Abstract
Molecular epidemiological tools for genotyping clinical isolates of Mycobacterium tuberculosis have been developed and used to help track and contain transmission of tuberculosis. We identified 87 short sequence repeat loci within the genome of the M. tuberculosis H37Rv strain. Nine tandem repeats were found to be variable (variable-number tandem repeats [VNTRs]) in a set of 91 isolates. Fifty-seven of the isolates had only four IS6110 bands. The other 34 isolates were members of the Beijing strain family. The number of alleles of each these nine VNTRs was determined by examining each isolate. Six of the loci (Mtb-v1, -v4, -v10, -v15, -v18, and -v20) were able to differentiate the Beijing spoligotype identical isolates into seven distinct genotypes. Five of the loci (Mtb-v3, -v5, -v6, -v10, and -v15) were informative in discriminating the four-band IS6110 restriction fragment length polymorphism isolates from each other. The Nei's diversity values of each marker ranged from 0.02 to 0.59, with the number of alleles ranging from two to eight across the entire strain set. These nine loci provide a useful, discriminatory extension of VNTR typing methods for application to molecular epidemiologic studies of M. tuberculosis.
Tuberculosis (TB) is the primary cause of death due to a single microbial pathogen, accounting for 2 to 3 million deaths and an estimated 8 to 10 million new cases per year worldwide (1). It is estimated that one-third of the world's population is infected with Mycobacterium tuberculosis. The global epidemic is growing, concomitant with human immunodeficiency virus coinfection and the increasing prevalence of drug-resistant strains. Genotyping clinical isolates of M. tuberculosis has the potential to significantly impact both clinical practice and public health. Clinically, determining strain relationships can distinguish treatment failure from potential laboratory cross-contamination and reactivation of drug-resistant disease from infection with another strain (23). In the public health arena, determining strain relationships during the course of a TB outbreak can focus public health resources. However, the most discriminatory M. tuberculosis genotyping method, IS6110 restriction fragment length polymorphism (RFLP) analysis, is time-consuming, technically demanding, and difficult to standardize between laboratories (3). In addition, IS6110 RFLP does not provide sufficient strain discrimination when fewer than six IS6110-hybridizing bands are present. A secondary genotyping method is required in these instances. A number of PCR-based genotyping methods have been developed but are limited in their ability to reproducibly provide a level of strain discrimination that matches associations defined by traditional epidemiology. In addition, the high degree of coding region sequence conservation between strains of M. tuberculosis virtually excludes the use of more commonly used population genetics methods like multilocus enzyme electrophoresis, restriction fragment length end labeling analysis, and gene sequencing (12).
Spoligotyping is currently the most widely used PCR-based method of M. tuberculosis genotyping due to the relative technical ease of the protocol and the ability to compare results between laboratories using a standardized nomenclature system (6). However, the discriminatory power of this method is low. In a database of 3,319 spoligotypes from isolates collected in geographically distinct areas, seven spoligotype patterns were found for 37% of all clustered isolates (17). These spoligotype patterns are associated with strains having a much larger variety of IS6110 RFLP patterns. For instance, 18% of isolates in the spoligotype database had the Beijing spoligotype pattern, characterized by the presence of spacer oligonucleotides 39 through 43 (17). However, although the W-Beijing family of strains is recognized as one of the most homogeneous families of M. tuberculosis strains (18), the Beijing spoligotype is associated with at least 450 distinct strains as determined by IS6110 RFLP analysis (2).
Following the public release of the complete genomic sequences of M. tuberculosis strains H37Rv and CDC-1551, investigators began the process of identifying repetitive genetic elements to develop higher-resolution DNA typing systems. Variable-number tandem repeats (VNTRs) of 21- to 111-bp genetic elements have been identified (9, 14, 16, 19, 20). A set of 12 VNTRs, also known as mycobacterial interspersed repeat units (MIRUs), was more discriminatory than IS6110 RFLP analysis when used to analyze 180 M. tuberculosis isolates with six or fewer IS6110-hybridizing bands. Combining MIRU analysis with spoligotyping and IS6110 RFLP analysis provided maximum specificity (5).
In this report, we describe a multiple-locus VNTR analysis (MLVA) genotyping system in which independent, tandemly inserted repeated motifs in the M. tuberculosis genome are amplified using fluorescently labeled primers in multiplexed PCRs and electrophoretically separated on polyacrylamide gels for analysis. We identified and developed nine novel VNTR loci located throughout the H37Rv genome. These were informative across the two sets of clinical isolates of M. tuberculosis: a set of 34 Beijing strains and another set of 57 isolates having four IS6110 RFLP bands. The purpose of introducing this method using isolates of limited diversity was to establish the usefulness of MLVA for rapid genotyping of M. tuberculosis. The discriminatory power of the MLVA targets described in this report expands the overall ability to rapidly determine molecular relationships of M. tuberculosis isolates using a combination of VNTR and spoligotype analyses, particularly with respect to the globally prevalent homogeneous W-Beijing family of strains.
MATERIALS AND METHODS
M. tuberculosis strains.
As part of the Centers for Disease Control and Prevention National Tuberculosis Genotyping and Surveillance Network, the University of Texas Health Science Center at San Antonio (UTHSCSA) serves as a genotyping reference laboratory for M. tuberculosis isolates collected by the Texas Department of Health (TDH). From 1996 to present, isolates from 4,269 persons with TB were genotyped (4, 15). The institutional review boards of the TDH and the UTHSCSA approved culture collection and analyses (TDH-Centers for Disease Control and Prevention cooperative agreement; TDH protocol 980025 and UTHSCSA protocol 001-6000-105). Two sets of clinical isolates were analyzed in the study reported here. One set consisted of 57 isolates having four IS6110 RFLP bands. The second set included 34 isolates with an identical Beijing spoligotype pattern containing spacer oligonucleotides 36 to 43 and 14 different IS6110 RFLP patterns
DNA isolation.
M. tuberculosis isolates were grown on Lowenstein-Jensen medium slants (Becton, Dickinson, and Co., Franklin Lakes, N.J.) for 4 to 6 weeks at 37°C under 5% CO2. In a biosafety-level-3 facility, the bacterial mass was removed using a sterile inoculating loop, placed in a microcentrifuge tube containing 1 ml of double-distilled H2O, and heat-killed at 85°C for 20 min. The cells were pelleted by centrifugation and resuspended in a 10 mM Tris-HCl-1 mM EDTA buffer (pH 8.0). Cell walls were digested with lysozyme (10 mg/ml), proteinase K (10 mg/ml), and 10% sodium dodecyl sulfate. DNA was extracted using 0.3 M cetyltrimethylammonium bromide and 5 M NaCl, purified by chloroform-isoamyl alcohol separation, and precipitated using isopropanol (Sigma, St. Louis, Mo.).
Spoligotyping.
Spoligotype analyses (10) were done using locally synthesized and biotinylated PCR primers (UTHSCSA Advanced Nucleic Acids Technology Core Facility, San Antonio, Tex.) and commercially available spoligotyping membranes (Isogen, Maarssen, The Netherlands) as previously described (15).
IS6110 RFLP analysis.
IS6110-based RFLP analysis was performed by Southern blotting of PvuII-digested genomic DNA using a 523-bp right-handed IS6110 probe (22), the ECL detection system (Amersham, Piscataway, N.J.), and a BioImage Whole Band analyzer (version 3.4.2; Genomic Solutions, Ann Arbor, Mich.) as previously described (15).
VNTR identification and primer design.
The complete genome sequence of the M. tuberculosis H37Rv strain was downloaded from the Sanger Centre (www.sanger.ac.uk). Potentially polymorphic repetitive sequences were identified using the DNAstar software program Genequest (Lasergene, Inc., Madison, Wis.). Selection criteria for repetitive sequences were set for nucleotide repeat motifs of more than 8 bp within 100 bp of each other. Primers were designed around 84 repetitive sequences identified using the DNAstar software program Primer Select (Lasergene, Inc.). Complementary primers were designed around interspersed repeats to minimize risk of mobile DNA targets. Fifteen of these repetitive sequences were found to be polymorphic, i.e., VNTR loci. Nine loci were chosen for use in this study (Table 1). Since our MLVA development, marker Mtb-v20 has been developed and published (13), and Mtb-v15 was found to be previously identified by Frothingham (9). The other five markers were excluded from this study for eclectic reasons; lack of diversity across this low-diversity collection, shadow band intensity, and primer failure due to poor design.
TABLE 1.
Name, location, and sequence of primers
| Primer | Location (kb)a | Primer sequence
|
|
|---|---|---|---|
| Forward | Reverse | ||
| Mtb-v1 | 55.481 | GTCGAACGAGACTTTCCCCAAACCGAC | GACCGTGGGCTGGATGACGGTCTC |
| Mtb-v3 | 241.423 | GATGACGGATCGTCGGGGGCGGGAAC | GCAACGCGAGGGCGATCAGTACTGCCAACA |
| Mtb-v4 | 472.658 | GCTGTGGCGCAGCTACACAGTACGACTC | GATTGCGCAGCGCCCAACAGC |
| Mtb-v5 | 566.196 | GGAGGCGTTGGGTACGGTCGCATC | GATTCGGAGCCCGACTACTTCTGGGT |
| Mtb-v6 | 1122.852 | CGCCGACGAGGCCGATGCCGAAGC | CCGCGGCGGCAGAGCCAACCAGGAT |
| Mtb-v10 | 2604.134 | CGAGGCGCCCAGCCCCACAA | CACCCGCGCTTTAGGATCGACACCTGA |
| Mtb-v15b | 3239.432 | GCGCCGCACCACCTCGACTT | CCGGGCAAAACCTCCGCCTAAC |
| Mtb-v18 | 4226.924 | ACAACGGCGAGGCCCGAATCTACGAA | GTCGACGCCGCCGATGACC |
| Mtb-v20c | 23.420 | CCCGGAGGGCCAGAGGGCACATAGC | TGGCGCAGAACCAGGAGTAGCACCAATGAG |
Location of VNTR locus is given by the 5′ of the forward primer in accordance with the H37Rv reference strain sequenced in 1998.
Mtb-v15 was previously identified as ETR-F by Frothingham.
Mtb-v20 was recently identified by Le Fleche et al. (13), and given the name Mtub01.
PCR amplification of VNTR loci.
PCRs were performed using a total volume of 10 μl. All PCR reagents used were obtained from Invitrogen (Madison, Wis.) unless otherwise indicated. Each PCR mixture contained the following: 1× PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl); 2 mM MgCl2; a 200 μM concentration of each of the four deoxynucleoside triphosphates; 0.4 U of Platinum Taq Polymerase (Gibco-Life Technologies) per μl; 0.2 μM forward primer; 0.2 μM reverse primer R110, R6G, or TAMRA phosphoramide fluorescence-labeled oligonucleotides (Perkin-Elmer Biosystems); 1 μl of DNA template; and double-distilled H2O, bringing the total volume to 10 μl. Each 10-μl PCR mixture was then denatured for 5 min at 95°C. Following the denaturing, the samples were cycled 35 times through the following program: 95°C for 20 s, 55°C for 25 s, and 72°C for 20 s. These 35 cycles were followed by a final extension at 72°C for 5 min, and the samples were then stored at −20°C until genotyped. Reactions that failed under the above conditions were then repeated with 2 μl of DNA in a 10-μl reaction mixture, with all other concentrations and conditions remaining the same.
Genotyping.
Detailed automated genotyping methods have been described previously (11). Briefly, fluorescence-labeled amplicons were sized and scored using the ABI software program Genotyper. An independent party has verified all sizes presented.
Statistical analysis.
Genetic distances were determined by calculating the percentage shared alleles pairwise among all isolates. The clustering method used was the unweighted pair group method using arithmetic averages (UPGMA). UPGMA clustering analysis was performed using the software program NTSYS (version 2.1; Exeter Software, Setauket, N.Y.). There was no missing data across the nine VNTR loci and 91 isolates. The diversity index of each VNTR marker was calculated using the formula [1 − Σ(allele frequencies)2] (from the Genetic Data Analysis II package; Sinauer Associates, Inc., Sunderland, Mass.).
RESULTS
MLVA analysis of M. tuberculosis isolates.
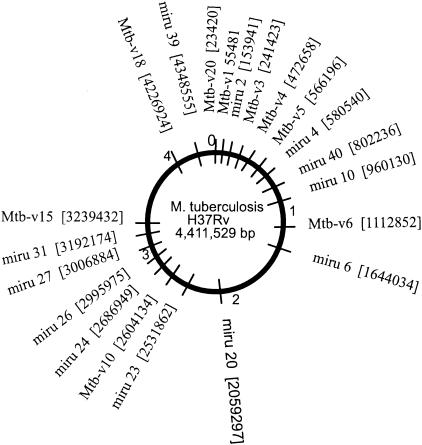
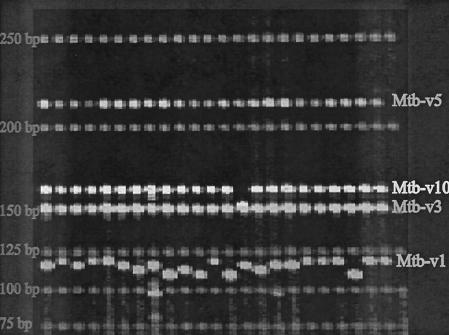
We identified 84 regions containing tandem repeat sequences as potential VNTRs from the H37Rv sequence. Nine of the loci were polymorphic across the isolates included in this study, and their genomic location is illustrated (Fig. 1). At least one VNTR marker was discovered in every Mbp region, and five were clustered in the first Mbp. Our approach focuses on short sequence repeats that can be readily PCR amplified and molecularly sized with automated fluorescence-based instrumentation (Fig. 2). Of the six VNTR not included in this study, four loci were found to be polymorphic across a more diverse worldwide isolate set but were not informative across this data set. The other two markers, although polymorphic across this data set, gave strong secondary amplification products and, hence, were not included in this study. The amplicon size range for 7 of the loci used in this study is 107 to 290 bp. This size range allows for electrophoresis runs of roughly 2 h on the ABI377 system (Applied Biosystems Incorporated, Foster City, Calif.). Two markers (Mtb-v15 and Mtbv-20) have larger ranges from 278 to 591 bp (Table 2). The number of alleles across these nine loci ranged from 2 to 8 (Table 3).
FIG. 1.
Genomic survey location of MLVA and MIRU markers. The locations of nine MLVA and 12 MIRU markers are shown on the H37Rv genome. The number in brackets indicates the location of the 5′ forward primer for each marker. All accession numbers of forward primer locations are given using the H37Rv sequence.
FIG. 2.
Electrophoresis of MLVA fragments. Fluorescent image of markers Mtb-v1,-v3,-v5, and -v10 on an ABI377 electrophoresis gel. Each marker allele is identified through a unique size and color combination, allowing adequate discrepant ability across comparable-sized fragments from sundry alleles. Actual sizes of these alleles can be seen in Table 4. Sizes are shown in nucleotide bases.
TABLE 2.
Characteristics of Mtb VNTRsd
| VNTR locus | Amplicon size | Size rangea | Motif × repeat(s)b | VNTR motif within gene |
|---|---|---|---|---|
| Mtb-v1 | 113 | 107-119 | 3 × 7 (+1) | PonA (penicillin resistance gene) |
| Mtb-v3 | 152 | 140-152 | 11 × 3 | Rv0203 (hypothetical gene) |
| Mtb-v4 | 146 | 137-164 | 9 × 4 (+2) | None (5′ adjacent to Rv0393) |
| Mtb-v5 | 212 | 196-230 | 18 × 3 (−1) | hbhA (heparin-binding hemagglutin gene) |
| Mtb-v6 | 284 | 278-290 | 6 × 4 (+2) | Rv0996 (hypothetical gene) |
| Mtb-v10 | 164 | 155-181 | 9 × 3 (+6) | IppP (probable lipoprotein-cell wall metabolism) |
| Mtb-v15 | 415 | 278-591 | 55 × 2 (−18), 79 × 3 (−13)c | None (5′ adjacent to ribonuclease III gene) |
| Mtb-v18 | 136 | 133-136 | 3 × 4 (+2) | Rv3780 (unknown gene) |
| Mtb-v20 | 560 | 560-578 | 18 × 2 (−4) | Rv0019c (conserved hypothetical gene) |
Size range given corresponds to range seen across all collections.
Numbers with a + or − sign in front represent the number of addition nucleotides present/absent in a complete repeat.
Nontandem polymorphic repeats include 8 × 6 and 12 × 2.
All table data are representative of findings from the H37Rv strain.
TABLE 3.
Allelic number and diversity of VNTR across data set
| VNTR name | No. of alleles | % Diversity
|
||
|---|---|---|---|---|
| All | Beijing | Four-band IS6110 | ||
| Mtb-v1 | 5 | 0.59 | 0.25 | 0.23 |
| Mtb-v3 | 2 | 0.06 | 0.0 | 0.09 |
| Mtb-v4 | 2 | 0.08 | 0.16 | 0.03 |
| Mtb-v5 | 2 | 0.02 | 0.0 | 0.03 |
| Mtb-v6 | 2 | 0.02 | 0.0 | 0.03 |
| Mtb-v10 | 4 | 0.48 | 0.16 | 0.30 |
| Mtb-v15 | 4 | 0.14 | 0.20 | 0.06 |
| Mtb-v18 | 2 | 0.12 | 0.20 | 0.03 |
| Mtb-v20 | 2 | 0.02 | 0.06 | 0.0 |
All of the polymorphic markers, except Mtb-v20, contained three or more copies of a tandem repeat sequence in the H37Rv sequence. Of these, seven had repeat motifs of three base multiples (Table 2), consistent with in-frame insertion and deletion mutational events. Seven loci were within hypothetical or known ORFs, while two were adjacent to genes (Table 2).
VNTR marker diversity.
In this relatively homogenous set, diversity across the nine loci varied greatly, as measured by allele number range and diversity value range (Table 3). Six of the loci (Mtb-v1, -v3, -v5, -v6, -v10, and -v15) were variable across the collection of isolates having four IS6110 bands, and six loci (Mtb-v1, -v4, -v10, -v15, -v18, and -v20) were variable across the Beijing collection. In contrast, three VNTR loci (Mtb-v1, -v10, and -v15) had four alleles and diversity values ranging from 0.14 to 0.59 across all samples.
When the 34 Beijing family strains were considered separately, the number of alleles and diversity values dropped. Six marker loci were informative within Beijing strains and three (Mtb-v1, -v15, and -v18) were still relatively diverse, with diversity values of 0.25, 0.20, and 0.20, respectively. Markers Mtb-v4 and -v15 and were more diverse across the Beijing collection than across the four-band set (Table. 3). Diversity values for collection of isolates having four IS6110 bands ranged from 0.0 to 0.3. The diversity values for Mtb-v5, -v6, and -v20 correspond to only one allele variation, in one isolate, across the entire collection set (Table 4).
TABLE 4.
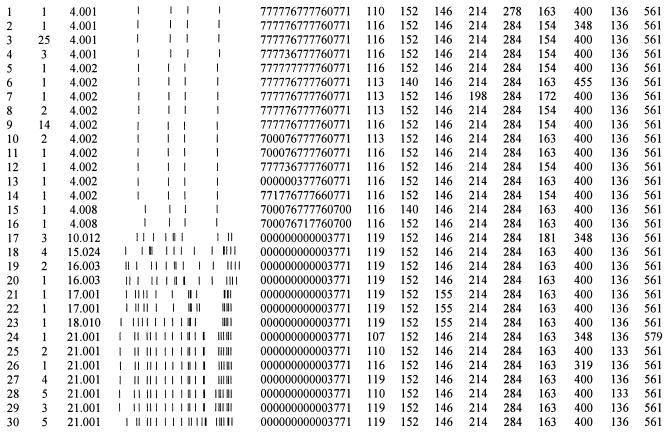
Clustering defined by IS6110 RFLP, spoligotype, and VNTR analysesa
Cluster designates a group of isolates with a distinct genotype defined by IS6110 RFLP analysis spoligotype analysis and MLVA; n is the number of isolates within the cluster; the IS6110 RFLP is numerically designated and the image is shown with high to low molecular weights oriented from left to right; spoligotype nomenclature is as defined by Dale et al. (6); and Mtb-v1 through Mtb-v20 indicate the amplicon size observed for the corresponding VNTR amplicons.
The diversity values of Mtb-v1 and Mtb-v10 across the combined strain set were greater than for either strain set alone. This was due to differential allele frequency, as opposed to unique allele number, across each data set. The Mtb-v1 size of 116 bp had a frequency of 88% within the set of strains having four IS6110 bands. In contrast, the allele size of 116 bp was seen on one occasion across the Beijing strain set, a 2.9% frequency. For Mtb-v10, a similar pattern of allele bias can be seen. The allele size of 154 bp is seen in 84% of the isolates having four IS6110 bands. In contrast, the allele size of 154 is not present at all in the Beijing strains. Twenty-nine (85%) of the Beijing strains had an allele size of 163, whereas eight (14%) of the isolates having four IS6110 bands had an allele size 163. Hence, the diversity value across Mtb-v1 and Mtb-v10 is higher when the data are combined.
As observed in Table 3, a larger number of alleles did not always correspond to a greater diversity value. In one sense, allele number represents potential discrimination power while the diversity value represents the realized discrimination in a given set of samples. Both values are useful for understanding the genetic loci and the isolate sets being studied.
Spoligotyping genotypes.
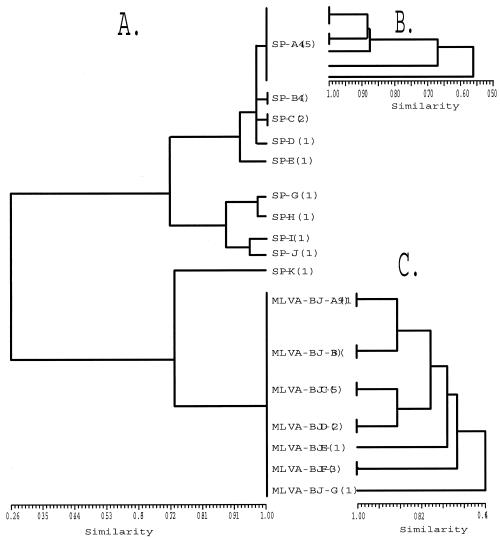
Spoligotyping differentiated the 91 isolates into nine unique genotypes (Fig. 3A). The spoligotyping dendrogram revealed the deepest branch at 0.26, which would concur with the number of loci considered in UPGMA, which is 43. By definition, the Beijing strains share spoligotype pattern designation 000000000003771 (6).
FIG. 3.
Dendrograms based upon UPGMA analysis of spoligotyping (A) and MLVA (B and C) data. (A) Clustering using spoligotyping data across all 91 isolates. (B) The group SP-A is broken into five genotypes using MLVA. (C) MLVA resolves the Beijing cluster into seven genotypes, designated MLVA-BJ-A through MLVA-BJ-G.
MLVA genotypes.
Among all 91 isolates, the nine-marker MLVA identified 17 unique genotypes (not shown); 12 unique MLVA genotypes are shown in Fig. 3, which were identified from two spoligotype clusters, SP-A and SP-L. Spoligotype genotype cluster A was represented by five MLVA genotypes (Fig. 3B), while the Beijing family isolates (34 total) were subdivided into seven unique types (Fig. 3C) based upon six informative marker loci (Table 2). Two of the isolates were unique, with the other isolates in clusters of 19, 5, 3, 3, and 2 (Fig. 3C).
DISCUSSION
This MLVA typing system is a PCR-based method for genotyping M. tuberculosis using nine novel independent VNTR loci. This PCR-based typing has advantages present in other PCR systems: rapid turnaround, amplification with crudely isolated or minute amounts of DNA, and the ability to use nonviable samples which can be transported across various laboratories with greater ease than viable ones. Here we have presented a rapid typing system using nine VNTR loci across a collection of 57 isolates of limited diversity, in that they all had four IS6110 bands, and 34 isolates from the Beijing strain family. This study introduces the use of smaller repeat motifs <50 bp as credible additions to the molecular typing of M. tuberculosis. The fluorescent labeling and size of each allele allow for unique and easy identification of each VNTR. The technology and protocols are becoming increasingly used and available. Automated gel analysis allows for a high-throughput system that reduces error and time in obtaining results, though at some loci, stutter peaks created by strand slippage during PCR can complicate results (5). Two additional variable markers (not included in Table 1) were removed from this data set because of such problems. High-resolution electrophoresis and discrete repeats allows for the use of standardized data sets and facilitates central databases (e.g., Table 4).
The nine VNTRs presented here can also contribute to the MIRUs recently developed (14). These primers are labeled with the same fluorescent dyes used in the MIRU set and could be analyzed on the same electrophoretic run as the MIRUs because of their smaller size and allele composition. It should also be possible to incorporate MIRU and Mtb markers in the same PCR multiplex reactions, although some optimization will be needed. This would function as a legitimate and helpful extension of the VNTR typing already taking place, without much additional time or labor, and would increase the discriminatory ability of the present VNTR-typing, as has been noted previously (5).
Molecular typing of M. tuberculosis using VNTR may produce a useful molecular clock. In MLVA, we have the ability to observe different rates of evolution for each VNTR locus. This gives us the flexibility to use VNTR markers that match the type of epidemic we are tracking. For example, cases of M. tuberculosis incidence in areas of endemicity would intuitively have little variation among markers of low diversity. The antithesis of that would be that more-diverse markers would be poor for establishing phylogeny across a diverse isolate collection, which is akin to similar findings with IS6110 elements (7). However, with MLVA we are able to pick and choose what markers would be most appropriate for a given epidemic. For example, we could deduce that VNTR loci with a low diversity index across a worldwide set would have an even lower or nonexistent value across a local outbreak. The “clocking” of a VNTR could be a credible methodology for time-geographical correlation. This ability to create a wide gamut of markers of sundry diversity is helpful in a geographically limited area where a particular genetic clone may be more prevalent (5). The intrinsic value of this concept supports the idea that some VNTRs are not suitable for possible transmission routes but are better as informers of plausible evolutionary scenarios, and vice versa (11, 21). Further MLVA work would determine if such “clocks” are really consistent with incidence demographics.
Some VNTR markers demonstrate an affinity between actual genetic distance and the expansion or contraction of a repeat motif. The dendrogram demonstrates this cascading trend with Mtb-v1. Within the Beijing collection, any clusters with a 0.11 difference (one allele) from each other had a repeat motif that differed by only one repeat or allele. For example, MLVA-BJ-A and -BJ-B differ by one marker score in Mtb-v10. Yet MLVA-BJ-C and BJ-D, which differ from MLVA-BJ-A and BJ-B by more than one marker allele, have an Mtb-v1 allele size of 110, as opposed to 119 in MLVA-BJ-A and BJ-B. This seems to suggest that phylogeny could be determined further by the difference in repeat motifs of any particular marker. We then could postulate that each marker gives two types of data: the raw score of allele size and a relative allele relationship. This relative relationship is possible with MLVA, where the diversity within one marker can be greater than 0.5, whereas binary data can give a maximum diversity of 0.5 for any one locus.
The dominance of a particular marker allele could prove to be a useful tool in quickly referencing a clonal dispersion. In our study, this distinct allele frequency is seen in both Mtb-v1 and Mtb-v10. This allele bias has been seen in Francisella tularensis as well, indicating two distinct biovars (8). Although this clonal referencing would be unhelpful in discrimination within an outbreak, it could prove to be a helpful tool in phylogeny studies. This idea of a distinctive allele type would only be viable when multiple loci are combined in analyzing the data. By doing so, we are able to curtail the effect of convergent evolution in the analysis.
The most immediate public health application of this typing method will be its use during an outbreak. Determination of strain relationship between infected individuals could help in deducing routes of transmission. This information could save time and money for clinics and health care-related professionals involved in eradication of M. tuberculosis. It is anticipated that this methodology will be a constructive addition to the molecular epidemiological methods currently used to track M. tuberculosis.
REFERENCES
- 1.Anonymous. 2002. Global tuberculosis control report, p. 6. World Health Organization, Geneva, Switzerland.
- 2.Bifani, P. J., M. B., Kurepina, N. E., Kreiswirth, B. N. 2002. Global dissemination of the Mycobacterium tuberculosis W family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 3.Braden, C. R., J. T. Crawford, and B. A. Schable. 2002. Assessment of Mycobacterium tuberculosis genotyping in a large laboratory network. Emerg. Infect. Dis. 8:1210-1215. [DOI] [PMC free article] [PubMed]
- 4.Castro, K. G., and H. W. Jaffe. 2002. Rationale and methods for the national tuberculosis genotyping and surveillance network. Emerg. Infect. Dis. 8:1188-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowan, L. S., L. Mosher, L. Diem, J. P. Massey, and J. T. Crawford. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 40:1592-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dale, J. W., D. Brittain, A. A. Cataldi, D. Cousins. J. T. Crawford, J. Driscoll, H. Heersma, T. Lillebaek, T. N. Quitugua, N. Rastogi, D. van Soolingen, V. Wright. 2001. Spacer oligonucleotide typing of Mycobacterium tuberculosis: recommendations for standardized nomenclature. Int. J. Tuberc. Lung Dis. 5:216-219. [PubMed] [Google Scholar]
- 7.Fang, Z., D. T. Kenna, C. Doig, D. N. Smittipat, P. Palittapongarnpim, B. Watt, and K. J. Forbes. 2001. Molecular evidence for independent occurrence of IS6110 insertions at the same sites of the genome of Mycobacterium tuberculosis in different clinical isolates. J. Bacteriol. 183:5279-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisella tularensis strain typing using multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 10.Kamerbeek J., L. S., A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, J. D. A van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Fleche P., M. Fabre, F. Denoeud, J. L. Koeck, G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 37:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quitugua, T. N., B. J. Seaworth, S. E. Weis, J. Taylor, J. Gillette, I. Rosas, K. C. Jost, Jr., D. M. Magee, R. A. Cox. 2002. Transmission of drug-resistant tuberculosis in Texas and Mexico. J. Clin. Microbiol. 40:2716-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519-528. [DOI] [PubMed] [Google Scholar]
- 17.Sola, C., I. Filliol, M. C. Gutierrez, I. Mokrousov, V. Vincent, and N. Rastogi. 2001. Spoligotype database of Mycobacterium tuberculosis: biogeographic distribution of shared types and epidemiologic and phylogenetic perspectives. Emerg. Infect. Dis. 7:390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sola, C., I. Filliol, E. Legrand, I. Mokrousov, and N. Rastogi. 2001. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR, and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J. Mol. Evol. 53:680-689. [DOI] [PubMed] [Google Scholar]
- 19.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991-1003. [DOI] [PubMed] [Google Scholar]
- 21.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 22.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Soolingen, D. 2001. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Int. Med. 249:1-26. [DOI] [PubMed] [Google Scholar]