Abstract
Staphylococci are an increasing cause of bloodstream infections. Rapid reliable identification of these organisms is essential for accurate diagnosis and prompt effective treatment. We evaluated the ability of the VITEK 2 system (bioMérieux, Inc, Hazelwood, Mo.) to identify these organisms rapidly and accurately. A total of 405 clinically relevant nonduplicate staphylococcal isolates (Staphylococcus aureus, n = 130; coagulase-negative staphylococci, n = 275) collected from blood cultures were tested. VITEK 2 results were considered correct when they were identical to those furnished by the comparison method based on the ID 32 STAPH system (bioMérieux, Marcy l'Etoile, France) plus supplementary manual testing. When discrepancies occurred, isolate identity was verified by molecular typing. The VITEK 2 correctly identified 387 (95.6%) isolates at the species level: 379 (including all but one [99.2%] of 130 S. aureus isolates and 249 of 275 [90.5%] coagulase-negative isolates) were identified by the automated reading; for the other eight, supplemental tests suggested by the manufacturer had to be used. Only one strain (0.2%) was misidentified (Staphylococcus hominis as Staphylococcus epidermidis), and four (1%), all S. epidermidis, were not identified. For the remaining 13 strains (including 10 S. hominis), the VITEK 2 system was unable to discriminate among two species, and no supplemental tests were suggested for conclusive identification. Over 90% of results were obtained within 4 h. These results suggest that the VITEK 2 system can provide rapid, accurate, and reliable species-level identification of staphylococci responsible for bloodstream infections, although there is room for improvement in the identification of certain coagulase-negative species, especially S. hominis.
Bloodstream infections are a major cause of morbidity and mortality. The frequency, etiology, and epidemiology of these infections have changed over the years. Staphylococcus aureus is now recognized as an important cause of both hospital and community-acquired bloodstream infections (1, 3, 8, 9, 27, 30). During the last 2 decades, the increased use of invasive procedures and broad-spectrum antibiotics, together with the growing number of immunocompromised and/or seriously ill patients, has led to the emergence of coagulase-negative staphylococci, particularly Staphylococcus epidermidis, and these organisms plays a prominent role in nosocomial bloodstream infections (1, 2, 3, 8, 9, 17, 18, 29). Rapid and reliable species identification of these organisms is essential for accurate diagnosis and prompt effective treatment of these infections (6, 7, 10, 16). Several manual and commercial identification methods have been developed and are now routinely used (4, 5, 15, 23, 24, 25, 26, 31). The fully automated VITEK 2 system (bioMérieux, Inc, Hazelwood, Mo.) can provide identification results for gram-positive cocci in a few hours thanks to the improved sensitivity of its fluorescence-based technology, and this feature represents a major improvement over earlier versions of the same system (21).
The present study was designed to evaluate the reliability of the VITEK 2 system in the identification of staphylococci responsible for bloodstream infections.
(Partial results of this study were presented at the 12th European Congress of Clinical Microbiology and Infectious Disease, Milan, Italy, 25 to 27 May 2002.)
MATERIALS AND METHODS
Design of the study.
The strains examined in this study were drawn from a consecutive series of staphylococcal isolates recovered from blood cultures between January 1999 and July 2001 in the Microbiology Laboratory of the “A. Gemelli” Medical Center of the Catholic University of the Sacred Heart, a 1,900-bed tertiary-care center in Rome. All 405 strains that were tested came from patients who met the Centers for Disease Control criteria for diagnosis of bacteremia (14). Each isolate was identified by using the VITEK 2 system as well as the comparison method described below. When discordant results emerged, the strains were retested with both methods, and the identity was confirmed with a nucleic acid-based procedure (described below).
Strain identification. (i) ID-GPC card and VITEK 2 system.
The VITEK 2 system was used according to the manufacturer's instructions; ID-Gram Positive Cocci cards (ID-GPC cards; bioMérieux) were used for identification. The ID-GPC card is a 64-well plastic card containing 18 empty wells and 46 wells for fluorescent and inhibitory tests that include pH change tests and derivatives to detect aminopeptidases and -osidases. Substrates used for detection of aminopeptidases are usually coupled with 7-amino-methylcoumarin (7AMC); substrates used for detection of -osidases are coupled with 4-methylumbelliferone (4MU). The 21 test substrates are as follows: 4MU-α-l-arabinofuranoside, 4MU-α-d-galactoside, 4MU-α-d-glucoside, 4MU-α-d-N-acetylneuraminic acid, 4MU-β-d-galactoside, 4MU-β-d-glucoside, 4MU-β-d-glucuronide, 4MU-β-d-mannoside, 4MU-n-acetyl-β-d-glucosaminide, 4MU-phosphate, alanine-7AMC, arginine-7AMC, aurease (butiloxicarbonyl-Val-Pro-Arg-AMC), histidine-7AMC, α-glutamic acid-7AMC, threonine-7AMC, lysine-7AMC, phenylalanine-7AMC, proline-7AMC, pyroglutamic acid-7AMC, and tyrosine-7AMC. In addition, the card includes a total of 16 fermentation tests (for amygdaline, arbutine, d-galactose, d-glucose, d-maltose, d-mannitol, d-melibiose, d-raffinose, d-sorbitol, d-trehalose, d-xylose, glycerol, lactose, l-arabinose, N-acetyl-glucosamine, and salicin), two decarboxylase tests (for arginine and ornithine), and six other tests (for urease, pyruvate, optochin, novobiocin, polymyxin B sulfate, and 6.5% NaCl).
The ID-GPC database of the VITEK 2 system includes the following taxa: S. aureus, Staphylococcus auricularis, Staphylococcus capitis, Staphylococcus chromogenes, Staphylococcus cohnii subsp. cohnii, S. cohnii subsp. urealyticus, S. epidermidis, Staphylococcus haemolyticus, Staphylococcus hominis, Staphylococcus hyicus, Staphylococcus intermedius, Staphylococcus kloosii, Staphylococcus lentus, Staphylococcus lugdunensis, Staphylococcus saprophyticus, Staphylococcus schleiferi, Staphylococcus sciuri, Staphylococcus simulans, Staphylococcus warneri, and Staphylococcus xylosus.
Each organism suspension was prepared from the growth of pure cultures of bacteria cultivated on plates containing Trypticase soy agar with 5% sheep blood (bioMérieux, Marcy l'Etoile, France) and incubated overnight at 35°C. Bacterial cells were suspended in 2.5 ml of a 0.45% sodium chloride solution. The suspension used in the VITEK 2 system was adjusted to a McFarland standard of 0.5 by using a Densicheck (bioMérieux).
In some cases, when the automated reading is inconclusive (i.e., low discrimination between two or more species), the manufacturer suggests one or more simple manual tests to resolve the identity of the test strain (i.e., hemolysis, coagulase tube test, clumping factor reaction, susceptibility to polymyxin B, or colony pigmentation). All of these tests were carried out according to standard procedures (19).
(ii) Comparison method.
The comparison method was based on the use of ID 32 STAPH strips (bioMérieux), which were read automatically after 24 h by using an ATB Expression instrument and ATB Plus software (version 2.7; bioMérieux). Both the assay strips and the reading equipment were used in accordance with the manufacturer's instructions.
When the automatic reading yielded ambiguous results (i.e., two or more different species), one or more of the following supplementary tests were performed (as appropriate) to resolve the identity of the strain: colony pigmentation, presence of hemolysins, anaerobic growth, coagulase testing (i.e., tube coagulase and clumping factor tests), and novobiocin and polymyxin B resistance. Coagulase assays were carried out with commercial kits (tube coagulase method using rabbit plasma and the Staph Plus latex agglutination test, both from bioMérieux). All other supplementary assays were carried out according to standard procedure (19).
Verification and analysis of identification results.
When the identity of a test strain supplied by the automated reading of the VITEK 2 system differed from that indicated by the comparison method, the strain was retested in triplicate with both methods. In addition, their identities were confirmed with the molecular typing method described by Yugueros et al. (32), which uses the staphylococcal gap gene as a target. Briefly, a colony for each strain was suspended in 100 μl of double-distilled water and incubated at 95°C for 15 min. A 1-μl aliquot of this solution was subjected to PCR using GF-1 and GR-1 primers and conditions as previously described (32). The obtained amplicons were digested with AluI restriction enzyme (Roche Molecular Diagnostics) at 37°C for 4 h and electrophoresed on agarose gel (3% MetaPhor agarose [FMC Bioproducts] in 1× Tris-acetate-EDTA [TAE]) at 80 V for 2 h. Restriction patterns were analyzed with a Fluor-S Max 2 instrument (Bio-Rad, Hercules, Calif.) and identified by comparison with those obtained from reference strains (see below).
Based on their concordance with the comparison method results (confirmed by molecular typing), the identification results of the VITEK 2 system were classified as follows: (i) correct identification (i.e., the species of the strain indicated by the comparison method was identical to that obtained with VITEK 2 based on the automatic reading alone or with supplemental tests suggested by the manufacturer); (ii) misidentification (i.e., the automated VITEK 2 reading indicated a single species that was different from that furnished by the comparison method); (ii) low discrimination (i.e., the automatic VITEK 2 reading indicated two or more possible species-level identities for the strain, and no supplemental tests were suggested to discriminate between these possibilities); and (iii) no identification (i.e., no identification was provided by the VITEK 2 system). Results are expressed in numbers and percentages.
Quality control.
In each testing session, the quality control strains S. aureus subsp. aureus ATCC 12600, S. capitis subsp. capitis ATCC 35661, S. cohnii subsp. cohnii ATCC 35662, S. epidermidis ATCC 14990, S. haemolyticus ATCC 29970, S. hominis ATCC 27844, S. lugdunensis ATCC 700328, S. simulans ATCC 27851, S. warneri ATCC 49454, and S. xylosus ATCC 29971 were tested with both the VITEK 2 identification system and the comparison method.
RESULTS
Comparison method results.
Based on the results of the comparison method, the 405 staphylococcal isolates from bloodstream infections were identified as follows: S. aureus, n = 130; S. capitis, n = 18; S. cohnii subsp. cohnii, n = 3; S. cohnii subsp. urealyticum, n = 1; S. epidermidis, n = 140; S. haemolyticus, n = 50; S. hominis, n = 32; S. lugdunensis, n = 8; S. schleiferi, n = 2; S. simulans, n = 8; S. warneri, n = 11; S. xylosus, n = 2. In all cases in which the sample was subjected to molecular typing (see above), the identity of the strain yielded by the comparison method was confirmed.
VITEK 2 system results.
Table 1 shows the rates of concordance between typing results provided by the VITEK 2 system and those of the comparison method for the 405 staphylococcal strains tested. Overall, the VITEK 2 system furnished correct species-level identification for 387 (95.6%) of the strains. The 379 (93.6%) strains that were correctly identified by the automated reading included all but one (99.2%) of the S. aureus and 249 of 275 (90.5%) coagulase-negative isolates. For the remaining eight strains (1 S. aureus, 3 S. epidermidis, and 4 S. hominis strains), the results of the automated reading were ambiguous, and the correct identity was revealed by supplemental tests suggested by the manufacturer (Table 2).
TABLE 1.
Identification results obtained with the ID-GPC card of the VITEK 2 system for 405 Staphylococcus sp. isolates
| Taxonb | No. (%)a of isolates
|
|||||
|---|---|---|---|---|---|---|
| Tested | Correctly identified
|
Identified with low discrimination | Misidentified | Not identified | ||
| Without supplemental testsc | With supplemental testsc | |||||
| S. aureus | 130 | 129 (99.2) | 1 (0.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| S. capitis | 18 | 17 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| S. cohnii subsp. cohnii | 3 | 3 | 0 | 0 | 0 | 0 |
| S. cohnii subsp. urealyticum | 1 | 1 | 0 | 0 | 0 | 0 |
| S. epidermidis | 140 | 133 (95.0) | 3 (2.2) | 0 (0.0) | 0 (0.0) | 4 (2.8) |
| S. haemolyticus | 50 | 48 (96.0) | 0 (0.0) | 2 (4.0) | 0 (0.0) | 0 (0.0) |
| S. hominis | 32 | 17 (53.1) | 4 (12.5) | 10 (31.3) | 1 (3.1) | 0 (0.0) |
| S. lugdunensis | 8 | 8 | 0 | 0 | 0 | 0 |
| S. schleiferi | 2 | 2 | 0 | 0 | 0 | 0 |
| S. simulans | 8 | 8 | 0 | 0 | 0 | 0 |
| S. warneri | 11 | 10 (90.9) | 0 (0.0) | 1 (9.1) | 0 (0.0) | 0 (0.0) |
| S. xylosus | 2 | 2 | 0 | 0 | 0 | 0 |
| Total | 405 | 379 (93.6) | 8 (2.0) | 13 (3.2) | 1 (0.2) | 4 (1.0) |
Percentages were calculated only when more than 10 strains were evaluated.
Definitive identification obtained with the comparison method (see Materials and Methods) and confirmed when necessary by the molecular method described in reference 32.
Supplemental tests suggested by the VITEK 2 manufacturer.
TABLE 2.
Staphylococcal strains correctly identified by the VITEK 2 system only after supplementary tests suggested by the manufacturer
| Taxona (n) | VITEK 2 automated reading | Supplementary test(s) usedb |
|---|---|---|
| S. aureus (1) | S. aureus/S. hyicus | Clumping factor reaction, hemolysis |
| S. epidermidis (3) | S. epidermidis/S. chromogenes | Pigmentation |
| S. hominis (4) | S. hominis/S. epidermidis | Susceptibility to polymyxin B |
Definitive identification obtained with the comparison method (see Materials and Methods) and confirmed by the molecular method described in reference 32.
Supplemental tests suggested by the manufacturer were performed according to standard procedures (19).
The VITEK 2 failed to provide correct identification of 18 of 405 (4.4%) strains. The results provided for these strains are summarized in Table 3.
TABLE 3.
Results of the VITEK 2 system for the 18 strains that were not identified correctly
| Taxona | VITEK 2 result | No. of strains with result/no. tested (%) |
|---|---|---|
| S. epidermidis | No identification data furnished | 4/130 (3.1) |
| S. hominis | Incorrectly identified as S. epidermidis | 1/32 (3.1) |
| Low discrimination: S. hominis vs S. warneri | 10/32 (31.3) | |
| S. warneri | Low discrimination: S. warneri vs S. hominis | 1/11 (9.1) |
| S. haemolyticus | Low discrimination: S. haemolyticus vs S. cohnii subsp. urealyticus | 2/50 (4.0) |
Definitive identification obtained with the comparison method (see Materials and Methods) and confirmed by the molecular method described in reference 32.
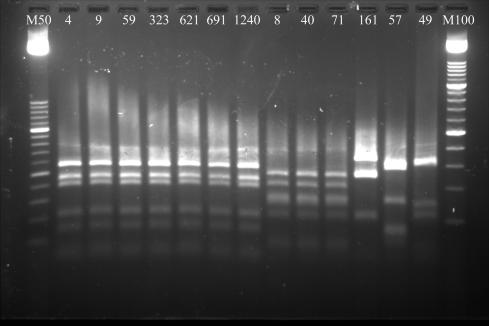
All four of the isolates that could not be identified at all by the VITEK 2 system (1% of the total series) were S. epidermidis, and the single case of misidentification (0.2%) involved an S. hominis strain. The other 13 failures (10 S. hominis, 2 S. haemolyticus, and 1 S. warneri strain; 2.7% of all strains tested) were associated with ambiguous results in the automated reading, and no supplemental tests were suggested for more precise discrimination. For all 18 of these isolates, as well as the eight that were correctly identified after supplemental testing, the identities indicated by the comparison method were confirmed by PCR-restriction fragment length polymorphism (RFLP) patterns of the staphylococcal gap gene. Some of these patterns are shown in Fig. 1.
FIG. 1.
PCR-RFLP patterns of 13 staphylococcal isolates identified with low discrimination or misidentified by VITEK 2. Lanes: 4, 9, 59, 323, 621, 691, and 1240, S. hominis; 8, 40, and 71, S. epidermidis; 57, S. aureus; 161, S. haemolyticus; 49, S. warneri; M50, 50-bp ladder; M100, 100-bp ladder.
All quality control strains were correctly identified at the species level by the VITEK 2 system.
For 90% of the strains, results were obtained within 2 to 3 h. For two isolates of S. aureus and 27 coagulase-negative staphylococcal strains, the time required was 4 h, and the remaining six isolates, all coagulase-negative, were identified after 6 h.
DISCUSSION
The incidence of staphylococcal bloodstream infections has increased over the past few decades due to several factors, primarily the growing number of patients at risk for these infections. Coagulase-negative staphylococci, which were once considered simple commensal organisms of human skin and mucous membranes, are now common opportunistic pathogens in intensive care units and oncology wards (1, 3, 8, 9, 27, 29). Although S. epidermidis still accounts for the vast majority of staphylococci (65 to 90%) recovered from blood cultures, the number of infections caused by other coagulase-negative staphylococci is increasing (2). In clinical laboratories that use screening tests alone, the organism responsible for the infection can be misidentified. The manual method developed by Kloos and Schleifer is highly reliable and accurate, but it is also time-consuming and labor-intensive, and 5 days of incubation is sometimes required for strain identification (20). For this reason, a number of automated commercial systems have been evaluated for routine laboratory use (4, 15, 23, 24, 25, 26, 31). The primary advantage of these systems, according to their manufacturers, is a significant savings in terms of the time needed for species-level identification. In this sense, the new fluorescence-based technology of the VITEK 2 system is undoubtedly a positive contribution.
Rapid results are meaningless, however, if they cannot be considered reliable. In the present study, we evaluated the performance of VITEK 2 compared with that of another automated system, the ID 32 STAPH system, and in all cases of discordant results, the results of the comparison method were fully confirmed by PCR-RFLP of the staphylococcal gap gene (Fig. 1). The ID 32 STAPH system is widely used for routine testing and also as a comparison method for the evaluation of other phenotypic identification systems (11). Nonetheless, cases of misidentification and low probability (i.e., <85%) of species identification have been reported (15, 27). For all strains that yielded discordant results in the VITEK 2 and ID 32 STAPH systems, the species-level identification furnished by the comparison method was confirmed with molecular typing. In contrast, when VITEK 2 results were concordant with the comparison method, molecular confirmation was not obtained, which means that, in theory, some of the strains “correctly identified” by the new system might actually have been misidentified by both the VITEK 2 and the ID 32 STAPH systems.
The overall agreement between VITEK 2 results and those of the comparison method was 95.6% (Table 1). The new system proved to be highly reliable for the staphylococcal species most commonly isolated in our laboratory from patients with bloodstream infections, i.e., S. aureus and S. epidermidis, which were ultimately identified in 100 and 97.2% of cases, respectively. However, while the vast majority of correct results were based on the automated reading alone, eight strains were correctly identified only after supplemental tests suggested by the manufacturer (Table 2). None of the supplementary tests used to resolve these cases of low discrimination was particularly complicated or expensive, but they did reduce (to some extent) the benefits of rapid automated testing. In fact, for the isolates correctly identified in the automated reading, results were most often available within 2 to 3 h, while up to 24 additional hours or even more were needed for identification based on supplementary testing.
The lowest percentage of correct results (65.6%) was recorded for the 32 S. hominis isolates. This species also proved to be a problem with the previous version of the VITEK system. Bannerman et al., who tested the original VITEK system in the identification of coagulase-negative staphylococci, found that it was the species identified with the lowest accuracy (63% of all strains tested) (4). Of 37 S. hominis isolates examined in their study, seven were incorrectly identified as either S. epidermidis (43%), S. saprophyticus/S. epidermidis (43%), or S. warneri (14%), and no identification was provided for seven other strains (43%) (4). Our experience with the VITEK 2 system, as well as that of others (A. Bassel et al., Abstr. 8th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. P255, 1997), indicates that this species continues to represent a weak point in the new system. The system was ultimately able to identify only 21 (65.6%) of the 32 isolates of this species that we tested, including four that were identified only after supplementary tests suggested by the manufacturer. The other 11 were either ambiguously identified (n = 10) or misidentified (as S. epidermidis; n = 1) (Table 3). However, with respect to the findings of Bannerman et al. with the original VITEK, our results suggest that some progress may have been made: the new system provided some type of identification data (correct or incorrect) for all our S. hominis isolates, in all but one case S. hominis was at least one of the possibilities indicated, and none were misidentified as possible S. saprophyticus strains.
The main cause of failure with S. hominis isolates was low discrimination, in particular the inability to exclude the possibility of S. warneri. Ligozzi et al. (21) recently evaluated the VITEK 2 system in the identification of a large series of gram-positive cocci, including 100 isolates of coagulase-negative staphylococci. Although only seven strains of S. hominis were included in this study, identification failures were recorded for three (42.8%), and in all cases, the problem was one of low discrimination. The details of these ambiguities were not provided. Those investigators hypothesized that the inferior performance of the VITEK 2 system with coagulase-negative staphylococci (compared with S. aureus) might be related to the relatively low rates of metabolism displayed by these species. Similar problems encountered with VITEK 2 identification of certain gram-negative bacteria (13, 22) have also been attributed to this factor. The possibility of false-negative reactions within the short incubation times used by this system may well have contributed to the 10 low-discrimination results (S. hominis versus S. warneri) that we observed with S. hominis. The biochemical profiles used by the VITEK 2 system to identify these two species are largely overlapping. The only differences are that S. hominis is expected to produce acid from lactose (but not from mannitol), while the reverse is expected from S. warneri. Since all 10 of the S. hominis isolates in question failed to produce acid either from lactose or mannitol in the VITEK 2 system, they could not be conclusively identified as either S. hominis (due to the lack of acid production from lactose) or as S. warneri (due to the absence of acid production from mannitol). The fact that all of these displayed an ability to produce acid from lactose (but not mannitol) with the ID 32 STAPH strips, which were read automatically after 24 h of incubation, suggests that the lactose negativity reported by the VITEK 2 system might indeed be an example of false negativity caused by its short incubation times. On the other hand, in conventional manual testing, some strains ultimately identified as S. hominis are indeed lactose negative (5, 19). As Kloos and Bannerman (19) have pointed out, however, the results of conventional carbohydrate tests may be slightly different from those obtained with rapid commercial test systems. For this reason, it is difficult to say whether the main problem in the cases described above is a false negativity for acid production from lactose (due to excessively short incubation times) or the weight placed on this characteristic by the system's database, which considers acid production from lactose a typical feature of S. hominis. Regardless of its specific cause(s), incorrect identification of S. hominis is undoubtedly a weakness of the VITEK 2 system (Bassel et al., 8th Eur. Congr. Clin. Microbiol. Infect. Dis.). Furthermore, when the system fails to distinguish this species from S. warneri, no supplemental tests are suggested by the manufacturer to resolve the issue, although this could easily be accomplished by tests for anaerobic growth and β-glucosidase activity (12, 19).
On a more positive note, the VITEK 2 system is capable of identifying S. lugdunensis, which is strongly recommended because it can produce infections that are just as severe as those caused by S. aureus. In clinical laboratories that use screening tests alone, S. lugdunensis can be misidentified as S. aureus or other coagulase-negative staphylococci (28). In the older VITEK system, whose database did not include this species, 79% of S. lugdunensis isolates were misidentified and the remaining 21% were unidentified (4). In our study the VITEK 2 system correctly identified all eight of the S. lugdunensis isolates tested.
In conclusion, accurate identification of staphylococcal isolates is crucial for the correct management of staphylococcal bloodstream infections, but it is also essential for a better understanding of the pathophysiological factors affecting the clinical outcome and for epidemiological surveillance (2, 6). As infections of this type become more common, clinical laboratories will have an increasing need for rapid and reliable methods of identifying the causative organisms at least at the species level. On the whole, the VITEK 2 system provided accurate and reliable results for the staphylococcal isolates that we tested, with considerable savings in terms of time and work. This experience suggests that the system can be used in clinical laboratories for routine identification of staphylococci responsible for bloodstream infections, although for some coagulase-negative species, particularly S. hominis, there is clearly room for improvement.
Acknowledgments
This work was supported by a grant from the Ministero dell'Università, Ricerca Scientifica e Tecnologica (2001-2002; ex MURST, 60%).
We thank Marian Kent for editorial assistance.
REFERENCES
- 1.Anonymous. 1999. National Nosocomial Infection Surveillance (NNISS) System report, data summary from January 1990-May 1999, issued June 1999. Am. J. Infect. Control 27:520-532. [DOI] [PubMed] [Google Scholar]
- 2.Archer, G. L. 1995. Staphylococcus epidermidis and other coagulase-negative staphylococci, p. 1777-1783. In G. L. Mandell, J. E. Bennet, and R. Dolin (ed.), Principles and practice of infectious diseases, 4th ed. Churchill Livingstone, New York, N.Y.
- 3.Banerjee, S. N., T. G. Emori, D. H. Culver, R. P. Gaynes, W. R. Jarvis, T. Horan, J. R. Eduards, J. Tolson, T. Henderson, and W. J. Martone. 1991. Secular trends in nosocomial primary bloodstream infections in the United States, 1980-1989. National Nosocomial Infection Surveillance System. Am. J. Med. 91:86S-89S. [DOI] [PubMed] [Google Scholar]
- 4.Bannerman, T. L., K. T. Kleeman, and W. E. Kloos. 1993. Evaluation of the Vitek Systems Gram-Positive Identification card for species identification of coagulase-negative staphylococci. J. Clin. Microbiol. 31:1322-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behme, R. J., R. Shuttleworth, A. McNabb, and W. D. Colby. 1996. Identification of staphylococci with a self-educating system using fatty acid analysis and biochemical tests. J. Clin. Microbiol. 34:3075-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamin, D. K., Jr., W. Miller, H. Garges, D. K. Benjamin, R. E. McKinney, Jr., M. Cotton, R. G. Fisher, and K. A. Alexander. 2001. Bacteremia, central catheters and neonates: when to pull the line. Pediatrics 107:1272-1276. [DOI] [PubMed] [Google Scholar]
- 7.Bryan, C. S. 1989. Clinical importance of positive blood cultures. Clin. Microbiol. Rev. 2:329-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diekema, D. J., M. A. Pfaller, F. J. Schimtz, J. Smayevsky, J. Bell, R. N. Jones, and M. Beach. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe and Western Pacific region for the Sentry Antimicrobial Surveillance program, 1997-1999. Clin. Infect. Dis. 32:S114-S132. [DOI] [PubMed] [Google Scholar]
- 9.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in the United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239-244. [DOI] [PubMed] [Google Scholar]
- 10.Emori, T. G., and R. P. Gaynes. 1993. An overview of nosocomial pathogens, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 6:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fahr, A. M., U. Eigner, M. Armbrust, A. Caganic, G. Dettori, C. Chezzi, L. Bertoncini, M. Benecchi, and M. G. Menozzi. 2003. Two center collaborative evaluation of the performance of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of Enterococcus and Staphylococcus spp. J. Clin. Microbiol. 41:1135-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes, B. A., D. F. Sahm, and A. S. Weissfeld (ed.). 1998. Staphylococcus, Micrococcus and similar organisms, p. 607-618. In Diagnostic microbiology, 10th ed. Mosby, Inc., St. Louis, Mo.
- 13.Funke, G., D. Monnet, C. deBernadinis, A. von Graevenitz, and J. Freney. 1998. Evaluation of the VITEK 2 system for rapid identification of medically relevant gram-negative rods. J. Clin. Microbiol. 36:1948-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garner, J. S., W. R. Jarvis, T. G. Emori, T. C. Horan, and J. M. Hughes. 1988. CDC definitions for nosocomial infections. Am. J. Infect. Control. 16:128-140. (Erratum, 16:177.) [DOI] [PubMed]
- 15.Ieven, M., J. Verhoven, S. R. Pattyn, and H. Goossens. 1995. Rapid end economical methods for species identification of clinically significant coagulase-negative staphylococci. J. Clin. Microbiol. 33:1060-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, S. D., L. C. McDonald, W. R. Jarvis, S. K. McAllister, R. Jerris, L. A. Carson, and J. M. Miller. 2000. Determining the significance of coagulase-negative staphylococci isolated from blood cultures at a community hospital: a role for species and strain identification. Infect. Control Hosp. Epidemiol. 21:213-217. [DOI] [PubMed] [Google Scholar]
- 17.Kloos, W. E., and T. L. Bannerman. 1994. Update on clinical significance of coagulase-negative staphylococci. Clin. Microbiol. Rev. 7:117-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloos, W. E., C. G. George, J. S. Olgiate, L. Van Pelt, M. L. McKinnon, B. L. Zimmer, E. Muller, M. P. Weinstein, and S. Mirrett. 1998. Staphylococcus hominis subsp. novobiosepticus, subsp. nov., a novel trehalose- and N-acetyl-d-glucosamine-negative, novobiocin- and multiple-antibiotic-resistant subspecies isolated from human blood cultures. Int. J. Syst. Bacteriol. 48:799-812. [DOI] [PubMed] [Google Scholar]
- 19.Kloos, W. E., and T. L. Bannerman. 1999. Staphylococcus and Micrococcus, p. 264-282. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 20.Kloos, W. E., and K. H. Schleifer. 1975. Simplified scheme for routine identification of human Staphylococcus species. J. Clin. Microbiol. 1:82-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ligozzi, M., C. Bernini, M. G. Bonora, M. De Fatima, J. Zuliani, and R. Fontana. 2002. Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. J. Clin. Microbiol. 40:1681-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling, T. K. W., P. C. Tam, Z. K. Liu, and A. F. B. Cheng. 2001. Evaluation of VITEK 2 rapid identification and susceptibility testing system against gram-negative clinical isolates. J. Clin. Microbiol. 39:2964-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monsen, T., M. Ronnmark, C. Olofsson, and J. Wistrom. 1998. An inexpensive and reliable method for identification of staphylococcal species. Eur. J. Clin. Microbiol. Infect. Dis. 17:327-335. [DOI] [PubMed] [Google Scholar]
- 24.Perl, T. M., P. R. Rhomberg, M. J. Bale, P. C. Fuchs, R. N. Jones, F. P. Koontz, and M. A. Pfaller. 1994. Comparison of identification systems for Staphylococcus epidermidis and other coagulase-negative Staphylococcus species. Diagn. Microbiol. Infect. Dis. 18:151-155. [DOI] [PubMed] [Google Scholar]
- 25.Renneberg, J., K. Rieneck, and E. Gutschik. 1995. Evaluation of STAPH ID 32 system and Staph-Zym system for identification of coagulase-negative staphylococci. J. Clin. Microbiol. 33:1150-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhoden, D. L., and J. M. Miller. 1995. Four-year prospective study of STAPH-IDENT system and conventional methods for reference identification of Staphylococcus, Stomatococcus, and Micrococcus spp. J. Clin. Microbiol. 33:96-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaberg, D. R., D. H. Culver, and R. P. Gaynes. 1991. Major trends in the microbial etiology of nosocomial infection. Am. J. Med. 91:72S-75S. [DOI] [PubMed] [Google Scholar]
- 28.Schnitzler, N., R. Meilicke, G. Conrads, D. Frank, and G. Haase. 1998. Staphylococcus lugdunensis: report of a case of peritonitis and an easy-to perform screening strategy. J. Clin. Microbiol. 36:812-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szewczyk, F. M., A. Piotrowski, and M. Rozalska. 2000. Predominant staphylococci in the intensive care unit of a pediatric hospital. J. Hosp. Infect. 45:145-154. [DOI] [PubMed] [Google Scholar]
- 30.Waldvogel, F. A. 1995. Staphylococcus aureus, p. 1754-1777. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 4th ed. Churchill Livingstone, New York, N.Y.
- 31.Wieser, M., and H. J. Busse. 2000. Rapid identification of Staphylococcus epidermidis. Int. J. Syst. Evol. Microbiol. 50:1087-1093. [DOI] [PubMed] [Google Scholar]
- 32.Yugueros, J., A. Temprano, B. Berzal, M. Sanchez, C. Hernanz, J. M. Luengo, and G. Naharro. 2000. Glyceraldehyde-3-phosphate dehydrogenase-encoding gene as a useful taxonomic tool for Staphylococcus spp. J. Clin. Microbiol. 38:4351-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]