster analysis of PFGE pattern from SPV and EPV strains of serotype Paratyphi B.
TABLE 6.
Molecular properties of the S. enterica serotype Paratyphi B SARA reference strains
| Designation | PCR result for:
|
Diagnostic classification | ||||
|---|---|---|---|---|---|---|
| sopE1 | avrA | sopB | sopD | sptP | ||
| SARA 41 | + | − | + | + | + | SPV variant 1 |
| SARA 44 | + | − | + | + | + | SPV variant 1 |
| SARA 45 | + | − | + | + | + | SPV variant 1 |
| SARA 46 | + | − | + | + | + | SPV variant 1 |
| SARA 47 | − | − | + | + | + | EPV variant 2 |
| SARA 49 | − | − | + | + | + | EPV variant 2 |
| SARA 50 | − | − | + | + | + | EPV variant 2 |
| SARA 51 | − | − | + | + | + | EPV variant 2 |
| SARA 52 | − | − | + | + | + | EPV variant 2 |
| SARA 53 | − | − | + | + | + | EPV variant 2 |
| SARA 54 | − | − | + | + | + | EPV variant 2 |
| SARA 55 | − | − | + | + | + | EPV variant 2 |
| SARA 56 | − | + | + | + | + | EPV variant 1 |
| SARA 57 | +a | − | + | + | + | SPV variant 2 |
| SARA 59 | − | − | + | + | + | EPV variant 2 |
| SARA 62 | − | + | + | + | + | EPV variant 1 |
This strain differs by its sopE1 polymorphism (Fig. 3).
TABLE 7.
Pattern pathogenic and biological properties among 83 strains of serotype S. Paratyphi B from clinical and animal sources
| No. of strains identified | PCR result for:
|
Diagnostic classification | |||||
|---|---|---|---|---|---|---|---|
| sopE1 | avrA | sopB | sopD | sptP | MLEE class | ||
| 11 | − | + | + | + | + | 3, 9 | EPV variant 1 |
| 23 | − | − | + | + | + | 1, 2, 4, 6, 7, 8 | EPV variant 2 |
| 13 | − | + | + | − | + | 3 | EPV variant 3 |
| 6 | +b | + | + | + | + | 1a, 1b, 5 | EPV variant 4 |
| 28 | +b | − | + | + | + | 1 | SPV variant 1 |
| 2 | +b | − | + | + | + | 1c | SPV variant 2 |
These strains have been found only very rarely among clinical specimens.
For differences between the sopE1 determinants, see Fig. 3; the differences between these variants correspond to their different sopE1 RFLPs.
TABLE 8.
Presence of SopB, SopD, and SopE1 in culture supernatants of serotype S. enterica serotype Paratyphi B strains
Sixty-eight strains from both collections (Tables 1 and 2) fermented d-tartrate, and 32 strains did not, even after 3 days, which allowed us to classify them as the biovars S. enterica serotype Paratyphi B sensu stricto and S. enterica serotype Java, respectively (12). A broad range of phage types (Table 1 and 2) and drug resistance patterns (data not shown) were detected among strains of systemic as well as of enteric origins. Surprisingly, all strains of systemic origin belong to the same MLEE group (arbitrarily designated MLEE type 1, which corresponds to Pb1 according to the system of Boy et al. [6]), whereas the enteric strains cluster into different groups (MLEE types 1 to 13) (Tables 1 and 4). Ribotyping and PFGE pattern analysis (Fig. 1) as well as PCR and Southern blotting for the presence of the effector protein genes sopB, sopE1, sopD, sptP, and avr confirmed the clusters (Fig. 2; Tables 6 and 7): all S. enterica serotype Paratyphi B strains from systemic origins (now termed SPV of the serotype Paratyphi B) were positive for sopE1, sopB, sopD, and sptP but negative for avrA, whereas the strains from intestinal infections and from poultry were heterogeneous (Tables 6 and 7). About a third of them (termed EPV variant 2) do not carry sopE1 and avrA; another third (EPV variant 3) lack sopE1 and sopD but were PCR positive for sopB, sptP, and avrA. Another third of the strains appeared to be variable with respect to sopE1 and avrA (EPV variant 1 and 4) (Tables 6 and 7).
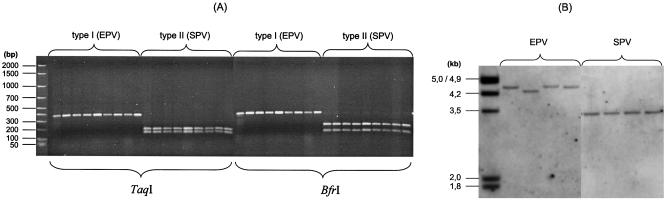
The sopE1 determinants identified among SPV and EPV strains are different according to their sopE1 RFLP patterns (Fig. 3A) and their hybridization patterns (Fig. 3B). These results indicate a different genetic background and vicinity of the sopE1 determinants. Moreover, upon analysis by a distinct PCR which starts in the sopE1 region and terminates in its upstream or its downstream neighboring sites, all sopE1-positive strains cluster according to the sopE1 polymorphism into two groups (Table 3): all the SPV strains harbor cluster II sopE1 determinants, whereas the EPV strains harbor sopE1 determinants in or related to cluster I (similar to SopEΦ). However, some of the enteric strains reveal a cluster I picture of sopE1 but together with a new or a variable adjacent DNA region (Table 3; e.g., see strain 99-08163 or 99-04814). The different adjacent DNA regions together with the different sopE1 genes among S. enterica serotype Paratyphi B strains might indicate heterogeneity in sopE1-carrying bacteriophages.
FIG. 3.
Polymorphism of sopE1 determinants among SPV and EPV strains of serotype Paratyphi B. (A) RFLP analysis with TaqI and BfrI; (B) Southern blot of genomic DNA digested with PstI using a sopE1 PCR probe.
Characterization of sopE1 carrying bacteriophages from S. enterica serotype Paratyphi B.
sopE1-positive S. enterica serotype Paratyphi B strains were subjected to mitomycin C induction, and plaques appeared on the bacteriophage-sensitive indicator strains A36 (S. enterica serotype Typhimurium) and 00-08652 (S. enterica serotype Paratyphi B). Single plaques were characterized by sopE1 PCR after several steps of lysis and lysogenization with the above-mentioned tester strains. A sopE1-carrying bacteriophage was isolated from SPV strain B309 of serotype Paratyphi B (originating from a paratyphoid infection), characterized by lysis and PCR properties, and designated ΦSopE309. As summarized in Table 3, ΦSopE309 does not resemble the previously described P2-like bacteriophage SopEΦ from S. enterica serotype Typhimurium (17, 27) with regard to its sopE1 RFLP pattern and its DNA regions neighboring sopE1 but represents another type of sopE1-carrying bacteriophage similar to the previously described sopE1 cluster II strains (17). As seen from Table 3, all SPV strains from S. enterica serotype Paratyphi B carry bacteriophages identical or very similar to ΦSopE309.
In contrast, the isolation of sopE1-carrying bacteriophages from EPV strains with the same technique was not successful.
Presence of SopB, SopE1, and SopD proteins in cultural supernatants.
The classification of S. enterica serotype Paratyphi B strains into EPV or SPV strains according to their genetic patterns of virulence properties (Tables 3, 6, and 7) was confirmed by testing the protein profiles of SopB, SopE1, and SopD (SopE2 was not considered, although it could be identified with α-SopE) (Table 8; see Discussion).
The results of Western blotting for the presence of SopB, SopE1, and SopD in the culture supernatants of the respective Salmonella strains are summarized in Table 8 and Fig. 4. The EPV strains express quantitatively more SopB and SopD proteins than strains from the SPV (Fig. 4B and C); in contrast, SopD- and SopB-negative variants have often been identified among SPV strains, although the respective genes were present. Moreover, SPV strains revealed a high production of SopE1, whereas SopE1 protein production among the rarely occurring sopE1-positive EPV variants remained reduced (Table 8; Fig. 4D).
DISCUSSION
S. enterica is a very diverse species, currently with 2,480 serotypes according to its serological classification (20). Earlier, the serotypes of S. enterica were regarded as independent species because they often show unique clinical and epidemiological manifestations or behaviors (13) and a serotype-associated distinct pathogenic makeup (15, 17, 21). Exceptions have been observed mainly with the serotype O1,4,5,12:Hb:1,2, designated S. enterica serotype Paratyphi B (21). Strains belonging to this serotype were found to be heterogeneous in their clinical outcomes: some of the strains seem to be associated primarily with typhoid or systemic infections, while others are associated with self-limiting gastroenteritis (2, 7, 8, 12, 18). Kauffmann (12) tried to correlate the clinical heterogeneity with their fermentative properties, e.g., d-tartrate fermentation, and he designated fermenting strains biovar Java and nonfermenting strains biovar Paratyphi B. However, the question of why a particular fermentative property is so closely correlated to the clinical and pathogenic potency of the strains remains to be answered.
In this communication we summarize data on several pathogenic and molecular properties which might help to distinguish between serotype Paratyphi B strains with more systemic or typhoid outcomes of infections (designated as SPV of S. enterica serotype Paratyphi B) and strains that are “restricted” to enteric infections (designated EPV).
First, all SPV strains contain a particular new sopE1-carrying bacteriophage (designated here ΦSopE309) with a cluster II sopE1 RFLP pattern (Fig. 3) and a high level of SopE1 protein expression (Fig. 4D; Table 8). ΦSopE309 resembles the previously described cluster II type of S. enterica serotype Hadar and S. enterica serotype Gallinarum (17). All SPV strains lack the avrA determinant (shown by PCR and Southern blots) common among S. enterica strains and have reduced or absent SopD protein production (Fig. 4C; Table 8). Moreover, they are clonally related (MLEE type, PFGE type, ribotype, and IS200 type) (Fig. 1 and 2; Table 4) irrespective of their different geographical and temporal origins (with the exception of three outbreak strains from Turkey [Table 2]).
Second, in contrast, EPV strains appeared to be heterogeneous: 40% of them are sopB and sopD positive by PCR and Southern blotting but avrA negative (EPV variant 2); 60% of them are also avrA positive, some with sopE1 (EPV variant 4) and some lacking sopD (no hybridization signal) (Table 7). These rare variants do carry sopE1 determinants resembling cluster I strains, e.g., SopEΦ from S. enterica serotype Typhimurium or from S. enterica serotype Typhi. It is interesting that the clonal, identical 11 EPV strains from poultry (Fig. 2) originated from different flocks in different geographical locations (Table 2).
The data summarized here allow us to conclude that the effector protein SopE1 but not AvrA seems to play an important part within the systemic phase of salmonellosis, probably with some other as-yet-unknown effector proteins (11), whereas SopD together with SopB is essential for the enteric outcome (28). The effector protein SopE2 was not considered throughout the study and might be of less importance for differentiating between SPV and EPV strains. Both pathovars reveal SopE2-producing and non-SopE2-producing variants; however, the reproducibility under our standard culture conditions was low, although α-SopE1 allowed us to detect SopE2 easily due to its different molecular mass (SopE1, 29.5 kDa; SopE2, 28.0 kDa).
The need for a broad range of Sop proteins to carry out enteric or systemic infection was also discussed earlier (29).
Since SPV and EPV strains have quite different clinical and epidemiological relevance, it is of great importance from a public health standpoint to have easy and reliable tests to distinguish between them (9, 14, 16, 19). Therefore, it is proposed here that the PCR-based testing for the presence of the virulence genes sopE1 and avrA be applied as a diagnostic tool: sopE1 is present and avrA is absent in all systemic variants of S. enterica serotype Paratyphi B, and sopE1 is absent and avrA is present among the EPV strains, with some exceptions (Tables 6 and 7).
The patterns of genetic properties of S. enterica serotype Paratyphi B strains summarized throughout this study, which help to distinguish between strains of systemic and enteric origins, are surprisingly in good correlation with their ability to ferment d-tartrate, which cannot be explained as of now. Therefore, the d-tartrate-fermenting property might be regarded as sufficient for clinical diagnostic purposes, as suggested earlier (12); however, the test for d-tartrate fermentation has often been found to be ambiguous and is sometimes difficult to read (3, 9). Consequently, d-tartrate fermentation alone is not reliable for discrimination between SPV and EPV strains.
Acknowledgments
We acknowledge the helpful suggestions and critical reading of the manuscript by W. D. Hardt, and we thank Ute Strutz, Gerlinde Bartel, Anette Weller, Ute Siebert, and Brigitte Tannert for their technical help as well as Erika Kleindienst for preparing the manuscript.
REFERENCES
- 1.Anderson, A. E. S. 1964. The phage typing of Salmonellae other than S. typhi, p. 89-110. In E. van Oye (ed.), The world problem of salmonellosis. Junk Publishers, The Hague, The Netherlands.
- 2.Barker, R. M. 1985. Utilisation of d-tartaric acid by Salmonella paratyphi B and Salmonella java: comparison of anaerobic plate test, lead acetate test, and turbidity test. J. Hyg. 95:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker, R. M., G. M. Kearney, P. Nicholson, A. L. Blair, R. C. Porter, and P. B. Crichton. 1988. Types of Salmonella paratyphi B and their phylogenetic significance. J. Med. Microbiol. 26:285-293. [DOI] [PubMed] [Google Scholar]
- 4.Beltran, P., S. A. Plock, N. H. Smith, T. S. Whittam, D. C. Old, and R. K. Selander. 1991. Reference collection of strains of the Salmonella typhimurium complex from natural populations. J. Gen. Microbiol. 137:601-606. [DOI] [PubMed] [Google Scholar]
- 5.Bollang, D. M., and S. J. Edelstein. 1991. Protein methods. A. J. Wiley and Sons, Inc., New York, N.Y.
- 6.Boy, E. F., F. K. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serotypes of subspecies. Int. J. Gen. Microbiol. 139:1125-1132. [DOI] [PubMed] [Google Scholar]
- 7.Breitenfeld, V., and D. Aleraj. 1967. Klinische und bakteriologische Eigenschaften der durch Salmonella java verursachten Salmonellose. Zentbl. Bakteriol. 204:89-99. [PubMed] [Google Scholar]
- 8.Claus, H., C. Cuny, B. Pasemann, and W. Witte. 1998. A database system for fragment patterns of genomic DANN of Staphylococcus aureus. Zentbl. Bakteriol. 287:105-116. [DOI] [PubMed] [Google Scholar]
- 9.Dorn, C., A. Schroeter, A. Miko, D. Protz, and R. Helmuth. 2001. Gehäufte Einsendungen von S. Paratyphi B-Isolaten aus Schlachtgeflügel an das Nationale Referenzlabor für Salmonellen. Berl. Münch. Tieraerztl. Wochenschr. 114:179-183. [PubMed] [Google Scholar]
- 10.Ezquerra, E., A. Burnens, C. Jones, and J. Stanley. 1993. Genotypic and phylogenetic analysis of Salmonella paratyphi B and S. java with IS200. J. Gen. Microbiol. 139:2409-2414. [DOI] [PubMed] [Google Scholar]
- 11.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015-1023. [DOI] [PubMed] [Google Scholar]
- 12.Kauffmann, F. 1955. Zur Differentialdiagnose und Pathogenität von Salmonella java und Salmonella paratyphi B. Z. Hyg. 141:546-550. [PubMed] [Google Scholar]
- 13.Kelterborn. E. 1967. Salmonella species. S. Hirzel Verlag, Leipzig, Germany.
- 14.Miko, A., B. Guerra, A. Schroeter, C. Dorn, and R. Helmuth. 2002. Molecular characterization of multiresistant d-tartrate-positive S. enterica serovar Paratyphi B isolates. J. Clin. Microbiol. 40:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, S. I., E. L. Hohmann, and D. A. Pegues. 1995. Salmonella (including Salmonella typhi). Churchill Livingstone, New York, N.Y.
- 16.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschäpe, H. Rüssmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE1 from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirold, S., W. Rabsch, H. Tschäpe, and W. D. Hardt. 2001. Transfer of the Salmonella type III effector sopE between unrelated phage families. J. Mol. Biol. 312:7-16. [DOI] [PubMed] [Google Scholar]
- 18.Murphy, J. B., J. Feldman, and H. Vogel. 1968. Epidemic of febrile gastroenteritis due to Salmonella java traced to smoked whitefish. Am. J. Public Health 58:114-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsen, S. J., R. Bishop, F. W. Brenner, T. H. Roels, N. Bean, R. V. Tauxe, and L. Slutsker. 2001. The changing epidemiology of Salmonella: trends in serotypes isolated from humans in the United States, 1987-1997. J. Infect. Dis. 183:753-761. [DOI] [PubMed] [Google Scholar]
- 20.Popoff, M. Y. 2001. Antigenic formulas of the Salmonella serovars. W. H. O. Collaborating Centre for Reference and Research on Salmonella. Institut Pasteur, Paris, France.
- 21.Prager, R., E. Tietze, S. Mirold, W. Rabsch, W. D. Hardt, and H. Tschäpe. 2000. Prevalence and polymorphism of genes encoding translocated effector proteins among clinical strains of Salmonella enterica. Int. J. Med. Microbiol. 290:605-617. [DOI] [PubMed] [Google Scholar]
- 22.Prager, R., A. Liesegang, W. Rabsch, B. Gericke, W. Thiel, W. Voigt, R. Helmuth, L. Ward, and H. Tschäpe. 1999. Clonal relationship of Salmonella enterica serovar Typhimurium phage type DT104 in Germany and Austria. Zentbl. Bakteriol. 289:399-414. [DOI] [PubMed] [Google Scholar]
- 23.Rische, H., and K. Ziesché. 1973. Salmonella paratyphi B, p. 65-86. In H. Rische and K. Ziesché (ed.), Infektionskrankheiten und ihre Erreger. Lysotypie, Band 14. G. Fischer Verlag, Jena, Germany.
- 24.Schmieger, H. 1999. Molecular survey of the Salmonella phage typing system of Anderson. J. Bacteriol. 181:1630-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selander, R. K., P. Beltran, N. H. Smith, R. M. Barker, P. B. Crichton, D. C. Old, J. M. Musser, and T. S. Whittam. 1990. Genetic population structure, clonal phylogeny, and pathogenicity of Salmonella paratyphi B. Infect. Immun. 58:1891-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seltmann, G., W. Voigt, and W. Beer. 1994. Application of physico-chemical typing methods for epidemiological analysis of Salmonella enteritidis strains of phage type 25/17. Epidemiol. Infect. 113:411-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Pelt, W., H. van der Zee, W. J. B. Wannet, A. W. van der Giessen, D. J. Mevius, N. M. Bolder, R. E. Komijn, and Y. T. H. P. van Duynhoven. 2002. Explosieve toename van Salmonella Java in pluimvee. Infect. Bull. 13:260-265. [PubMed] [Google Scholar]
- 28.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 26:1005-1011. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, S., R. L. Santos, R. M. Tsolis, S. Stender, W. D. Hardt, A. J. Bäumler, and L. G. Adams. 2002. The Salmonella enterica serovar Typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect. Immun. 70:3843-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]