Abstract
We describe an outbreak, in a community of men who have sex with men, of serogroup C meningococcal disease caused by a genetic variant of the serotype 2a ET-15 Neisseria meningitidis characterized by a point mutation in the gene coding for the serotype 2a antigen. A microbiological characterization of the outbreak strain is presented in this report.
Invasive meningococcal disease (IMD) is a notifiable disease under surveillance in Canada (9). The disease is endemic in the city of Toronto, Ontario, Canada, with a 5-year average incidence rate of 0.7 cases per 100,000 persons per annum over the years 1996 to 2000 (surveillance data from Toronto Public Health; authors' unpublished observation). Ongoing surveillance and a detailed case-by-case investigation by Toronto Public Health's Communicable Disease Control investigators identified a cluster of cases as well as a rise in the incidence of IMD between early May and mid-July 2001. Six cases of IMD caused by a unique strain of Neisseria meningitidis were diagnosed during this 10-week period. N. meningitidis was isolated from all six cases; five patients had a positive blood culture, and in one case the organism was isolated from cerebrospinal fluid.
All six cases were found in the community of men who have sex with men (MSM) and five of these were known or suspected to have had direct or indirect contact with bathhouses for males during the incubation period. No such meningococcal outbreak has been reported to date in the literature. The patients were not known to have had direct contact with one another; however, detailed and accurate histories were difficult to obtain from patients, due both to the anonymous nature of bathhouse contact and to the fulminant medical sequelae of the illness. Two cases were rapidly fatal, yielding a case fatality rate of 33%. Throughout the outbreak period, five additional cases of IMD occurred in Toronto but were not linked epidemiologically to the outbreak. Strains from these five patients were found to be different from the outbreak strain by both antigenic and pulsed-field gel electrophoresis analysis (see description below). All six of the outbreak patients were male, ranging in ages from 23 to 39 years old. Because sexual preference and practice of IMD cases are not risk factors that are routinely elicited by public health investigators, the baseline incidence of IMD in the MSM community prior to the outbreak is unknown.
Three of the patients were known to have attended at least one of three bathhouses for males in downtown Toronto (venues for sexual activity among MSM) during their incubation period, and an additional two patients had indirect contact (i.e., through close friends) with bathhouses during their incubation period. Bathhouse attendance was therefore believed to have been a likely site of transmission and acquisition of IMD. No other risk factors for IMD were identified. Persons in close contact with patients, including household and potential bathhouse contacts, were advised to receive chemoprophylaxis. In response to the outbreak, Toronto Public Health, in cooperation with the MSM community, several community care providers, and the Ontario Ministry of Health and Long-Term Care, organized and implemented an immunization campaign to prevent further cases of IMD. More than 50 immunization clinics were held in a variety of locations, including bathhouses, an MSM community center, a well-reputed sexually transmitted disease clinic, and hospital-based immunodeficiency clinics. Between 25 July and 18 August 2001, more than 3,850 doses of quadrivalent polysaccharide vaccine (Menomune; Connaught Laboratories Ltd., North York, Ontario, Canada) were administered free of charge to eligible persons (homosexual or bisexual men having one or more of the following risk factors: is 20 to 44 years old, has sex with more than one man, frequents bathhouses or gay bars, or has a condition that seriously affects their immune system). Subsequently, no further cases of IMD were reported in the MSM community.
Microbiology of the Neisseria meningitidis outbreak strains.
All six strains isolated from patients involved in this outbreak gave typical biochemical reactions that identified them as N. meningitidis, and all belonged to serogroup C based on bacterial agglutination with specific antisera to the different capsular polysaccharide groups. Besides the grouping based on their capsular polysaccharide structures, meningococci can be further subdivided or typed by detection of serologically distinct epitopes present on their outer membrane proteins (OMPs). Although there are five different classes of OMPs expressed by N. meningitidis, most epidemiological studies are based on antigens expressed on three of these five classes of proteins. Serotype antigens of meningococci are found on their class 2 or class 3 PorB porin OMPs, which are expressed in a mutually exclusive fashion, i.e., a strain will express either a class 2 PorB or a class 3 PorB but not both together. Serosubtype antigens of meningococci are found on their class 1 PorA OMP. Since both PorB and PorA are surface proteins, their specific epitopes are usually determined by a panel of specific monoclonal antibodies reacting with the meningococcal whole cells.
The serotype and serosubtype antigens of the outbreak strains were determined by indirect whole-cell enzyme-linked immunosorbent assay method (1) using monoclonal antibodies against the serotype antigens 1, 2a, 2b, 4, 14, and 15 and monoclonal antibodies against serosubtype antigens P1.1, P1.2, P1.4, P1.5, P1.6, P1.7, P1.9, P1.10, P1.12, P1.13, P1.14, P1.15, and P1.16 (Rijksinstituut voor Volksgezondhei en Milieu, National Institute of Public Health, The Netherlands). All six strains were identified as nonserotypeable (NT) with the serosubtype antigen of P1.2.
All six strains were sensitive to penicillin, chloramphenicol, ceftriaxone, and rifampin when determined by agar dilution (3) and were β-lactamase negative when the enzyme was assayed by a chromogenic procedure using the DrySlide beta-lactamase test (Becton Dickinson Microbiology Systems, Sparks, Md.).
Multilocus enzyme electrophoresis demonstrated that all six outbreak strains belonged to the ET-15 clone in the ET-37 complex showing the characteristic fumarase enzyme allele 2 (2, 5).
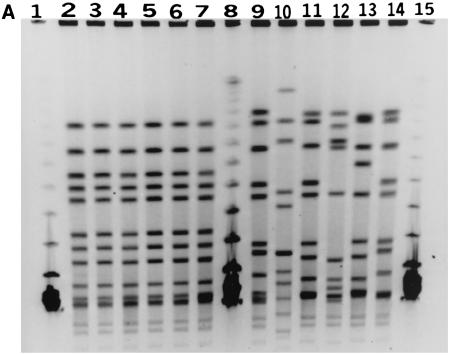
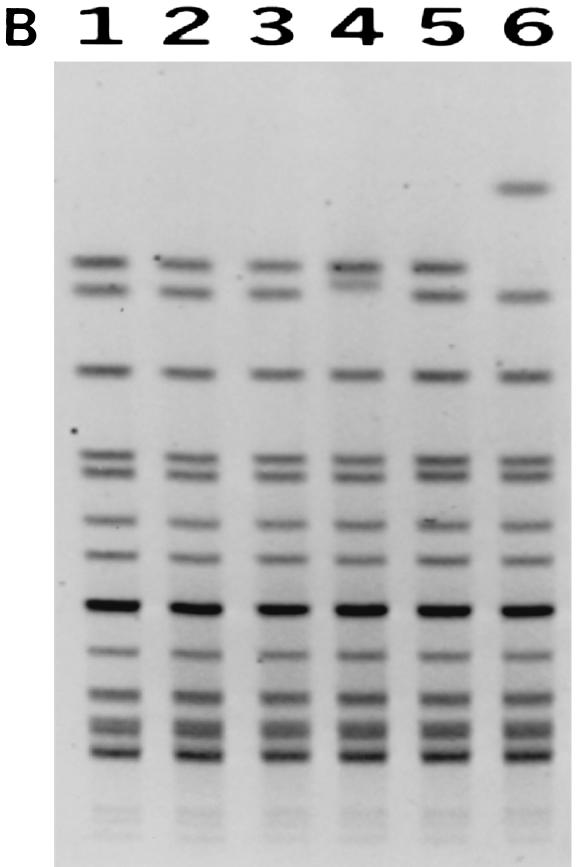
Pulsed-field gel electrophoresis (PFGE) analysis of restriction enzyme-digested genomic DNA from all six strains confirmed that these strains were related. When the bacterial DNA was restricted with the NheI enzyme, all six strains were genetically almost indistinguishable (Fig. 1a). With the restriction enzyme SpeI, the digested DNA from these six strains demonstrated three closely related patterns (Fig. 1b) that differed from one another by only one band, which could be accounted for by a single genetic event such as an insertion or deletion of genetic material (8). As a control, six other unrelated serogroup C meningococcal strains isolated from IMD cases in the Toronto area in 2001 were also analyzed together with the outbreak strains. The unrelated serogroup C meningococci gave PFGE patterns demonstrating differences of more than seven bands from the patterns obtained with the outbreak strains (Fig. 1a).
FIG. 1.
(A) PFGE profiles of NheI-digested DNA from the outbreak and control strains. Lanes 1, 8, and 15, molecular mass markers (lambda phage DNA ladder from New England BioLabs, Beverly, Mass.); lanes 2 to 7, outbreak strains (C:NT:P1.2); lanes 9 to 14, serogroup C meningococcal strains (C:2a:P1.5,16; C:NT:P1.6; C:2a:P1.1,7; C:2a:P1.2; C:2a:P1.2; and C:2a:P1.5,16) unrelated to the outbreak and isolated from the Toronto area in 2001. (B) PFGE profiles of SpeI-digested DNA from the outbreak strains. Lanes 1 to 6, outbreak strains.
DNA sequence analysis of the porB genes of N. meningitidis strains has revealed four variable regions within the PorB proteins which can be correlated to their serotype or serological specificities (6). To further understand the serotype nature of the outbreak strains, partial sequences of their porB genes encoding the variable regions VR1, VR2, VR3, and VR4 were determined from PCR-amplified products of porB by using primers described by Sacchi et al. (6). Prior to sequencing, PCR products were purified by using the QIAquick PCR purification kit (Qiagen Inc., Mississauga, Ontario, Canada). Sequencing was performed on an ABI 377 DNA sequencer by using the Prism Dye Termination Cycle Sequencing kit (Applied Biosystems, Inc., Foster City, Calif.), and the data were compiled by using software from DNASTAR, Inc., Madison, Wis. All six outbreak strains gave identical porB DNA sequences. Their PorB VR types in the surface-exposed OMP loops I, V, VI, and VII (which correspond to VR1, VR2, VR3, and VR4, respectively) were either identical or highly similar to the VR types of typical serogroup C serotype 2a ET-37 meningococci. In the typical serotype 2a strain, the VR types according to the variable regions 1 to 4 have been determined to be C, Eb, 2a, and C (6), while in the outbreak strains the corresponding VR types were found to be C, Eb, and 2a(a) (the PorB VR type designation is according to the scheme developed by Sacchi et al. [6]), and C. The only difference between the porB genes of the outbreak strains and that of the typical serotype 2a strain was a single nonsynonymous nucleotide substitution at position 700 in loop VI (i.e., VR3), which codes for the serotyping antigenic specificity. This single base change from G to A led to a change in the encoded amino acid from a negatively charged glutamic acid, which has an acidic side chain, to the positively charged lysine, which has a basic side chain. Currently we do not have any data to show whether this single amino acid substitution in the VR3 serotype antigen coding region may contribute to any change in the serological specificity of the outbreak strain. Further experiments, such as raising a specific antiserum to this mutated form, may allow us to understand the serological specificities of the mutated serotype 2a strains. Nevertheless, the fact that the amino acid substitution occurred in an antigen-coding region that is surface exposed coupled with the fact that a monoclonal antibody to the native form failed to react with it suggests that the loss of the outbreak strains' ability to react with the serotype 2a monoclonal antibody may be related to this mutation.
In 2001, there was an increase in meningococcal disease activity in several provinces in Canada, caused mostly by N. meningitidis serogroup C. As a result of this increased disease activity, there were altogether 173 strains of serogroup C meningococci isolated from sterile body sites of individual patients in that year (as submitted to the National Microbiology Laboratory). In Canada, most serogroup C meningococci involved in both endemic disease and outbreaks belong to the electrophoretic type of ET-15, a new variant within the ET-37 clonal complex (2, 5). ET-15 meningococci have been characterized by multilocus enzyme electrophoresis to have a fumarase housekeeping enzyme showing a different allele (2) from the allele 1 found in strains of the ET-37 complex (2, 5). At the genetic level, the difference between the fumarase locus alleles 1 and 2 seems to be in only one base pair change from G to A at position 640 of their fumC gene (12). Of the 173 strains of serogroup C meningococci isolated from IMD cases in 2001, 166 belonged to ET-15 and 4 were ET-37 but not ET-15. Only 3 (1.7%) did not belong to the family of ET-37/ET-15 clonal complex.
Like most ET-37 serogroup C N. meningitidis in other parts of the world, the most common serotype of serogroup C meningococci in Canada is serotype 2a, with 149 (86.1%) of the 173 strains from IMD cases in 2001 belonging to this type. There was only one strain which was serotype 2b, and 23 strains were nonserotypeable by monoclonal antibodies that recognize the serotype antigens 1, 2a, 2b, 4, 14, and 15. Of the 23 nonserotypeable group C N. meningitidis isolates, 8 had the serosubtype antigen of P1.2, 6 were P1.2,5, and 7 strains showed no reaction with the panel of serosubtyping antibodies.
Based on analysis of 2001 data, C:NT:P1.2 strains were not common in Canada. Of the 173 strains analyzed, only 8 (4.6%) belonged to this phenotype and 7 of them were isolated in the Province of Ontario, where this strain was responsible for causing this cluster of meningococcal disease in an MSM community in Toronto. Among this cluster of six IMD cases, two were fatal, leading to a case fatality rate of 33%. Whether this mortality rate was affected by human immunodeficiency virus infection could not be determined. However, bathhouses for males are now recognized to be settings of potential IMD transmission.
In the outbreak that occurred in the Ottawa-Carleton region in Ontario a decade ago (4), which affected mainly high school students, the case fatality rate recorded was four deaths among the 10 confirmed and clinically diagnosed cases. Of the four fatal cases, three were caused by N. meningitidis serogroup C, serotype 2a ET-15 and one was due to a serogroup C, nontypeable ET-15 meningococcus. In a more recent serogroup C meningococcal outbreak in Edmonton, Alberta, Canada, which involved 61 cases due to serogroup C, serotype 2a ET-15 meningococci, only two deaths were recorded (10).
Partial DNA sequencing of the porB genes in this group of N. meningitidis strains revealed that they all belonged to the serotype 2a family, with only one base pair change in the VR3 region of their genes. This resulted in a single amino acid change in the stretch of the polypeptide that corresponds to the surface-exposed loop VI of their PorB OMPs. Antibodies to PorB and PorA proteins are bactericidal (7), and phylogenetic evidence suggests that PorB protein is under strong immune selection (11). Although there are no data to support that human antibodies developed to the PorB protein of serogroup C meningococci during natural infection are directed against the serotype 2a epitope on the VR3 region, the hypothesis that the porB gene mutation seen in the outbreak strains is a result of immune selection by bactericidal antibodies is not unreasonable. Since it was first recognized in the mid-1980s to cause a significant percentage of IMD cases in Canada, N. meningitidis serogroup C (mostly represented by serotype 2a organisms) continued to increase in the 1990s in its contribution to the percentage of IMD cases that it caused. By the years 2000 and 2001, there were more serogroup C IMD cases than those caused by serogroup B organisms (9). Considering the prevalence of this organism in the population, it is possible that serum antibodies developed in the population to the surface components of this organism, including the serotype 2a antigen, are exerting an immune selection on the meningococcci. Therefore, we are speculating that N. meningitidis serotype 2a strains alter their PorB OMPs in order to evade the host defense and escape the natural immunity in order to have an increased ability to cause disease.
In summary, this report documents a cluster of IMD cases in an MSM community in Toronto, Ontario, and identifies bathhouses for males as potential venues for IMD transmission. The outbreak was caused by a genetic variant of the C:2a:P1.2 meningococci presenting with the phenotype of C:NT:P1.2. The nature of the serotyping antigens on their PorB OMPs was characterized by DNA sequencing and shown to contain a single-base-pair nonsynonymous mutation that led to a change in the amino acid composition in the PorB surface-exposed loop VI. The significance of this finding was discussed.
Nucleotide sequence accession numbers.
The partial sequences of porB genes showing the novel mutations described in this study have been deposited in GenBank (NCBI) with the assigned accession numbers AY234206, AY234207, AY234208, AY234209, AY234210, and AY234211.
Acknowledgments
We thank the staff at the Toronto Public Health and the Ontario Ministry of Health and Long Term Care (Public Health Branch and Central Public Health Laboratory) for their contribution in the investigation and management of the outbreak and the DNA core facility at Health Canada's NML for the DNA sequencing work. Vaccines for the control of this outbreak were provided by the Ontario Ministry of Health and Long-Term Care and administered by staff of Toronto Public Health.
The molecular characterization of strains was performed with financial support from Health Canada's Genomics R&D Fund.
REFERENCES
- 1.Abdillahi, H., and J. T. Poolman. 1987. Whole-cell ELISA for typing Neisseria meningitidis with monoclonal antibodies. FEMS Microbiol. Lett. 48:367-371. [PubMed] [Google Scholar]
- 2.Ashton, F. E., J. A. Ryan, A. Borczyk, D. A. Caugant, L. Mancino, and D. Huang. 1991. Emergence of a virulent clone of Neisseria meningitidis serotype 2a that is associated with meningococcal group C disease in Canada. J. Clin. Microbiol. 29:2489-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, S., G. Riley, and F. Jamieson. 2001. Neisseria meningitidis with decreased susceptibility to penicillin in Ontario, Canada 1997-2000. Can. Commun. Dis. Rep. 27:73-75. [PubMed] [Google Scholar]
- 4.Gemmill, I. 1992. An outbreak of meningococcal disease in Ottawa-Carleton December 1991-February 1992. Can. J. Public Health 83:134-137. [PubMed] [Google Scholar]
- 5.Jelfs, J., R. Munro, F. E. Ashton, and D. A. Caugant. 2000. Genetic characterization of a new variant within the ET-37 complex of Neisseria meningitidis associated with outbreaks in various parts of the world. Epidemiol. Infect. 125:285-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sacchi, C. T., A. P. S Lemos,. A. M. Whitney, C. A. Solari, M. E. Brandt, C. E. A. Melles, C. E. Frasch, and L. W. Mayer. 1998. Correlation between serological and sequence analyses of the PorB outer membrane protein in the Neisseria meningitidis serotyping scheme. Clin. Diagn. Lab. Immunol. 5:348-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saukkonen, K., H. Abdillahi, J. T. Poolman, and M. Leinonem. 1987. Protective efficacy of monoclonal antibodies to class 1 and class 3 outer membrane proteins of Neisseria meningitidis B:15:P1.16 in infant rat infection model: new prospects for vaccine development. Microb. Pathog. 3:261-267. [DOI] [PubMed] [Google Scholar]
- 8.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsang, R. S. W., S. G. Squires, W. D. Zollinger, and F. E. Ashton. 2002. Distribution of serogroups of Neisseria meningitidis and antigenic characterization of serogroup Y meningococci in Canada, January 1, 1999 to June 30, 2001. Can. J. Infect. Dis. 13:391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyrrell, G. J., L. Chui, M. Johnson, N. Chang, R. P. Rennie, J. A. Talbot, and the Edmonton Meningococcal Study Group. 2002. Outbreak of Neisseria meningitidis, Edmonton, Alberta, Canada. Emerg. Infect. Dis. 8:519-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urwin, R., E. C. Holmes, A. J. Fox, J. P. Derrick, and M. C. J. Maiden. 2002. Phylogenetic evidence for frequent positive selection and recombination in the meningococcal surface antigen PorB. Mol. Biol. Evol. 19:1686-1694. [DOI] [PubMed] [Google Scholar]
- 12.Vogel, U., H. Claus, M. Frosch, and D. A. Caugant. 2000. Molecular basis for distinction of the ET-15 clone within the ET-37 complex of Neisseria meningitidis. J. Clin. Microbiol. 38:941-942. [DOI] [PMC free article] [PubMed] [Google Scholar]