Abstract
PCR tests were used to assign genomovar status to 39 non-cystic fibrosis (non-CF) and 11 CF Burkholderia cepacia complex isolates from patients in hospitals in Recife, Brazil. Non-CF isolates were assigned to genomovar IIIA (71.8%), genomovar I (15.4%), B. vietnamiensis (7.7%), and B. multivorans (5.1%). CF isolates were assigned to genomovar IIIA (18.2%), B. vietnamiensis (18.2%), and genomovar I (9.1%). Six CF isolates sharing recA PCR-restriction fragment length polymorphism (RFLP) and randomly amplified polymorphic DNA (RAPD) patterns could not be assigned to a genomovar. 16S rDNA sequence obtained from these isolates indicated a closest relationship to B. anthina, but the recA sequence was equally divergent from several genomovars. PCR screening indicated the presence of cblA in only two isolates, whereas the B. cepacia epidemic strain marker was found in 22 of 28 genomovar IIIA isolates. A type III secretion gene was detected in all but genomovar I isolates. RAPD and PCR-RFLP assays, targeting both recA and fliC, indicated a large amount of genetic variability among the isolates, with many novel patterns being observed. Nine genomovar IIIA isolates from different non-CF patients and clinical sources had identical genotypes, indicating the presence of a common clone.
Burkholderia cepacia is an important opportunistic respiratory pathogen, particularly in patients with cystic fibrosis (CF) (7) and chronic granulomatous disease (23). In addition, B. cepacia causes catheter-associated urinary tract infections, wound infections, intravenous catheter-associated bacteremia, and endocarditis (25, 26).
What is now known as the B. cepacia complex was subdivided by DNA-DNA hybridization, whole-cell protein pattern similarity, and phenotypic markers into five genomic species or genomovars (28), including B. multivorans (formerly genomovar II), B. stabilis (formerly genomovar IV), and B. vietnamiensis (formerly genomovar V). Genomovar III, which includes most CF epidemic strains, can be further subdivided into two groups on the basis of recA sequences (groups IIIA and IIIB) (15). More recently, four new members of the B. cepacia complex have been identified: genomovar VI (4), B. ambifaria (genomovar VII [5]), B. pyrrocinia (genomovar IX [27]), and B. anthina (genomovar VIII [27]). At present there is no major phenotypic difference to differentiate strains of genomovars I, III, and VI. PCR assays based on variations in the recA sequence are available for the specific detection of genomovar I, B. multivorans, genomovars IIIA and IIIB, B. stabilis, B. vietnamiensis, and B. ambifaria (15). Recently, a test for B. anthina has also been published (27).
There have been a number of studies aimed at determining the distribution of genomovars among isolates from CF and non-CF patients (1, 2, 13, 24). In this paper, we report the application of PCR tests to the identification of 39 non-CF and 11 CF B. cepacia isolates from patients in hospitals in Brazil. We further report the results of PCR-based fingerprinting to determine genetic variability among the isolates and data concerning the distribution of genetic markers associated with cable pili, type III secretion (TTS), the B. cepacia epidemic strain marker (BCESM), and a putative polysaccharide export gene cluster.
MATERIALS AND METHODS
Bacterial strains.
Thirty-nine non-CF isolates came from inpatients at neurological wards or intensive care units of Hospital Português, a large hospital facility in Recife, Brazil. Most patients were old debilitated individuals with respiratory problems and were undergoing ventilation. The CF isolates were recovered from 11 outpatients younger than 14 years, who were attending the CF unit of Instituto Materno Infantil de Pernambuco. All isolates were cultured on B. cepacia selective agar (9) at 35°C for 48 h and preliminarily identified as belonging to the B. cepacia complex by using a panel of conventional phenotypic tests (10).
PCR assays.
One or more colonies of bacteria were taken from growth on nutrient agar plates and resuspended in 20 μl of sterile distilled water. The suspensions were boiled for 5 min at 95°C to obtain a lysed bacterial suspension. Cell debris was removed by centrifugation, and the crude DNA preparation was used immediately in PCR amplification using the Eppendorf MasterTaq system under the conditions recommended by the supplier. Previously described oligonucleotide primers and annealing temperatures were used for the genomovar-specific (15), B. anthina-specific (27), 16S rDNA (15), cblA (6), BCESM (18), fliC (8), and bcscQ (19) PCR assays. Control strains were taken from the published representative panel of strains (17). PCR assays for detection of putative exopolysaccharide genes with homology to the wcbB and wzm2 genes of the Burkholderia pseudomallei capsule production gene cluster were carried out as described previously (20).
Molecular typing.
recA PCR-restriction fragment length polymorphism (RFLP) patterns were generated using the restriction enzyme HaeIII as described previously (15). fliC PCR amplicons were digested with HaeIII and MspI as described previously (8, 30). Randomly amplified polymorphic DNA (RAPD) fingerprinting and macrorestriction analysis using pulsed-field gel electrophoresis (PFGE) were carried out using published procedures (16, 31).
Nucleotide sequence analysis.
The nucleotide sequence of recA and 16S rRNA PCR amplicons was obtained by Lark Technologies Inc., using the same oligonucleotide primers employed in the PCR amplification and internal primers. Sequences were edited and aligned using the GCG sequence analysis software package (Genetics Computer Group, University of Wisconsin). BLASTN searches were conducted using the site http://www.ncbi.nlm.nih.gov.
RESULTS
Determination of genomovar designations.
Fifty B. cepacia clinical isolates from Brazil were subjected to recA-based PCR tests (15) to assign them to a genospecies. The strains consisted of both CF (n = 11) and non-CF (n = 39) isolates and were obtained from various clinical sources (Table 1). A positive control strain, with a known genomovar status, from the B. cepacia complex was used in each case. The distribution of the 39 non-CF isolates screened using genomovar-specific PCR tests was as follows: 28 (71.8%) genomovar IIIA isolates, 6 (15.4%) genomovar I isolates, 3 (7.7%) B. vietnamiensis isolates, and 2 (5.1%) B. multivorans isolates. Of the 11 CF isolates, 2 (18.2%) were PCR positive using genomovar IIIA-specific primers, 2 (18.2%) were PCR positive using B. vietnamiensis-specific primers, and 1 (9.1%) was PCR positive using genomovar I-specific primers. None of the remaining six CF isolates were PCR positive using tests for genomovar I, B. multivorans, genomovar IIIA and IIIB, B. stabilis, B. vietnamiensis, or B. ambifaria. These strains were subjected to PCR with B. anthina-specific primers. On one occasion, this resulted in the production of a single amplicon. The reaction was repeated several times, and on each subsequent occasion the same strains gave two-banded PCR products. Unfortunately, a positive control strain for B. anthina was not available.
TABLE 1.
Distribution of isolates according to source
| Source | No. of isolates of genomovar:
|
||||
|---|---|---|---|---|---|
| IIIAa | I | B. multivorans | B. vietnamiensis | No ID | |
| Non-CF (n = 39) | |||||
| Sputum (n = 21) | 15 (5) | 6 | 0 | 0 | 0 |
| Blood (n = 6) | 3 (1) | 0 | 1 | 2 | 0 |
| Urine (n = 4) | 4 (1) | 0 | 0 | 0 | 0 |
| Trachea (n = 5) | 3 (1) | 0 | 1 | 1 | 0 |
| Pleura (n = 1) | 1 (1) | 0 | 0 | 0 | 0 |
| Intravenous catheter (n = 1) | 1 (0) | 0 | 0 | 0 | 0 |
| Skin (n = 1) | 1 (0) | 0 | 0 | 0 | 0 |
| CF (n = 11) | 2 (0) | 1 | 0 | 2 | 6 |
Numbers for isolates sharing RAPD profiles, and recA and fliC RFLP types equivalent to the ET12 lineage, are indicated in parentheses.
recA PCR-RFLPs.
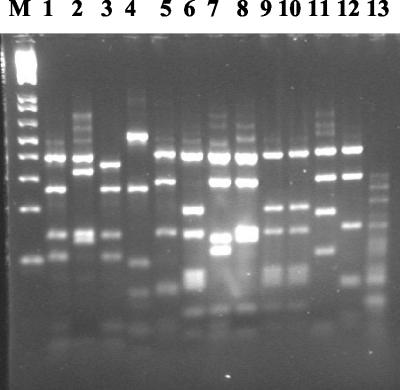
The isolates were subjected to recA PCR amplification using the oligonucleotide primers BCR1 and BCR2 (15). All isolates yielded an amplicon of 1,043 bp, with the exception of one of the CF isolates, which was identified as B. vietnamiensis. Digestion of the recA amplicon with the endonuclease HaeIII generated a number of patterns revealing considerable variability (Fig. 1). recA RFLP patterns from the Brazilian isolates were compared with those from the previously published representative panel of the B. cepacia complex (17), where HaeIII patterns designated A to J were reported. Figure 1 shows all the different recA RFLP patterns obtained from the Brazilian isolates. Twenty-six of the Brazilian isolates gave recA RFLP patterns that had been reported previously for the representative panel of strains. However, nine new patterns were observed (Fig. 1).
FIG. 1.
recA HaeIII RFLP patterns of Brazilian B. cepacia CF and non-CF isolates. Pattern E (lane 1), pattern D (lane 3), pattern G (lane 6), and pattern B (lane 10) have been reported previously (13). Other patterns (lanes 2, 4, 5, 7, 8, 9, 11, 12, and 13) are novel patterns not found in the representative panel of strains reported previously. Lane 13 contains the pattern derived from a representative of the unidentified CF isolates. M, 1-kb ladder (Helena Biosciences).
Of the seven genomovar I CF and non-CF isolates, three gave recA pattern E and one gave pattern D, patterns that had been reported previously (15). Two of the isolates generated the same novel pattern (Fig. 1, lane 2). The two non-CF isolates identified as B. multivorans both revealed novel patterns (lanes 4 and 5). Genomovar IIIA isolates produced five different RFLP patterns. The majority of the isolates (20 of 28) gave recA pattern G, indicative of genomovar IIIA (15). All other genomovar IIIA isolates gave RFLP patterns not reported by Mahenthiralingam et al. (15). Four isolates gave the pattern shown in lane 8, three gave the pattern shown in lane 7, and two shared a pattern with one of the B. multivorans isolates (lane 5). One of two genomovar IIIA CF isolates gave a novel pattern (lane 9), whereas the other gave recA pattern G. Two RFLP patterns were obtained for the B. vietnamiensis isolates. Specifically, of the five isolates identified as B. vietnamiensis, two gave the previously observed pattern B (15) and two had the same novel RFLP pattern (Fig. 1, lane 11). All six of the unassigned CF isolates gave the same novel recA RFLP pattern (lane 13).
fliC PCR-RFLPs.
When the 50 Brazilian isolates were subjected to fliC PCR amplification using primers BC4 and BCR12, only 26 isolates yielded a PCR product. Of the 26 fliC PCR-positive isolates, 25 showed a product of 1.0 kb, indicative of type II flagellins. One B. multivorans isolate gave a PCR product of 1.4 kb, indicative of type I flagellins (8). Twenty-four of the isolates did not generate a PCR product. Five of these were B. vietnamiensis isolates, a genospecies that has been shown not to amplify with the specific primers in a previous study (30). The PCR test was repeated at least twice to confirm the results, and the isolates were assumed not to amplify with the fliC primers.
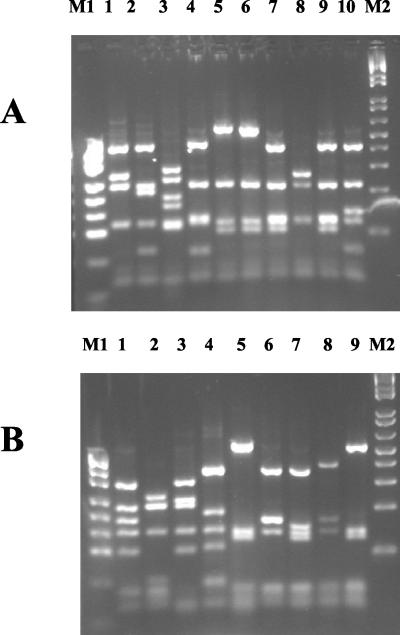
The 22 isolates that generated a fliC amplicon were subjected to RFLP analysis using the endonucleases HaeIII and MspI (Fig. 2). A number of RFLP patterns not observed in previous studies were found. In all, nine HaeIII and nine MspI RFLP patterns were identified. Of these, seven HaeIII and eight MspI patterns were novel. When HaeIII and MspI patterns were considered in combination, only one RFLP type corresponded to a previously reported RFLP group: 12 genomovar IIIA isolates belonged to RFLP group I as defined previously (8,31) and including members of the electrophoretic type 12 (ET12) lineage.
FIG. 2.
fliC RFLP patterns generated using MspI (A) and HaeIII (B). All patterns observed in this study are shown. Lane 4 in panel A and lane 5 in panel B are RFLP patterns indicative of fliC RFLP group I. Lanes 5 and 6 in panel A are from two isolates with the same MspI RFLP pattern. M1, pUC19/MspI (Helena Biosciences); M2, 1-kb ladder (Helena Biosciences).
The five genomovar I isolates could be subdivided into three RFLP groups, with three non-CF isolates sharing the same patterns for both endonucleases. The HaeIII pattern generated from these three isolates had been observed previously in genomovar I, but the MspI pattern was novel. Genomovar IIIA isolates could be subdivided into six RFLP groups, but only two of them contained more than one isolate. The largest group comprised the 12 isolates belonging to RFLP group I. The second largest group comprised four non-CF isolates. The one B. multivorans isolate that amplified with the fliC primers produced novel HaeIII and MspI patterns. Twenty of the isolates, including both fliC PCR-positive and PCR-negative isolates, were tested for motility, and only one was found to be nonmotile.
RAPD and PFGE profiles.
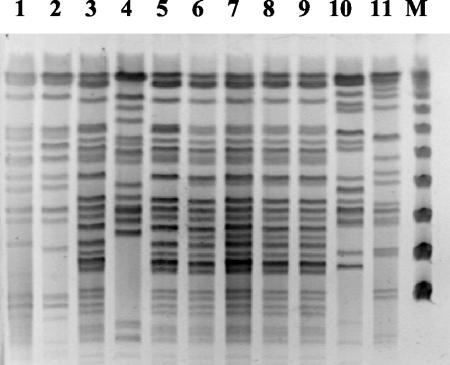
Twenty-four different RAPD profiles were obtained from the Brazilian isolates. The 20 genomovar IIIA isolates giving recA RFLP pattern G could be subdivided into five RAPD profiles. A total of nine isolates shared recA RFLP pattern G, were in fliC RFLP group I, and had the same RAPD profile. This RAPD profile differed from that of a control strain representing the ET12 lineage. The unidentified CF isolates were indistinguishable by RAPD analysis. These relationships were confirmed using PFGE (Fig. 3).
FIG. 3.
PFGE profiles of B. cepacia isolates. Restriction digestion profiles generated using XbaI are shown for ET12 strain K56-2 (lane 1), ET12 strain J2315 (lane 2), isolates of fliC RFLP group I and recA RFLP pattern G that share a RAPD profile (lanes 3, 5, 6, 7, 8, and 9), an isolate of a different fliC RFLP group but with recA RFLP pattern G (lane 4), and isolates of fliC RFLP group I and recA RFLP pattern G that share a different RAPD profile (lanes 10 and 11). Isolates used for the PFGE profiles in lanes 3, 5, 6, 7, 8, and 9 are representatives of the common clone sharing features with but differing from the ET12 lineage. M, pulse marker, 50 to 1,000 kb (Sigma-Aldrich).
Distribution of genetic markers.
PCR amplification was used to screen the Brazilian isolates for the presence of cblA, BCESM, bcscQ, and genes from a putative polysaccharide export cluster. Only one of the non-CF genomovar I isolates, BC50, and one non-CF genomovar IIIA isolate, BC32, were PCR positive for cblA. The rest of the isolates, including all of the CF isolates, were PCR negative for cblA. Of the 28 non-CF and 2 CF isolates identified as genomovar IIIA, 22, including both CF isolates, were PCR positive for the BCESM. All isolates attributed to fliC RFLP group I were PCR positive for BCESM. All unassigned isolates and isolates identified as genomovar I, B. multivorans, or B. vietnamiensis tested negative for BCESM.
TTS systems are implicated in the pathogenicity of a number of gram-negative bacterial pathogens, including the CF pathogen Pseudomonas aeruginosa (11). In a previous study, a cluster of TTS genes was identified in strain J2315 of the ET12 lineage (19). Using two conserved structural TTS genes (bcscV) and (bcscQ) as targets in PCR assays and Southern blot hybridizations, the distribution of TTS genes among members of the B. cepacia complex was determined. The results suggested that all members of the B. cepacia complex carried the TTS genes with the exception of genomovar I (19). A PCR amplification assay for the detection of bcscQ was performed on all the Brazilian isolates. All seven CF and non-CF isolates identified as genomovar I failed to give a product. All other isolates were PCR positive for bcscQ.
PCR amplification assays for the detection of putative exopolysaccharide genes were carried out on isolates identified as belonging to genomovar I or IIIA. Detection involved the use of two primer sets. In accordance with the findings in our previous study (20) comparing PCR-based and hybridization methods for detection, isolates that were found to amplify with either primer set were considered to be PCR positive. Genomovar I isolates did not amplify with either set of primers. Seven genomovar IIIA isolates were PCR positive, but only two closely related isolates yielded amplicons with both primer sets. The remaining genomovar IIIA isolates were PCR negative for both sets of primers. The seven PCR-positive strains fell into four RAPD types and two recA RFLP groups.
Unassigned isolates.
Six CF isolates could not be assigned to a genomovar by using the genomovar-specific PCR tests. PCR-RFLP and RAPD data indicated that the six unassigned CF isolates were indistinguishable. The 16S rRNA sequence was obtained from amplicons derived from a representative of these isolates. The sequence revealed a closest match with B. anthina (GenBank accession number AJ420880), with a difference of only 1 nucleotide over a 972-bp sequence.
The recA PCR amplicon sequence was also obtained from a representative of the unassigned CF isolates. The sequence had 96 to 97% identity to recA sequences from several genospecies including genomovar I, B. multivorans, genomovar III (A and B), B. stabilis, genomovar VI, B. ambifaria, and B. pyrrocinia. B. anthina recA sequences were obtained from E. Mahenthiralingam (University of Cardiff). The recA sequences of BC14 had 96% sequence identity to the recA sequence of B. anthina. Thus, B. anthina was not the closest match for the recA sequence of the unidentified isolates. Interestingly, the recA sequence data indicated a 1-nucleotide mismatch with the sequence of one of the B. anthina-specific primers. This probably explains the presence of multiple bands when the B. anthina PCR test is applied. Our data indicate that results obtained with the B. anthina-specific primers should be treated with caution. recA and 16S rDNA sequences for representatives of the unassigned isolates have been deposited in GenBank under the accession numbers AY228543 and AF429992.
DISCUSSION
Molecular typing.
Overall, we found the genomovar-specific PCR tests to be an effective tool for assigning the majority of isolates to a genomovar. The products could be seen as distinct bands on gels, although nonspecific binding of primers was frequently observed, with weak bands of incorrect size occurring in strains that were not of the genomovar being tested. A recent evaluation of the genomovar-specific tests concluded that they were 92% sensitive and 100% specific for genomovar III and 100% sensitive and specific for B. multivorans (29). It has been reported that the test for genomovar I can lead to misidentification due to cross-reaction with B. pyrrocinia. However, the results obtained using recA and fliC PCR-RFLPs and a PCR assay for bcscQ led us to conclude that most, if not all, of the isolates identified as genomovar I in this study can be regarded as belonging to genomovar I. In a previous study using genomovars I to VII, only genomovar I isolates were PCR negative for bcscQ (19). In our study, there is an exact correlation between genomovar I designation and a PCR-negative test for bcscQ. However, B. pyrrocinia has not been tested by this assay; therefore, misidentification cannot be ruled out entirely. Another limitation to the species-specific recA PCR tests is the absence of specific primers for genomovar VI.
Genomovar IIIA isolates were clearly dominant among non-CF isolates. Interestingly, none of the isolates were identified as genomovar IIIB. The high prevalence of genomovar III isolates has been observed in various previous studies of CF isolates (1, 13, 24) and environmental isolates (2). In CF patients, the high prevalence of genomovar III isolates is a major concern since genomovar III infections are more likely to be chronic and patients with genomovar III infections face the highest mortality (14).
The isolates in this study were all recovered from hospitals in Recife, Brazil, and included isolates identified as genomovar I (14%), B. vietnamiensis (10%), and B. multivorans (4%). None of the isolates were identified as B. stabilis, B. ambifaria, or genomovar VII. Interestingly, in a study including 23 non-CF isolates, 13 (57%) were identified as B. stabilis (1). In other studies examining the molecular epidemiology of B. cepacia isolates, the distributions of genomovars other than genomovar III vary. In a recent study of CF patients from whom isolates phenotypically related to the B. cepacia complex had been obtained, 80% were infected with genomovar III, 9.6% were infected with B. multivorans, and 3.8% were infected with B. stabilis. The remaining genomovars were present at low frequency (<2%) or were not detected (24). In another study involving a panel of B. cepacia clinical and environmental isolates from Italy, 37.3% of environmental isolates were identified as B. ambifaria. Genomovar I (2.9%), B. stabilis (7.4%), and B. pyrrocinia (2.9%) were found among clinical isolates, and genomovar I (1.3%) and B. pyrrocinia (8%) were identified among environmental isolates (2). However, genomovar III was the dominant genomovar among both clinical (86.8%) and environmental (53.4%) isolates.
It has been demonstrated that whereas strains of genomovar IIIA predominate among genomovar III CF isolates in Canada and Italy, genomovar IIIB is predominant in the United States (14). We found no genomovar IIIB isolates among the non-CF or CF isolates in this study. This suggests either that the population in Brazil resembles those in Canada and Italy rather than the United States or that non-CF isolates may be distributed differently from CF isolates. We found only two genomovar III CF isolates in our study. Although both were identified as genomovar IIIA, a larger collection of isolates is required to reach a meaningful conclusion about the relevant proportions of genomovars IIIA and IIIB among CF isolates in Brazil.
Although the rDNA sequence suggested that the unidentified CF isolates may belong to the species B. anthina, recA sequence data indicated that these isolates were equally divergent from several of the other genomovars and that they may belong to a new, as yet uncharacterized genospecies. Unidentified isolates have been present in previous epidemiological studies (1, 13, 24), and it seems likely that the number of genospecies constituting the B. cepacia complex will continue to rise.
PCR-RFLP analysis of the recA gene revealed a number of patterns not matching any of those published previously. Only five of the known recA RFLP types (15) were recognized. The types all correlated with the genomovars to which the isolates had been assigned. In general, isolates with the same recA RFLP patterns also shared other characteristics. It has been reported that more than 50 B. cepacia recA RFLP patterns have been identified using the enzyme HaeIII (14). Unfortunately, data from all of these RFLP types are not available. Therefore, it is not possible to state whether any of the new types in our study have been found previously. Such a large number of RFLP types would also make it difficult to use this method for genomovar typing, since single genomovars can have multiple recA RFLP patterns.
fliC PCR-RFLP has been used in three previous studies to identify variation between strains in the B. cepacia complex (8, 30, 31), including the representative panel proposed for the B. cepacia complex genomovars I to V. In the previous studies, use of the oligonucleotide primers BC4 and BCR12 led to successful PCR amplification from all but the B. vietnamiensis strains. Since then, we have also observed that strains of genomovar VI and B. ambifaria fail to yield PCR amplicons with these primers (unpublished data). The fact that PCR amplification failed in this study with a number of isolates from genomovars that have not previously led to such failures suggests that variations are occurring in primer binding sequences. When it was possible to obtain a fliC genotype, there was good correlation between recA and fliC genotyping using PCR-RFLPs.
Cable pili mediate specific binding to mucin carbohydrates and to epithelial cells (21, 22) and are associated with genomovar IIIA and epidemic strains of the transmissible ET12 lineage, exemplified by strain J2315 (12). However, it is clear that not all genomovar IIIA strains carry the cblA gene. In addition, some strains lacking cblA, primarily from B. multivorans, have been associated with epidemic spread. The 1.4-kb open reading frame known as the BCESM has also been associated with CF epidemic strains (18), although it has been reported that sporadic strains of B. cepacia can also carry this marker (3).
In our study, only one of the isolates identified as genomovar IIIA was PCR positive for the cblA gene. However, 20 of the genomovar IIIA non-CF isolates, including the cblA-positive isolate, possessed the BCESM. Both genomovar IIIA CF isolates also contained the BCESM but lacked the cblA gene. The findings correlate with observations from other studies (2, 13, 24). It has been suggested that the absence of such markers from some epidemic strains indicates that the BCESM and cblA markers may not be reliable indicators of transmissibility (13). In keeping with previous observations, we found the BCESM only in genomovar III isolates.
In a previous study, the genomic island carrying the polysaccharide genes was detected in the ET12 lineage and in some but not all other representatives of genomovar IIIA, as well as in some strains of genomovar I and B. multivorans (20). The present study confirms the observation that the genes can be found in genomovar IIIA, but only in some isolates. The PCR assay approach used correlates well with results obtained using DNA-DNA hybridization for genomovars I and IIIA (20) and was therefore restricted to these genospecies.
Evidence for the presence of a clone among non-CF isolates.
We identified 12 non-CF isolates of recA RFLP group G and fliC RFLP group I, both indicative of the ET12 lineage. All of these isolates were PCR positive for BCESM, although all but one was PCR negative for cblA. The cblA-positive strain showed no evidence for the formation of cable pili when viewed using electron microscopy (data not shown). In addition, these isolates lacked the putative polysaccharide production gene cluster found in the ET12 lineage. RAPD analysis indicated that only 9 of these 12 isolates had the same RAPD profile, with the other 3 having a different RAPD profile. Neither of these matched either the RAPD or PFGE profiles of ET12 control strains. It should be noted that this and previous studies have demonstrated the fliC gene to be extremely variable within the B. cepacia complex, even within genomovars (30, 31). The fact that 12 non-ET12 isolates produce fliC RFLP patterns identical to ET12 strains indicates that the isolates may have acquired a fliC gene by gene transfer from the ET12 lineage.
The nine isolates that had the same genotype for all of the molecular typing methods were isolated from a number of different clinical sources and patients within the same hospital (Table 1). This suggests that these isolates may constitute an epidemic strain, which is either transmissible or particularly well adapted to survival in the hospital environment.
Acknowledgments
C.W. and C.A.H. acknowledge funding from the United Kingdom Cystic Fibrosis Trust.
REFERENCES
- 1.Agodi, A., E. Mahenthiralingam, M. Barchitta, V. Giannino, A. Sciacca, and S. Stefani. 2001. Burkholderia cepacia complex infection in Italian patients with cystic fibrosis: prevalence, epidemiology, and genomovar status. J. Clin. Microbiol. 39:2891-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bevivino, A., C. Dalmastri, S. Tabacchioni, L. Chiarini, M. L. Belli, S. Piana, A. Materazzo, P. Vandamme, and G. Manno. 2002. Burkholderia cepacia complex bacteria from clinical and environmental sources in Italy: genomovar status and distribution of traits related to virulence and transmissibility. J. Clin. Microbiol. 40:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clode, F. E., M. E. Kaufmann, H. Malnick, and T. L. Pitt. 2000. Distribution of genes encoding putative transmissibility factors among epidemic and nonepidemic strains of Burkholderia cepacia from cystic fibrosis patients in the United Kingdom. J. Clin. Microbiol. 38:1763-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coenye, T., J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroecke, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. E vol. Microbiol. 51:271-279. [DOI] [PubMed] [Google Scholar]
- 5.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51:1481-1490. [DOI] [PubMed]
- 6.Goldstein, R., L. Sun, R. Z. Jiang, U. Sajjan, J. F. Forstner, and C. Campanelli. 1995. Structurally variant classes of pilus appendage fibers coexpressed from Burkholderia (Pseudomonas) cepacia. J. Bacteriol. 177:1039-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Govan, J. R., J. E. Hughes, and P. Vandamme. 1996. Burkholderia cepacia: medical, taxonomic and ecological issues. J. Med. Microbiol. 45:395-407. [DOI] [PubMed] [Google Scholar]
- 8.Hales, B. A., J. A. W. Morgan, C. A. Hart, and C. Winstanley. 1998. Variation in flagellin genes and proteins of Burkholderia cepacia. J. Bacteriol. 180:1110-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry, D., M. Campbell, C. McGimpsey, A. Clarke, L. Louden, J. L. Burns, M. H. Roe, P. Vandamme, and D. P. Speert. 1999. Comparison of isolation media for recovery of Burkholderia cepacia complex from respiratory secretions of patients with cystic fibrosis. J. Clin. Microbiol. 37:1004-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henry, D., E. Mahenthiralingam, P. Vandamme, T. Coenye, and D. P. Speert. 2001. Phenotypic methods for determining genomovar status of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:1073-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, W. M., S. D. Tyler, and K. R. Rozee. 1994. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J. Clin. Microbiol. 32:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell III, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 14.Mahenthiralingam, E., A. Baldwin, and P. Vandamme. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51:533-538. [DOI] [PubMed] [Google Scholar]
- 15.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahenthiralingam, E., M. E. Campbell, D. A. Henry, and D. P. Speert. 1996. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J. Clin. Microbiol. 34:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahenthiralingam, E., D. A. Simpson, and D. P. Speert. 1997. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J. Clin. Microbiol. 35:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsons, Y. N., K. J. Glendinning, V. Thornton, B. A. Hales, C. A. Hart, and C. Winstanley. 2001. A putative type III secretion gene cluster is widely distributed in the Burkholderia cepacia complex but absent from genomovar I. FEMS Microbiol. Lett. 203:103-108. [DOI] [PubMed] [Google Scholar]
- 20.Parsons, Y. N., R. Banasko, M. G. Detsika, K. Duangsonk, L. Rainbow, C. A. Hart, and C. Winstanley. 2003. Suppression-subtractive hybridisation reveals variations in gene distribution amongst the Burkholderia cepacia complex, including the presence in some strains of a genomic island containing putative polysaccharide production genes. Arch. Microbiol. 179:214-223. [DOI] [PubMed] [Google Scholar]
- 21.Sajjan, U., Y. Wu, G. Kent, and J. Forstner. 2000. Preferential adherence of cable-piliated Burkholderia cepacia to respiratory epithelia of CF knockout mice and human cystic fibrosis lung explants. J. Med. Microbiol. 49:875-885. [DOI] [PubMed] [Google Scholar]
- 22.Sajjan, U. S., F. A. Sylvester, and J. F. Forstner. 2000. Cable-piliated Burkholderia cepacia binds to cytokeratin 13 of epithelial cells. Infect. Immun. 68:1787-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speert, D. P., M. Bond, R. C. Woodman, and J. T. Curnutte. 1994. Infection with Pseudomonas cepacia in chronic granulomatous disease: role of nonoxidative killing by neutrophils in host defense. J. Infect. Dis. 170:1524-1531. [DOI] [PubMed] [Google Scholar]
- 24.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speller, D. C. 1973. Pseudomonas cepacia endocarditis treated with co-trimoxazole and kanamycin. Br. Heart J. 35:47-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speller, D. C., M. E. Stephens, and A. C. Viant. 1971. Hospital infection by Pseudomonas cepacia. Lancet i:798-799. [DOI] [PubMed] [Google Scholar]
- 27.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 28.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 29.Vermis, K., T. Coenye, E. Mahenthiralingam, H. J. Nelis, and P. Vandamme. 2002. Evaluation of species-specific recA-based PCR tests for genomovar level identification within the Burkholderia cepacia complex. J. Med. Microbiol. 51:937-940. [DOI] [PubMed] [Google Scholar]
- 30.Winstanley, C., M. G. Detsika, K. J. Glendinning, Y. N. Parsons, and C. A. Hart. 2001. Flagellin gene PCR-RFLP analysis of a panel of strains from the Burkholderia cepacia complex. J. Med. Microbiol. 50:728-731. [DOI] [PubMed] [Google Scholar]
- 31.Winstanley, C., B. A. Hales, J. A. Morgan, M. J. Gallagher, S. D. Puthucheary, M. F. Cisse, and C. A. Hart. 1999. Analysis of fliC variation among clinical isolates of Burkholderia cepacia. J. Med. Microbiol. 48:657-662. [DOI] [PubMed] [Google Scholar]