Abstract
Carbonic anhydrase (CA) IV is a glycosylphosphatidylinositol-anchored isozyme expressed on plasma membranes of capillary endothelial cells and certain epithelial cells of the nephron, the colon, and the genitourinary tract. CA IVs purified from bovine and rabbit lungs are high-activity enzymes, like human CA IV, while CA IV from mouse and rat lungs had only 10–20% as much catalytic activity. To explain the molecular basis for these differences in activity, we isolated and characterized the full-length cDNAs for bovine and rabbit CA IVs and compared their sequences to those we previously reported for human, murine, and rat CA IVs. These comparisons led us to postulate that a Gly-63 → Gln substitution adjacent to His-64 in the rodent enzymes accounts for their lower activity. To test this hypothesis, we made the Gly-63 → Gln mutants of bovine and rabbit CA IVs and the Gln-63 → Gly mutant of murine CA IV by site-directed mutagenesis, and compared the activities of mutant and wild-type CA IVs expressed in COS-7 cells. In addition, we produced recombinant cDNAs expressing secretory forms of the Gly-63 and Gln-63 forms of each of the three enzymes and compared the activities of the enzymes purified from transfected COS-7 cell secretions with the activities of CA IVs purified from lungs. These studies demonstrated that Gly-63 is important for the high activity of bovine and rabbit CA IVs, and they showed that the low activity of murine CA IV could be improved by the Gln-63 → Gly substitution. We suggest that the lower activity of the rodent CA IVs can be largely explained by the Gln-63 substitution which reduces the efficiency of proton transfer by the adjacent His-64.
The carbonic anhydrases (CAs), which catalyze the
reversible hydration of CO2 in the reaction
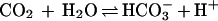 ,
are a family of zinc metalloenzymes found in nearly all
organisms (1, 2). To date, at least seven isozymes have been described
in mammals. These include cytosolic (CA I, II, III, VII) (3, 4),
membrane-associated (CA IV) (5, 6, 7, 8, 9), mitochondrial (CA V) (10), and
secretory (CA VI) forms (11, 12). In addition, three additional
isoforms designated CAs VIII–X have been discovered and characterized
recently (13).
,
are a family of zinc metalloenzymes found in nearly all
organisms (1, 2). To date, at least seven isozymes have been described
in mammals. These include cytosolic (CA I, II, III, VII) (3, 4),
membrane-associated (CA IV) (5, 6, 7, 8, 9), mitochondrial (CA V) (10), and
secretory (CA VI) forms (11, 12). In addition, three additional
isoforms designated CAs VIII–X have been discovered and characterized
recently (13).
CA IV, the membrane-associated CA, was originally purified from bovine lung and found to be a glycoprotein of 52 kDa and a high-activity enzyme like CA II (5). However, subsequent studies on CA IVs from nine mammalian species revealed that the CA IVs have wide variation in specific activity from 300 to 3000 enzyme units (EU)/mg of protein (5, 6, 7, 8, 9). Murine and rat CA IVs were among the isozymes with lowest activity, while human, bovine, and rabbit CA IVs were at the high end of the activity spectrum. The molecular mechanisms of this variation have not been determined.
The kinetics of enzyme catalysis have been studied extensively.
Considerable evidence suggests that the catalytic mechanism can be
divided into two steps (14, 15), conversion of CO2 to
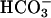 , leaving water as a ligand on
the zinc (Eq. 1), and transfer of proton to solvent buffer
molecules (B) through a proton shuttle group, histidine-64 (His-64)
(Eq. 2).
, leaving water as a ligand on
the zinc (Eq. 1), and transfer of proton to solvent buffer
molecules (B) through a proton shuttle group, histidine-64 (His-64)
(Eq. 2).
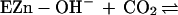 |
1 |
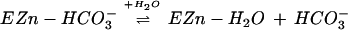 |
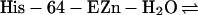 |
2 |
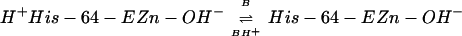 |
Previous studies of CA II have revealed that His-64 functions as an efficient proton shuttle group transferring proton between the zinc-bound water and surrounding buffer molecules, as indicated in Eq. 2 (16, 17). In fact, when His-64 is replaced by nonionizable amino acids, alanine (16) or glutamine (17), the maximal CO2 hydration activity is drastically reduced except when the measurements are performed in imidazole buffer or its derivatives.
In this study, we have cloned and characterized full-length CA IV cDNAs from bovine and rabbit libraries and compared the amino acid sequences with human (18), murine (19), and rat (20) CA IVs to detect differences that might explain differences in activity between rodent and other mammalian CA IVs. The survey of amino acid sequences around His-64 revealed two potentially important differences. First, bovine and rabbit CA IVs have a glycine residue at position 63, like the high-activity human CA IV, while the published sequence for murine and rat CA IVs reveals a glutamine in this position. Second, the bovine, rabbit, and human CA IVs have a methionine residue at position 67, while the murine and rat CA IVs both have glutamic acid at this position. A survey of other published sequences suggested that Gly-63 → Gln has the more significant difference. Met-67 is not present in any other human CAs, and there is considerable variation in the amino acid found at position 67 in other CAs. On the other hand, Gly-63 is highly conserved. Not only do all reported mammalian CAs have a glycine in this position, but every CA in this family, extending back to Chlamydomonas and Neisseria, has a glycine in this position (13). Perturbation of the structure around His-64 by the Gly-63 → Gln substitution might explain the differences in catalytic activity between bovine, rabbit, and human CA IVs and the murine and rat CA IVs. To test this hypothesis, we carried out expression experiments in COS-7 cells using wild-type CA IVs and CA IVs mutant at residue 63 (Gly-63 → Gln in bovine and rabbit CA IVs, and Gln-63 → Gly in murine CA IV). These studies indicated that Gly-63 is more favorable for catalytic activity than Gln-63 in CA IV from all three species that normally have glycine in this position, and they suggest that the reduced activity of the rodent CA IVs can be largely explained by Gly → Gln substitution at position 63.
MATERIALS AND METHODS
Materials.
The bovine kidney cDNA library in Uni-ZAP XR vector and the rabbit kidney cDNA library in Lambda ZAP II vector were purchased from Stratagene (catalog nos. 937713 and 936904, respectively). COS-7 cells were donated by Maurice Green of the Institute for Molecular Virology (St. Louis). Chemicals and reagents used in this study included Gene Amp RNA PCR kit from Perkin–Elmer, nitrocellulose filters from MSI, 32P-labeled nucleotides from ICN, 35S-labeled nucleotides from Amersham, random-primed DNA-labeling kit from Boehringer Mannheim, and Chameleon double-stranded site-directed mutagenesis kit from Stratagene.
Isolation and Characterization of cDNAs for Bovine and Rabbit CA IVs.
The bovine kidney cDNA library was screened by using a 0.4-kb PCR amplification product derived from the bovine kidney cDNA library. The PCR primers 5′-CAITTTGCCATGGAGATGCA-3′ and 5′-GGGCCTIACATTGTCCTTCA-3′, as forward and reverse primers, respectively, were designed from the published sequences of the human (18), murine (19), and rat (20) CA IV cDNAs. The rabbit kidney cDNA library was screened by using a 0.4-kb reverse transcriptase–PCR amplification product derived from mRNA from rabbit colon, because PCR amplification from the rabbit cDNA library directly was not successful. The primer used in reverse transcriptase reaction was oligo(dT)16 nucleotides, and primers used in PCR for rabbit screening were the same as those for bovine screening. The amplified DNA fragments from both bovine and rabbit templates were sequenced directly to verify the sequences similar to other mammalian CA IVs.
Plaque hybridization was carried out essentially as described by Maniatis and colleagues (21). The prehybridizations were carried out for 3 hr at 42°C in 2× 1,4-piperazinediethanesulfonic acid buffer, pH 7.0/50% formamide/0.5% SDS containing denatured herring sperm DNA at 100 μg/ml. The hybridizations were performed for 18 hr at 42°C after adding the 32P-labeled probe at ≅1 × 106 cpm/ml into the solution used for prehybridization. The filters were washed twice in 2× SSC/0.1% SDS at room temperature for 10 min, followed by a wash in 0.1× SSC/0.1% SDS at 50°C for 10 min. The filters were dried and autoradiographed at −70°C overnight.
The cDNA insert in pBluescript SK− phagemid was recovered from the Uni-ZAP XR vector or Lambda ZAP II vector by using the in vivo excision protocol supplied by the manufacturer. The double-stranded cDNA insert was sequenced at least once on both strands by the dideoxynucleotide chain-termination method using 35S-labeled dATP thio analog. The nucleotide and deduced amino acid sequences were analyzed using the dnasis-Mac Version 2.0 sequence analysis system (Hitachi Software Engineering).
Construction of Mutant cDNAs for Bovine, Rabbit, and Murine CA IVs.
The numbering system of human CA I (1) is used throughout the text. In this system, residues 63 and 267 in human CA I correspond to residues 69 and 268 in the numbering system of bovine, rabbit, and murine CA IVs, respectively (see Fig. 2). The missense mutants at residue 63 (G63Q in bovine and rabbit CA IVs and Q63G in murine CA IV), the truncated mutants at residue 267 which produced the secretory form of enzymes as described (22) (G267X in bovine CA IV, Q267X in rabbit CA IV, and H267X in murine CA IV), and the combination of both mutants at residues 63 and 267 was made by site-directed mutagenesis using mismatched primers. The procedures for generating mutants were based on the protocol by the manufacturer (23). The coding regions of constructed plasmids were sequenced to verify that no additional substitutions were introduced.
Figure 2.

Comparison of deduced amino acid sequences for CA IV from cow, rabbit, human, mouse, and rat. ▿, Sites at which the leader peptide is cleaved. This proteolytic cleavage site is tentative in murine and rat CA IVs. ▾, Sites at which the hydrophobic tail is cleaved, based on agreement with common features for glycosylphosphatidylinositol (GPI)-anchored proteins. Sixteen of the 17 amino acid residues, thought to be near the “active sites” and common to nearly all CAs, are boxed. ↓, Conserved histidine implicated in proton transfer. ∗, Three potential zinc-binding histidines. ○, Four cysteines that are involved in intramolecular disulfide bond formation. The putative N-glycosylation sites defined by the tripeptide consensus sequence of Asn-Xaa-Thr/Ser are underlined. The amino acid residues that were engineered by site-directed mutagenesis are indicated by boldface type.
Expression of CA IV cDNA in COS-7 Cells.
Both wild-type and mutant cDNAs of bovine, rabbit, and murine CA IVs were subcloned in the pCAGGS vector originally described by Miyazaki et al. (24). COS-7 cells in 60-mm dishes were transfected in 5 ml of DMEM with 10 μg of DNA per dish by the DEAE-dextran procedure for 8 hr (25) followed by a 3-hr treatment with 100 μM chloroquine (26). For wild-type CA IVs and CA IVs mutant at residue 63, the transfected cells were harvested 72 hr after transfection by scraping and homogenized in 50 mM Tris·H2SO4, pH 7.5, containing 1 mM benzamidine (buffer A). For the secretory forms of CA IVs, that is, those truncated in residue 267, with or without a substitution at residue 63, both media and cells were recovered. The expression experiments were done in duplicate, and the duplicate samples were combined before enzyme purification and/or assay.
Purification of CA IV.
To purify the secretory form of enzymes, the media from COS-7 cells transfected with cDNAs from bovine, rabbit, and murine CA IVs were applied to inhibitor-affinity chromatography media according to the procedure of Zhu and Sly (7) as described (22).
Isolation of CA IV from Lungs.
Homogeneous CA IVs were obtained from 200 g of lungs. Preparation of microsomes, extraction, and purification using inhibitor-affinity chromatography were carried out as described (7, 8).
CA Activity Assay.
CA activity was determined by the procedure of Maren (27) as described (28). SDS-resistant CA activity was measured on samples preincubated with 0.2% SDS at room temperature for 30 min prior to assay. The protein concentration was measured according to the Lowry procedure using bovine serum albumin as a standard (29). CA activity is expressed in EU/mg of cell protein for unpurified enzymes or in EU/mg of CA IV for affinity-pure enzymes.
N-Terminal Amino Acid Sequencing.
To determine the N-terminal amino acid sequences, Edman degradation of the affinity-pure bovine and rabbit CA IVs was carried out in an Applied Biosystems model A477 automatic protein sequencer as described (7).
RESULTS
Isolation of cDNAs for Bovine and Rabbit CA IVs.
Two 400-bp PCR or reverse transcriptase-PCR products, as described in Materials and Methods, were used as probes to isolate bovine or rabbit CA IV cDNA. Thirty-two positive clones were identified in 5.0 × 105 plaques in the bovine cDNA library, and two positive clones were identified in the 1.5 × 106 plaques in the rabbit cDNA library. One clone, selected arbitrarily from each library, was subcloned, sequenced, and found to represent a full-length cDNA.
Nucleotide Sequence, Deduced Amino Acid Sequence, and Direct N-Terminal Amino Acid Sequencing.
Fig. 1 a and b present the nucleotide sequences and the deduced amino acid sequences of bovine and rabbit CA IV cDNAs, respectively. Translation initiation sites were identified by homology with human, murine, and rat sequences and best agreement with initiation codon consensus (30). N-terminal sequences determined on CA IVs purified from transfected COS-7 cells agreed with the N-terminal amino acid sequences deduced from the cDNAs and indicated that the N termini of the mature bovine and rabbit CA IVs are highly homologous to the N terminus of the mature human CA IV (18). The bovine CA IV cDNA contains a 37-bp 5′ untranslated region, a 936-bp open reading frame, and a 119-bp 3′ untranslated region containing a polyadenylylation cleavage signal (ATTAAA) starting 24 bp upstream from the poly(A) tail. The rabbit CA IV cDNA contains a 41-bp 5′ untranslated region, a 924-bp open reading frame, and a 101-bp 3′ untranslated region containing a poly(A) signal (ATTAAA) starting 24 bp upstream from the poly(A) tail.
Figure 1.

Nucleotide sequences and deduced amino acid sequences of the bovine (a) and rabbit (b) CA IV cDNAs. The deduced amino acid sequences are numbered at the end of each row. The putative leader sequence is amino acids −18 to −1 in both bovine and rabbit CA IVs. An open triangle preceding amino acid +1 indicates the N-terminal amino acid found in the mature protein. The putative N-glycosylation sites defined by tripeptide consensus sequence of Asn-Xaa-Thr/Ser (32) are underlined. The putative polyadenylylation signals in the 3′ untranslated region are indicated by double underlines.
Comparison of Bovine and Rabbit CA IV Amino Acid Sequences with Those of Human, Murine, and Rat CA IVs.
The deduced amino acid sequences from bovine, rabbit, human, murine, and rat CA IVs are compared in Fig. 2.
Several points can be noted from the sequence comparison. (i) All five enzymes have 16 of the 17 highly conserved “active site” residues found in most other CAs (boxed in Fig. 2) (1, 2). The one exception is a highly conserved proline residue (Pro-202 in human CA I), which is replaced by other amino acid residues in CA IV (position 211 in Fig. 2). (ii) All five enzymes have a highly conserved histidine residue, which has been implicated as a proton shuttle group (His-64 in human CA I, position 70 in Fig. 2). However, the amino acid immediately upstream of this conserved histidine is a glutamine in murine and rat CA IVs, instead of a highly conserved glycine in bovine, rabbit, and human CA IVs (1, 2). This amino acid is at position 69 in Fig. 2, but will be referred to as Gln-63 or Gly-63 to retain the CA I numbering system (1). (iii) All five enzymes contain three potential zinc-binding histidine residues (2). (iv) All five enzymes contain four cysteine residues, which have been shown to be involved in intramolecular disulfide bond formation in human CA IV (7, 18). (v) All five enzymes have serine residues at position 267 (Fig. 2) (this amino acid will be referred to as Ser-266 in the CA I numbering system). This is the residue from which the hydrophobic C terminus is cleaved and to which the GPI anchor is attached post-translationally (18, 31). (vi) The numbers of N-glycosylation sites defined by the consensus tripeptide sequence (Asn-Xaa-Thr/Ser) (32) were four in bovine CA IV, two in rabbit, murine, and rat CA IVs, and none in human CA IV.
As noted above, the comparison of the amino acid sequences of the five CA IVs presented in Fig. 2 suggested a potentially important difference between the high-activity CA IVs (bovine, rabbit, and human) and the low-activity rodent enzymes. The residue immediately upstream of His-64, the residue involved in proton transfer, is Gly-63 in bovine, rabbit, and human CA IVs, and Gln-63 in the murine and rat CA IVs. To examine the possibility that the substitution of Gln for Gly in the rodent enzymes explains the striking difference in the enzyme activity, we engineered bovine and rabbit cDNAs with Gly-63 → Gln and murine cDNA with Gln-63 → Gly, and we compared their activities when expressed in transfected COS-7 cells with those of the respective wild-type CA IVs.
CA IV Activity in COS-7 Cells Transfected with Wild-Type and Mutant CA IV cDNAs.
From the results summarized in Table 1, a number of points can be made. First, the cDNAs for wild-type bovine and rabbit CA IVs both expressed higher levels of activity (17.7 and 10.7 EU/mg of cell protein, respectively) than did the wild-type murine cDNA (3.3 EU/mg of cell protein). Second, swapping Gln for Gly at position 63 in bovine and rabbit CA IVs reduced the activities expressed in COS-7 cells to 37% and 42%, respectively, of the activities of the prospective wild-type Gly-63 CA IV transfections. On the other hand, swapping Gly for Gln in the murine CA IV nearly tripled the activity expressed from the wild-type murine CA IV cDNA. We also measured their resistance to SDS. As had been reported for human CA IV (7), bovine and rabbit CA IVs were SDS-resistant. However, both the Gln-63 and Gly-63 forms of the murine CA IV were less resistant to SDS.
Table 1.
CA activity in homogenate of COS-7 cells transfected with vector pCAGGS only or pCAGGS with CA IV cDNA insert
| cDNA insert | EU/mg of cell protein
|
|
|---|---|---|
| No treatment | SDS (0.2%) | |
| None (vector pCAGGS only) | 0.7 | 0.1 |
| Bovine CA IV, wild type | 17.7 | 20.9 |
| Bovine CA IV, Gly-63 → Gln | 6.6 | 5.4 |
| Rabbit CA IV, wild type | 10.7 | 14.4 |
| Rabbit CA IV, Gly-63 → Gln | 4.5 | 3.8 |
| Murine CA IV, wild type | 3.3 | 1.4 |
| Murine CA IV, Gln-63 → Gly | 9.1 | 3.0 |
CA activity in the total cell homogenates reflects both the amount synthesized and the efficiency of processing of the precursor into an active CA IV molecule. To compare the activities of the purified mature CA IVs, we expressed secretory versions of both wild-type CA IVs and CA IVs mutant at residue 63 and purified the expressed enzymes from the media. The secretory forms were produced by introducing a stop codon in place of amino acid residue 267 in each cDNA, which results in removal of the C-terminal hydrophobic domain and the signal for GPI anchoring. The truncated enzymes are secreted as soluble forms of the respective enzyme. We previously showed that the secretory form of human CA IV produced in this way is not only fully active but is secreted in amounts greater than that which accumulates in cells expressing the wild-type, GPI-anchored CA IV (22). The enzyme purified from cells expressing these constructs allowed us to compare the specific activity of each of the respective recombinant CA IVs with that of the respective native enzymes purified from the lung tissues of the three different mammals (Table 2). These data show that purified secretory forms of the wild-type recombinant CA IVs are at least as active as those of the GPI-anchored enzymes purified from lung tissue. Furthermore, the results summarized in Table 2 show that the specific activities from affinity-pure CA IVs from bovine, rabbit, and murine cDNAs show the same effects of swapping of Gly-63 → Gln or Gln-63 → Gly were seen with the GPI-anchored enzymes expressed in COS-7 cells (Table 1). Thus, the differences in activity are properties of the enzymes themselves–not properties of the COS-7 cell expression system.
Table 2.
CA specific activity of affinity-pure, secretory form of wild type and mutant at residue 63 for bovine, rabbit, and murine CA IVs transfected with pCAGGS and cDNA insert in COS-7 cells, and of affinity-pure CA IVs isolated from the lung tissues
| Enzyme | EU/mg of pure CA IV
|
|
|---|---|---|
| From cDNAs in COS-7 cells | From lung tissues | |
| Bovine CA IV, wild type | 3024 | 2300 |
| Bovine CA IV, Gly-63 → Gln | 634 | |
| Rabbit CA IV, wild type | 2495 | 2200 |
| Rabbit CA IV, Gly-63 → Gln | 520 | |
| Murine CA IV, wild type | 436 | 350 |
| Murine CA IV, Gln-63 → Gly | 2200 | |
DISCUSSION
We have isolated and sequenced full-length cDNAs for CA IV from bovine and rabbit kidney cDNA libraries. The deduced amino acid sequences have many characteristics in common with those of other mammalian CA IVs. The bovine and rabbit CA IV proteins include N-terminal signal sequences for secretory proteins (33), central segments corresponding to the mature proteins, and C-terminal hydrophobic domains of the precursors of the membrane-associated proteins, which are cleaved to allow transfer to the GPI anchor (31). The bovine and rabbit CA IVs expressed from the cDNAs in COS-7 cells show similarity to CA IVs from bovine and rabbit lungs in their association with the plasma membrane via a GPI anchor, their relative insensitivity to inactivation by SDS (7, 8), and their relatively higher activity (compared with the rodent CA IVs).
To explain the higher activity in bovine and rabbit CA IVs relative to that of rodent (murine and rat) CA IVs, we compared the deduced amino acid sequences available for five mammalian CA IVs. All five enzymes have highly conserved residues found in most other CAs, including His-64, which has been implicated in the proton shuttle (14, 15, 16); His-94, His-96, and His-119, which bind the zinc ligand (14); and Glu-106 and Thr-199, which are involved in the hydrogen bonding network (34). One striking difference was found at residue 63 immediately upstream of the His-64. The glycine found at this position in rabbit and bovine CA IV is also found in human CA IV. In fact, it is conserved in all known CAs in the α CA gene family (13). This difference from the rodent sequence led us to examine the possibility that this substitution of Gln for Gly in the rodent CA IVs may explain their lower activity. CA activity assay on enzymes of wild-type CA IVs and CA IVs mutant at residue 63 have shown that the Gly-63 → Gln replacement in bovine and rabbit CA IVs reduces activity to approximately one-third that of the wild-type CA IV. By contrast, the replacement Gln-63 → Gly in murine CA IV increases its activity 2- to 3-fold over that of the wild-type murine CA IV. These results favor the interpretation that the substituted Gln residue at Gly-63 largely accounts for the lower activity of rodent CA IVs.
The purpose of constructing the secretory forms of CA IVs was to obtain higher amounts of fully active enzymes that could be purified and characterized. The purified secretory forms of enzymes of wild-type and mutant CA IVs showed the same effects of swapping of Gly-63 → Gln or Gln-63 → Gly on the CA IV activity. These results from affinity-pure enzymes enabled us to exclude the possibility that a lower amount of CA IV production in COS-7 cells expressing Gln-63 enzymes might explain their lower activity, or that differential enzyme stability might contribute to the lower activity of Gln-63-containing enzymes. We conclude that Gln-63 actually reduces the catalytic activity of the rodent enzymes to levels only 20–25% those seen with the higher-activity CA IVs that have Gly-63. We propose that this reduction in activity occurs because proton transfer by His-64 is impaired by the bulkier residue at position 63 in the rodent enzymes.
The Gly-63 → Gln substitution is unique to rodent CA IVs. The Gly residue in other CA structures, including human CA IV, the three-dimensional structure of which was reported recently (35), adopts backbone φ/ψ angles that would be disallowed for any other amino acid except glycine. Presumably the backbone of Gln-63 must undergo some type of change to accommodate proper φ/ψ angles, and this change must propagate to adjacent residues and perturb the conformation of the adjacent His-64. We speculate that it is the conformational shift by His-64 to accommodate Gln-63 which reduces its efficiency in catalytic proton transfer. This speculation assumes that His-64 is implicated in catalysis in CA IV as has been shown to be the case for CA II.
The emergence of a rodent CA IV with reduced catalytic function is of interest from an evolutionary standpoint. The members of the CA gene family are all presumed to have arisen from an ancient common precursor by gene duplication. Sequence evidence suggests that rodent CA IVs are the only descendants of this ancient common precursor that have Gln in place of Gly-63. Thus, the common precursor must have had Gly at this position. It has been suggested that the duplication giving rise to CA IV arose even before the emergence of vertebrates (13). At some subsequent point after the emergence of rodents, but before the divergence of mice and rats, Gln replaced Gly at position 63 in rodent CA IV and became fixed. Apparently, the 80% decrease in catalytic activity must not have been physiologically disadvantageous enough to be lost through selection. In fact, conservation of the Gln at this position over the millions of years since the divergence of mice and rats raises the interesting possibility that rodents differ from other mammals in their bicarbonate metabolism in such a way that the less efficient rodent enzyme confers some selective advantage.
Acknowledgments
We thank Dr. Jung Huang for amino acid sequencing, Dr. David Christianson for suggesting a structural interpretation of the findings, and Elizabeth Torno for editorial assistance. We also acknowledge receipt of a manuscript prior to its publication from C. A. Winkler, A. M. Kittelberger, and G. J. Schwartz, which independently reported the deduced amino acid sequence of rabbit CA IV (GenBank accession no. L48928L48928). The work reported here was supported by National Institutes of Health Grants DK40163 and GM34182.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
References
- 1.Tashian R E. Adv Genet. 1992;30:321–356. doi: 10.1016/s0065-2660(08)60323-5. [DOI] [PubMed] [Google Scholar]
- 2.Sly W S, Hu P Y. Annu Rev Biochem. 1995;64:375–401. doi: 10.1146/annurev.bi.64.070195.002111. [DOI] [PubMed] [Google Scholar]
- 3.Deutsch H F. Int J Biochem. 1987;19:101–113. doi: 10.1016/0020-711x(87)90320-x. [DOI] [PubMed] [Google Scholar]
- 4.Tashian R E. BioEssays. 1989;10:186–192. doi: 10.1002/bies.950100603. [DOI] [PubMed] [Google Scholar]
- 5.Whitney P L, Briggle T V. J Biol Chem. 1982;257:12056–12059. [PubMed] [Google Scholar]
- 6.Wistrand P J, Knuuttila K-G. Kidney Int. 1989;35:851–859. doi: 10.1038/ki.1989.63. [DOI] [PubMed] [Google Scholar]
- 7.Zhu X L, Sly W S. J Biol Chem. 1990;265:8795–8801. [PubMed] [Google Scholar]
- 8.Waheed A, Zhu X L, Sly W S. J Biol Chem. 1992;267:3308–3311. [PubMed] [Google Scholar]
- 9.Carter N D, Fryer A, Grant A G, Hume R, Strange R G, Wistrand P J. Biochim Biophys Acta. 1990;1026:113–116. doi: 10.1016/0005-2736(90)90340-t. [DOI] [PubMed] [Google Scholar]
- 10.Storey B T, Dodgson S J, Forster R E, II. Ann NY Acad Sci. 1984;429:210–211. doi: 10.1111/j.1749-6632.1984.tb12380.x. [DOI] [PubMed] [Google Scholar]
- 11.Feldstein J B, Silverman D N. J Biol Chem. 1984;259:5447–5453. [PubMed] [Google Scholar]
- 12.Murakami H, Sly W S. J Biol Chem. 1987;262:1382–1388. [PubMed] [Google Scholar]
- 13.Hewett-Emmett D, Tashian R E. Mol Phylogenet Evol. 1996;5:50–77. doi: 10.1006/mpev.1996.0006. [DOI] [PubMed] [Google Scholar]
- 14.Silverman D N, Lindskog S. Acc Chem Res. 1988;21:30–36. [Google Scholar]
- 15.Christianson D W. Adv Protein Chem. 1991;42:281–285. doi: 10.1016/s0065-3233(08)60538-0. [DOI] [PubMed] [Google Scholar]
- 16.Tu C K, Silverman D N, Forsman C, Jonsson B-H, Lindskog S. Biochemistry. 1989;28:7913–7918. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]
- 17.Engstrand C, Forsman C, Liang Z, Lindskog S. Biochim Biophys Acta. 1992;1122:321–326. doi: 10.1016/0167-4838(92)90412-7. [DOI] [PubMed] [Google Scholar]
- 18.Okuyama T, Sato S, Zhu X L, Waheed A, Sly W S. Proc Natl Acad Sci USA. 1992;89:1315–1319. doi: 10.1073/pnas.89.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamai S, Cody L B, Sly W S. Biochem Genet. 1996;34:31–43. doi: 10.1007/BF02396238. [DOI] [PubMed] [Google Scholar]
- 20.Fleming R E, Crouch C E, Ruzicka C A, Sly W S. Am J Physiol. 1993;265:L627–L635. doi: 10.1152/ajplung.1993.265.6.L627. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Okuyama T, Waheed A, Kusumoto W, Zhu X L, Sly W S. Arch Biochem Biophys. 1995;320:315–322. doi: 10.1016/0003-9861(95)90015-2. [DOI] [PubMed] [Google Scholar]
- 23.Deng W P, Nickoloff J A. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 24.Miyazaki J, Takaki S, Araki K, Tashiro F, Tominaga A, Takatsu K, Yamamura K. Gene. 1989;79:269–277. doi: 10.1016/0378-1119(89)90209-6. [DOI] [PubMed] [Google Scholar]
- 25.Lopata M A, Cleveland D W, Sollner-Webb B. Nucleic Acids Res. 1984;12:5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luthman H, Magnusson G. Nucleic Acids Res. 1983;11:1295–1308. doi: 10.1093/nar/11.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maren T H. J Pharmacol Exp Ther. 1960;130:26–29. [PubMed] [Google Scholar]
- 28.Sundaram V, Rumbolo P, Grubb J, Strisciuglio P, Sly W S. Am J Hum Genet. 1986;38:125–136. [PMC free article] [PubMed] [Google Scholar]
- 29.Lowry O H, Rosebrough N J, Farr A L, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 30.Kozak M. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 31.Cross G A M. Annu Rev Cell Biol. 1990;6:1–13. doi: 10.1146/annurev.cb.06.110190.000245. [DOI] [PubMed] [Google Scholar]
- 32.Marshall R D. Biochem Soc Symp. 1974;40:17–26. [PubMed] [Google Scholar]
- 33.Blobel G. Methods Enzymol. 1983;96:663–682. doi: 10.1016/s0076-6879(83)96056-1. [DOI] [PubMed] [Google Scholar]
- 34.Krebs J F, Ippolito J A, Christianson D W, Fierke C A. J Biol Chem. 1993;268:27458–27466. [PubMed] [Google Scholar]
- 35.Stams T, Nair S K, Okuyama T, Waheed A, Sly W S, Christianson D W. Proc Natl Acad Sci USA. 1996;93:13589–13594. doi: 10.1073/pnas.93.24.13589. [DOI] [PMC free article] [PubMed] [Google Scholar]


